Abstract
The Hedgehog Signaling Pathway Database is a curated repository of information pertaining to the Hedgehog developmental pathway. It was designed to provide centralized access to a wide range of relevant information in an organism-agnostic manner. Data are provided for all genes and gene targets known to be involved in the Hh pathway across various organisms. The data provided include DNA and protein sequences as well as domain structure motifs. All known human diseases associated with the Hh pathway are indexed including experimental data on therapeutic agents and their molecular targets. Hh researchers will find useful information on relevant protocols, tissue cell lines and reagents used in current Hh research projects. Curated content is also provided for publications, grants and patents relating to the Hh pathway. The database can be accessed at http://www.hedgehog.sfsu.edu.
INTRODUCTION
The secreted signaling molecule Hedgehog (Hh) and its associated transduction pathway are conserved across much of the animal kingdom, from insects to vertebrates (1). The Hh pathway plays a fundamental role in such diverse processes as brain development, endodermal tissue formation and axis formation, and has been implicated in a variety of human diseases.
The Hh signaling molecule has been found to act as both a mitogen and a morphogen (2,3). The pathway has been shown to have effects on cell proliferation, cell survival and cell fate determination and plays a key regulatory role in embryonic development (4,5). Mutations in the pathway often result in gross defects across species (6–9). Hedgehog's regulatory role in embryonic development is the subject of active investigation in the embryonic stem cell research community.
The adult human Hh pathway is thought to be involved in normal tissue maintenance and repair processes (10). Tissue damage activates the Hh pathway, which in turn stimulates adult stem cells to initiate mitosis and to begin production of growth factors. Given its role in cell-cycle regulation and growth factor production, it follows that misactivation of the Hh pathway may potentially have profound effects on a tissue's ability to control its own cell proliferation (11,12). It is not surprising that the Hh pathway has been implicated in a number of cancers particularly in tissues needing frequent repair such as the lungs and intestinal lining (13–15). Misactivation of the pathway has also been found to be involved in Basal Cell Carcinoma (16–19).
Given its broad range of involvement in disease and organism development, the Hh pathway is of interest to a wide variety of research communities. However, a complete mechanistic understanding of the pathway remains elusive. The immense number of avenues of active research and the wide range of sub-disciples involved has presented a challenge to researchers in the field. There exists a need for a central repository of information pertaining to the Hedgehog pathway. Such a repository would provide a means of collecting, collating and correlating data from disparate sources. Our goal is to provide such a resource.
In the current version of the database we provide a selection of high-quality Hh pathway annotation as well as a compilation of resources useful to investigators in the Hh field. Our emphasis is on providing access to data that may not be readily available without extensive literature or external resource searches. Much of the data found in the database relies heavily on human curation efforts and as such provides a unique aid to researchers. We also provide a central location from which to link to external sources of other relevant information.
DATABASE DESCRIPTION
The Hedgehog Signaling Pathway Database can be accessed at http://www.hedgehog.sfsu.edu. The public interface to the data is through a web portal with a familiar ‘drop down’ menu structure providing ready access to the various resources available on the site. Retrieved information is displayed largely in static, tabular form with limited sorting functionality. Future iterations of the database will include a more flexible and interactive user interface. What follows is a brief description of the major elements of the database.
Genes in the pathway
The database contains all major known genes in the pathway. Genes are sorted according to their roles in Hh production, Hh reception or transcriptional response. Upon selecting a gene the user is presented with a page displaying all known orthologs across six model organisms (rat, mouse, chicken, zebrafish, human, Drosophila). Each of the orthologs is linked to a gene summary page with information pertaining to the gene.
Gene summary
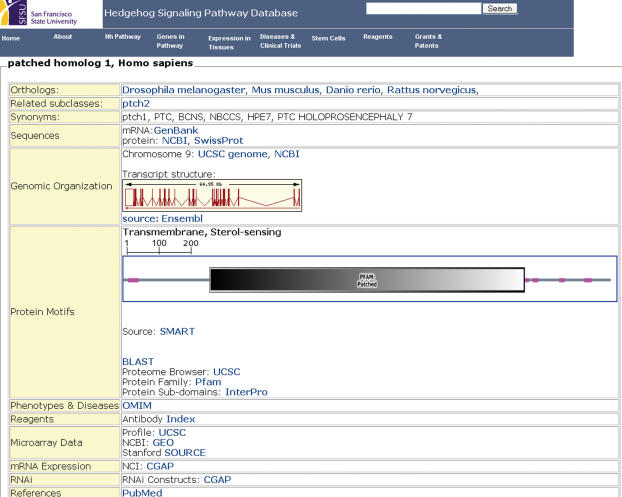
Each listed gene has an associated summary page (Figure 1). Displayed on the summary page is a collection of pertinent information such as orthologs, gene synonyms, sequence data, microarray data and computational evidence of pathway affiliation. In addition, SMART protein motifs (20) and ENSEMBL transcript structures (21) are displayed as visual gifs.
Figure 1.
Gene summary page for Patched 1.
Diseases
Extensive literature searches have been combined and collated into a list of diseases and human mutations known to be associated with the Hh pathway. Where available each disease entry includes information on implicated gene(s) as well as the affected tissue. Publication references and OMIM (22) links are also provided.
Human mutations
All available human mutation data pertaining to the pathway has been collected and sorted by the affected gene. Information with respect to the disease or defect associated with the mutation as well as specifics such as mutation type and nucleotide position can be viewed for each of the listed genes.
Hh investigator resources
We have collected a number of resources that active bench researchers in the field will find useful. These include a list and description of useful reagents along with the supplier catalog links, data related to embryonic stem cell research as it pertains to Hh pathway research, grants and patents related to the Hh pathway, RNAi gene screening data with links back to FLYBASE (23), and a zebrafish microarray based screen of vertebrate transcriptional targets (24).
Publications
A great deal of effort has gone into creating a hand-curated collection of Hh-related publications. Each publication was scanned for a variety of criteria such as genes studied, model organism used, pharmacological agents and cell lines employed, and target disease if any. In addition, text was extracted from each paper that summarizes the major results. These excerpts were hand selected and are intended as an adjunct to standard publication abstracts to aid the researcher in determining whether a particular paper warrants further investigation.
Our goal in creating this compendium of publications is to provide a comprehensive list of Hh-related research papers. Of course, we have included those papers that deal explicitly with the Hh pathway and are likely to be returned using standard searches of PubMed. However, we have also included references that may be of interest to Hh researchers but were generated from outside the Hh community and might otherwise be overlooked. Planned upgrades of the web interface to our database will allow easy searching and grouping of publications and will provide the opportunity for novel connections to be made across disparate fields of study.
FUTURE DEVELOPMENTS
The current database consists largely of a collection of hand-curated references supplemented by links to external data, all of which is supported by a simple SQL database schema. Our vision for the database includes a more interactive interface to the underlying data. Work is underway to implement a robust schema that will fully support complex queries. Using these queries users will be able to retrieve data from the Hedgehog Signaling Database according to any of a number of search criteria. A draft of the new schema has been prepared according to developing interoperative standards such as GO (25) and GMOD (26). We are also actively investigating tools such as Atlas Integrated Database Interface (27) and SeqHound (28) which will allow us to mine data in a more efficient and automated fashion. We are continuing to update and add additional data while expanding our base of informational sources. Comments and suggestions are welcome.
Acknowledgments
This work was supported in part by funding from grant (U56 CA 96217) from the National Cancer Institute, National Institutes of Health; and by funding from a grant (P20 MD000262) from the Research Infrastructure in Minority Institutions Program, National Center on Minority Health and Health Disparities; and from a grant (NIH MBRS SCORE S06 GM52588) from the National Institute of Health. We would also like to acknowledge the support provide by San Francisco State University. Funding to pay the Open Access publication charges for this article was provided by San Francisco State University.
Conflict of interest statement: None declared.
REFERENCES
- 1.Ingham P.W., McMahon A.P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Mehlen P., Mille F., Thibert C. Morphogens and cell survival during development. J. Neurobiol. 2005;64:357–366. doi: 10.1002/neu.20167. [DOI] [PubMed] [Google Scholar]
- 3.Vervoort M. Hedgehog and wing development in Drosophila: a morphogen at work? Bioessays. 2000;22:460–468. doi: 10.1002/(SICI)1521-1878(200005)22:5<460::AID-BIES8>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 4.Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 5.Shin S., Dalton S., Stice S.L. Human motor neuron differentiation from human embryonic stem cells. Stem Cells Dev. 2005;14:266–269. doi: 10.1089/scd.2005.14.266. [DOI] [PubMed] [Google Scholar]
- 6.Wallis D., Muenke M. Mutations in holoprosencephaly. Hum. Mutat. 2000;16:99–108. doi: 10.1002/1098-1004(200008)16:2<99::AID-HUMU2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Hayhurst M., McConnell S.K. Mouse models of holoprosencephaly. Curr. Opin. Neurol. 2003;16:135–141. doi: 10.1097/01.wco.0000063761.15877.40. [DOI] [PubMed] [Google Scholar]
- 8.Ming J.E., Roessler E., Muenke M. Human developmental disorders and the Sonic hedgehog pathway. Mol. Med. Today. 1998;4:343–349. doi: 10.1016/s1357-4310(98)01299-4. [DOI] [PubMed] [Google Scholar]
- 9.Chiang C., Litingtung Y., Lee E., Young K.E., Corden J.L., Westphal H., Beachy P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 10.Beachy P.A., Karhadkar S.S., Berman D.M. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 11.Kenney A.M., Rowitch D.H. Sonic hedgehog promotes G1 cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavine K.J., White A.C., Park C., Smith C.S., Choi K., Long F., Hui C.C., Ornitz D.M. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grachtchouk M., Mo R., Yu S., Zhang X., Sasaki H., Hui C.C., Dlugosz A.A. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nature Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 14.Watkins D.N., Berman D.M., Burkholder S.G., Wang B., Beachy P.A., Baylin S.B. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 15.Douard R, Moutereau S., Pernet P, Chimingqi M, Allory Y, Manivet P, Conti M, Vaubourdolle M, Cugnenc P.H., Loric S. Sonic Hedgehog-dependent proliferation in a series of patients with colorectal cancer. Surgery. 2006;139:665–670. doi: 10.1016/j.surg.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Blaess S., Corrales J.D., Joyner A.L. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- 17.Sukegawa A., Narita T., Kameda T., Saitoh K., Nohno T., Iba H., Yasugi S., Fukuda K. The concentric structure of the developing gut is regulated by Sonic hedgehog derived from endodermal epithelium. Development. 2000;127:1971–1980. doi: 10.1242/dev.127.9.1971. [DOI] [PubMed] [Google Scholar]
- 18.Schier A.F. Axis formation and patterning in zebrafish. Curr. Opin. Genet. Dev. 2001;11:393–404. doi: 10.1016/s0959-437x(00)00209-4. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz i Altaba A., Sanchez P., Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nature Rev. Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 20.Letunic I., Copley R.R., Pils B., Pinkert S., Schultz J., Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ada Hamosh A.F.S., Joanna A., Carol B., David V., Victor A.M. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grumbling G., Strelets V. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J., Srinivas B., Tay S.Y., Mak A., Yu X., Lee S.G., Yang H., Govindarajan K.R., Leong B., Bourque G., et al. Genome-wide expression profiling in the zebrafish embryo identifies target genes regulated by Hedgehog signaling during vertebrate development. Genetics. 2006;174:735–752. doi: 10.1534/genetics.106.061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah S.P., Huang Y., Xu T., Yuen M.M., Ling J., Ouellette B.F. Atlas—a data warehouse for integrative bioinformatics. BMC Bioinformatics. 2005;6:34. doi: 10.1186/1471-2105-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalickova K., Bader G.D., Dumontier M., Lieu H., Betel D., Isserlin R., Hogue C.W. SeqHound: biological sequence and structure database as a platform for bioinformatics research. BMC Bioinformatics. 2002;3:32. doi: 10.1186/1471-2105-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]