Abstract
The goal of the Encyclopedia Of DNA Elements (ENCODE) Project is to identify all functional elements in the human genome. The pilot phase is for comparison of existing methods and for the development of new methods to rigorously analyze a defined 1% of the human genome sequence. Experimental datasets are focused on the origin of replication, DNase I hypersensitivity, chromatin immunoprecipitation, promoter function, gene structure, pseudogenes, non-protein-coding RNAs, transcribed RNAs, multiple sequence alignment and evolutionarily constrained elements. The ENCODE project at UCSC website (http://genome.ucsc.edu/ENCODE) is the primary portal for the sequence-based data produced as part of the ENCODE project. In the pilot phase of the project, over 30 labs provided experimental results for a total of 56 browser tracks supported by 385 database tables. The site provides researchers with a number of tools that allow them to visualize and analyze the data as well as download data for local analyses. This paper describes the portal to the data, highlights the data that has been made available, and presents the tools that have been developed within the ENCODE project. Access to the data and types of interactive analysis that are possible are illustrated through supplemental examples.
INTRODUCTION
The goal of the ENCODE project is to identify all functional elements in the human genome sequence (1). The pilot phase of the project is focused on a specific 30 megabases (∼1%) of the human genome, with an international consortium of computational and laboratory-based scientists working to develop and apply high-throughput approaches for detecting all sequence elements that confer biological function. UC Santa Cruz is the main repository for sequence-based data, with microarray data being held at GEO and ArrayExpress. The roles of UC Santa Cruz are (i) to collect the experimental data and analyses, (ii) to perform basic quality assurance (QA) on the submitted data, (iii) to publicly release the data with comprehensive descriptions, (iv) to provide interactive displays for integrating the ENCODE data with existing genome-wide data and (v) to provide interactive tools for analysis. General details of the Genome Browser have been described previously (2,3), and are briefly reviewed here for clarity.
Within the Genome Browser, each dataset is represented as a track, which is a horizontal, graphical representation of the underlying data table. A complete description of each dataset is available on the description page for each track. The Table Browser has been previously described as a general purpose tool for analyzing data in the UCSC Genome Browser (4), possibly with integrated user-supplied data. Several features have been added to this platform in the context of the ENCODE project. In addition to interactive browsing and analysis tools that are only available at the ENCODE project at UCSC site (http://genome.ucsc.edu/ENCODE), the data are available for public download (http://hgdownload.cse.ucsc.edu/goldenPath/encode/).
Data from this project are made publicly available as quickly as possible after submission. All data on the UCSC Browsers, including the ENCODE data, pass through an extensive QA and documentation process before release. Biological validation criteria have been defined for each of the datasets and are the responsibility of the submitters to confirm before submission. Our developers and QA staff work with the data to provide fast, clear display and to confirm that the file formats and genomic coordinates are consistent.
It is expected that the ENCODE project will transition from the May 2004 human genome assembly (hg17; NCBI Build 35) to the newly released human genome assembly (hg18; NCBI Build 36) in early 2007. Following this, as the ENCODE project expands from the current 1% to the whole genome, UCSC is poised to support this growth. This paper describes the site and the tools that have been developed for viewing, retrieving and analyzing the data from the ENCODE project.
RESULTS
Portal and data
We have extended the UCSC Genome Browser (5) to include specialized support for the ENCODE project and its data. The ENCODE portal is accessible both through a link on the main Genome Browser site and directly at http://genome.ucsc.edu/ENCODE. This portal provides access to the ENCODE data and serves as a starting point for the computational analyses that are possible with the new data and analysis tools. It also contains announcements of new data releases and tool deployments, terms of use for the ENCODE data, and information about the contributors. The ‘Regions’ link opens a frames page allowing the user to quickly scan all ENCODE regions by selecting one region in the navigator frame, which opens a customizable view of that region in a display frame.
Track groups
We have added two levels of organization to reduce the complexity of accessing the data. Tracks of a similar type are collected into track groups, which provide high-level organization to the datasets. The six ENCODE-specific track groups roughly parallel the analysis working groups: Regions and Genes; Transcript Levels; Chromatin Immunoprecipitation; Chromosome, Chromatin and DNA Structure; Comparative Genomics; and Variation. The individual tracks are too numerous to list here and are frequently being updated with new results from the Consortium. The track status page at http://genome.cse.ucsc.edu/ENCODE/trackStatus.html provides a current snapshot of the data, including new datasets that are being developed, those that are in QA, and the fully released datasets.
Composite tracks
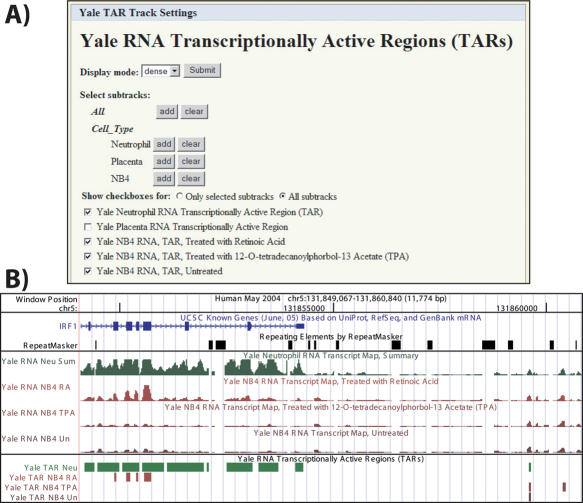
Sometimes one experiment will be run repeatedly with many different experimental conditions, producing the same data type but many parallel datasets, such as with the many combinations of cell lines, antibodies and stimulation conditions used in chromatin immunoprecipitation. For organizational simplicity, these composite tracks allow a set of similar data, usually from a single data provider, to be controlled through a single interface. On the track's user interface page, parameters that are common to all sub-tracks (e.g. visibility mode, track height, display range limits) are presented once. Just below those controls, a checkbox for each sub-track allows it to be individually included or excluded from the display. Experiments can be grouped into logical categories (e.g. cell type, transcription factor) with shared controls. Figure 1 shows the Yale RNA transcriptionally active regions (TARs) (6,7) track as an example of the streamlined interface and the resulting display of the composite tracks.
Figure 1.
Composite track control and display. (A) Controls for options that apply to all data in this track (top) with checkboxes to include or exclude individual sub-tracks as desired (bottom). (B) Example of a composite track display showing the IRF1 gene, repeats, Yale transcript maps and Yale transcriptionally active regions (6,7). The latter two are composite tracks, each containing multiple datasets. The Placenta RNA checkbox is deselected above, so that the data are not displayed in the image below.
Multiple sequence alignment display
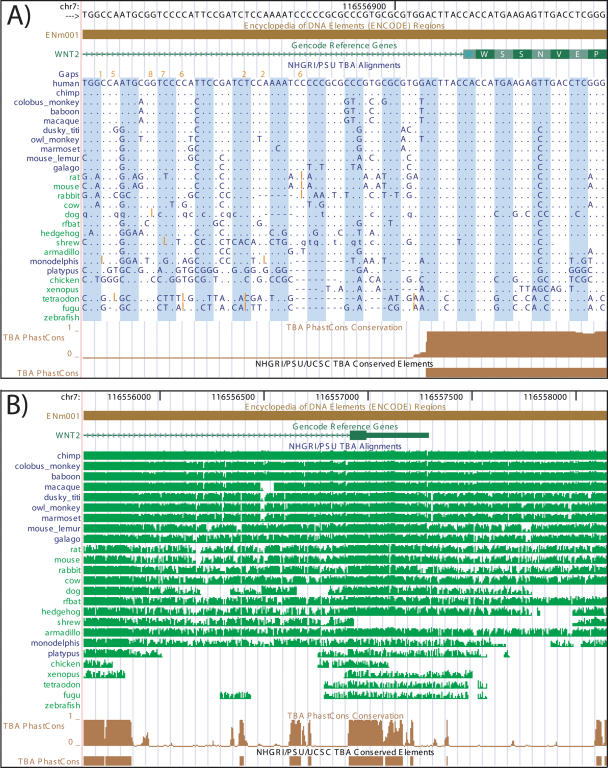
In addition to our data display and repository role, UCSC and collaborators have been developing algorithms for sequence alignment (8) and conservation analysis (9). As this produces extremely rich datasets and parallels the efforts of several other consortium members, we have created a special display that combines multiple species alignments and conservation scores in the same track, as shown in Figure 2. Alignments are projected onto a reference species for display in the browser by removing alignment columns in which the reference species is a gap. Additional enhancements include annotation of alignment gaps to indicate missing sequence and syntenic breaks, and translation in coding regions with user-selectable reading frames based on available gene annotations. When even more detail is necessary, full unprojected alignments are available on the details page for this track.
Figure 2.
Conservation display. (A) Conservation track at the base level shows details of a multiple sequence alignment, conservation scores and amino acid translations in coding regions. (‘.’: base is identical to human; ‘N’: missing sequence, ‘=’: sequence that does not align to reference is present in this species; orange numbers/lines: additional bases that are present in other species). (B) Conservation track zoomed out shows pairwise identity summary and conservation scores, highlighting non-coding elements in addition to exons.
The comparative genomics efforts within the ENCODE Consortium are also receiving special attention. The group is producing a common dataset of sequences from 23 mammals and 5 other vertebrates, which provides a rich dataset for the development and comparison of algorithms for multiple sequence alignment and detection of evolutionary constraint. Four separate alignment algorithms are being developed [MAVID (10), MLAGAN (11), PECAN (B. Paten and E. Birney, submitted for publication) and TBA (8)], and three separate conservation scoring methods [binCons (12), GERP (13,14) and phastCons (9)] are being applied to each of these alignments. Each alignment is presented in its own Alignment track, with two composite tracks to represent the real-valued Conservation scores and the predicted Elements.
Tools for analysis
The Table Browser has always provided summary statistics on a single dataset, and we have added tools for exploring correlation between genomic datasets. Data within composite tracks can be treated as a single set for simplified comparison against other tracks. An example of this is available in Supplementary Data, where promoters that are active in at least one cell line are joined to create a set of ‘functional’ promoters.
The correlation function calculates correlation coefficients, covariance, scatter plots, residuals and histograms on the fly for the selected datasets. Briefly, the data points from each table are projected down to the base level. The two datasets are intersected and only bases that contain values in both datasets are retained, resulting in datasets of equal length n. These two datasets (X,Y) are then used in a standard linear correlation function, computing the correlation coefficient:
where σX and σY are the standard deviations of the datasets X and Y, and σXY is the covariance, computed as follows:
The data values from a track are used in the calculations when available. For tracks that do not have data values, such as gene-structured tracks, the data value is 1.0 for bases that are covered by exons and 0.0 at all other positions in the region. Simple tracks that are neither gene structures nor have data values, e.g. BED tracks, are encoded as 1.0 over the extent of the item and 0.0 for all other positions in the region.
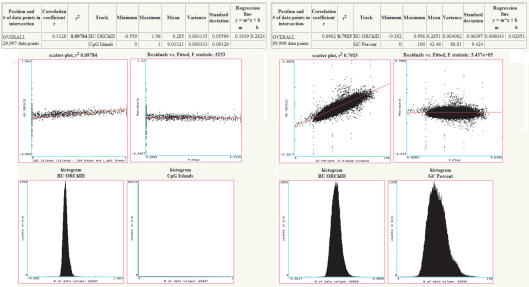
Figure 3 shows such correlation between the Boston University •OH Radical Cleavage Intensity Database (ORChID) (15–17) and the CpG Island and GC Percent tracks. The CpG Island histogram shows significant skew in the data due to many zero values, which obscures the correlation of ORChID values within CpG Islands. The correlation of ORChID values with GC Percent is very strong at r = 0.89, which reveals a potential confounding factor when comparing the ORChID values with other datasets. This method is further described in Supplementary Data.
Figure 3.
Track correlation in the Table Browser. Correlation of the Boston University. •OH Radical Cleavage Intensity Database (ORChID) (15–17) is shown with the CpG Island (left) and with the GC Percent (right) tracks. Statistical summaries (upper panels), scatter and residual plots (middle panels) and histograms (lower panels) are shown.
The hgLiftOver tool, accessible via the Genome Browser's ‘Utilities’ link, translates genomic coordinates within a species from one assembly version to another and also retrieves putative orthologous regions between species using UCSC's chained and netted alignments. These tools have been used to migrate the ENCODE regions from one assembly to another, and have also been used in the Multiple Species Alignment working group to provide orthology predictions for the preparation of the sequence datasets as described above.
DISCUSSION
The ENCODE project at UC Santa Cruz extends the powerful Genome Browser with datasets and tools to aid researchers in their quest to understand the functional elements in the genome. This extension of the Browser brings datasets on DNA replication, chromatin regulation, promoter function, gene models and multiple species comparisons together and makes them available for visualization, analysis and download. Integration of the datasets generate by the ENCODE Consortium, in addition to other genome-wide data, proves to be a rich source for addressing questions about functional elements in 1% of the human genome, and is poised to expand with the needs of the ENCODE project.
Extensions have been made to the display, providing capabilities such as composite tracks for better organization and increased customization. Analysis tools have been built into the Table Browser to simplify merging of related tables and to assess correlation between datasets. These build on the general usability, integration with genome-wide resources, ability to do online analyses and simplicity of exporting data for external analyses that have made the data analysis more accessible to biologists. Newer additions such as the Gene Sorter, In-Silico PCR and VisiGene (2,3) continue to add value by bringing resources together so that detailed analysis can proceed rapidly.
WEBSITES FOR REFERENCE
http://genome.ucsc.edu/; UCSC Genome Browser. http://genome.ucsc.edu/ENCODE/; ENCODE Portal http://hgdownload.cse.ucsc.edu/; Data downloads. http://genome.ucsc.edu/ENCODE/trackStatus.html; Status.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
This work was supported by grants from the National Human Genome Research Institute (NHGRI) for browser development and for the ENCODE project, the Howard Hughes Medical Institute (HHMI), and the National Cancer Institute. The authors like to thank many collaborators who have contributed annotation data to their project, as well as their users for their feedback and support. The authors also like to thank the dedicated system administrators who have provided an excellent computing environment: Jorge Garcia, Patrick Gavin, Chester Manuel, Victoria Lin and Paul Tatarsky. D.J.T., B.L.R., R.M.K., G.P.B., B.J.R., A.S.H., D.K., A.T., A.S.Z. and W.J.K. were funded by NHGRI. K.R.R., B.J.R., G.P.B. and K.E.S. were funded by NHGRI ENCODE. H.T., R.A.H. and H.C. were funded by NHGRI and NCI. D.H. was funded by HHMI. Funding to pay the Open Access publication charges for this article was provided by NHGRI.
Conflict of interest statement. All authors receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
REFERENCES
- 1.The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;322:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 2.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn B., the UCSC Genome Bioinformatics Group The UCSC Genome Browser Database: update 2007. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkl928. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P.A., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 8.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smith A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the Threaded Blockset Aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siepel A., Bejerano G., Pedersen J., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray N., Pachter L. MAVID multiple alignment server. Nucleic Acids Res. 2003;31:2525–2526. doi: 10.1093/nar/gkg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S. NISC Comparative Sequencing Program. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margulies E.H., Blanchette M., NISC Comparative Sequencing Program, Haussler D., Green E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper G.M., Brudno M., Stone E.A., Dubchak I., Batzoglou S., Sidow A. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. 2004;14:539–548. doi: 10.1101/gr.2034704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper G.M., Stone E.A., Asimenos G., NISC Comparative Sequencing Program, Green E.D., Batzoglou S., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balasubramanian B., Pogozelski W.K., Tullius T.D. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl Acad. Sci. USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price M.A., Tullius T.D. Using the hydroxy radical to probe DNA sequence. Methods Enzymol. 1992;212:194–219. doi: 10.1016/0076-6879(92)12013-g. [DOI] [PubMed] [Google Scholar]
- 17.Tullius T.D. Probing DNA structure with hydroxyl radicals. In: Beaucage S.L., Bergstrom D.E., Glick G.D., Jones R.A., editors. Current Protocols in Nucleic Acid Chemistry. Wiley; 2001. pp. 6.7.1–6.7.8. [DOI] [PubMed] [Google Scholar]