Abstract
The Stanford Microarray Database (SMD; http://smd.stanford.edu/) is a research tool and archive that allows hundreds of researchers worldwide to store, annotate, analyze and share data generated by microarray technology. SMD supports most major microarray platforms, and is MIAME-supportive and can export or import MAGE-ML. The primary mission of SMD is to be a research tool that supports researchers from the point of data generation to data publication and dissemination, but it also provides unrestricted access to analysis tools and public data from 300 publications. In addition to supporting ongoing research, SMD makes its source code fully and freely available to others under an Open Source license, enabling other groups to create a local installation of SMD. In this article, we describe several data analysis tools implemented in SMD and we discuss features of our software release.
INTRODUCTION
Since its inception in 1999, the Stanford Microarray Database [http://smd.stanford.edu/, (1)] has grown in its size and scope. It has become a research tool for a large scientific community that is no longer restricted to Stanford researchers and SMD now supports the research of over 1200 active users in close to 300 laboratories around the world. Our users study the biology of 43 organisms and have entered data generated from more than 60 000 microarrays. Data stored in SMD have led to the publication of over 300 research papers and all raw data related to these publications (including results from over 6000 Homo sapiens and 1000 Saccharomyces cerevisiae and several hundred Arabidopsis thaliana, Mus musculus and Caenorhabditis elegans microarrays) are made freely available via the SMD website. SMD stores gene expression as well as array CGH and chip-chIP experiments. SMD supports multiple microarray platforms (spotted cDNA or oligonucleotide arrays, Affymetrix, Agilent, Combimatrix and Nimblegen arrays) and the installation at Stanford holds data mostly from spotted arrays, Agilent arrays and Affymetrix arrays. In addition to being a research database, SMD also serves as a laboratory information management system (LIMS) for spotted microarrays. SMD provides extensive biological annotation for genes and sequences from over 40 organisms, as well as annotations from the Gene Ontology when they are available. SMD provides researchers with tools to make their data fully compliant to the Minimal Information About a Microarray Experiment standards (2,3). SMD also has tools to read and write MAGE-ML files (4) and has a data pipeline that can communicate published data directly to ArrayExpress (5) and GEO (6). The public data can be selected, viewed, downloaded and analyzed by the public using most of the data analysis and quality assessment tools that are available to registered SMD users.
The source code for SMD has been downloaded and installed at several academic and private locations. SMD software is released under the permissive MIT license and is free of charge. SMD's code was first released in April 2001 (version 0.1), and since mid-2003 new code releases have been made on a roughly quarterly basis. As of August 2006, SMD is using version 1.11, which is also publicly available. The Stanford installation of SMD uses Oracle [Oracle Server Enterprise Edition version 10g Release 2 (10.2.0.1)] as its database management system and is run on a Sun V880 server with Solaris 9 as its operating system. SMD's software has been successfully ported to use the freely available PostgreSQL database on a Linux platform (7) and is also able to run on the MacOSX. Most of SMD's software is written in Perl, although some of our most recently written programs use Java. In addition, some of our programs used for computationally intensive tasks such as clustering and image processing were written in C.
Installer feedback since SMD's first public release has resulted in a more simplified installation process. In each new release, SMD provides complete installation scripts to create all database tables and objects required to create a fully functional SMD schema de novo. In addition, the update scripts used to update the SMD schema as installed at Stanford are also included to help remote installers easily update their installations. The current schema in SMD consists of 173 tables and the complete schema including table and column definitions is posted on the web at http://smd.stanford.edu/schema/. To help SMD installers solve common problems, to provide a place for suggestions and to synchronize development by installers, we maintain an active developer forum at http://smdforum.stanford.edu/smdforum/index.php. Since the initial release, SMD software has been downloaded hundreds of times and it is actively used at a number of institutions, including Princeton University, University of North Carolina, University of Tennessee and St Jude's Children's Research Hospital in Memphis.
SMD TOOLS
SMD provides a wide array of web-accessible tools for processing, analyzing, visualizing and sharing microarray data in a research environment. Extensive online help documentation is available at http://smd.stanford.edu/help/index.shtml. Defining a set of microarrays of interest is the first step for any analysis process. Since most users have access to data from thousands of microarrays in the Stanford installation, they need effective tools to locate microarrays of interest. Accordingly, SMD provides two different search forms—called ‘Basic Search’ and ‘Advanced Search’—to find relevant data. The options presented in these forms give users great flexibility in their searches, allowing them to find data based on, for example, the researcher who entered the microarray data, keywords, text searches of experiment descriptions or pre-grouped selections of microarrays. As a shortcut, users can also select data from all the arrays they themselves entered in the database directly from the menu bar by selecting one of the options under the ‘My Data’ menu item. Once the microarrays are selected, users decide whether they intend to do an analysis that is applicable to one array at a time or to a group of arrays. In the former case users go to the ‘Data Display’ page, while in the latter they enter the ‘Data Retrieval pipeline’.
The ‘Data Display’ page is organized as a table, listing one microarray per row and providing links to tools available to explore data from that microarray. In addition to administrative tools, which allow an experimenter to view, edit or delete the annotations associated with data from a single microarray, SMD provides various analysis and quality assessment tools that can be applied on a per-array basis. For instance, since microarray data are known to be sensitive to several experimental factors, it is important to be able to assess the quality of the data collected from a microarray and to correct—normalize—the data appropriately. SMD has several tools that can be used for the assessment of microarray quality: an exploratory graphing tool produces histograms or scatter plots of user-selected fields to look at distribution of data points; an ANOVA analysis to detect spatial and print-tip bias on the array (called ‘Ratios on Arrays’) and a tool from BioConductor's ArrayQuality package to view diagnostic and doping control plots for HEEBO/MEEBO arrays. Some of these tools have been described in more detail in (8). Background correction and normalization methods are often used to correct experimental biases in microarray data and SMD accordingly provides access to several normalization and background correction methods using the marray and limma packages, respectively, from BioConductor (9).
When retrieving data from several related microarrays, users have a wealth of options to specify what to include in the retrieved dataset. We offer tools to filter data on a per-array basis based on the quality assessment of each microarray. Users can specify the data field that is retrieved for the analysis and can select the biological annotations retrieved for reporters on the microarrays. Once a dataset is retrieved, it can be analyzed either directly, or it can be saved in the Data Repository (1). The Data Repository is a central place in SMD where registered users of an SMD installation can save data files retrieved from the database or uploaded from their desktop machines and access tools to analyze these datasets. Because the Data Repository requires long-term storage of data in the database, the implementation of the SMD software at Stanford makes this feature available only to registered SMD users with a user name and password and is not available to public users. Registered users can also use the Data Repository to share their retrieved datasets, and analyses thereof with other users and laboratory groups. Analysis tools available from the Data Repository include hierarchical clustering, self-organizing maps, singular value decomposition (SVD) (described below), KNNImpute (described below) as well as other tools. A detailed description of the features of the Data Repository, with a description of the different analysis tools and methods to share data, is provided in the SMD Data Repository tutorial, freely available as a PowerPoint presentation at http://smd.stanford.edu/help/tutorials_subpage.shtml.
Here, we discuss some of the recent developments at SMD that have expanded the ability of SMD users to analyze microarray data. All of these tools were originally developed by people associated with SMD as standalone tools, were implemented as part of the SMD workflow and are available as web-accessible tools. As part of our extensive help documentation, we have a page (http://smd.stanford.edu/help/smd_tools.shtml) that lists all the tools available in SMD, gives a brief description of each tool, indicates how the user can find the tool and links to a help document that gives detailed instructions on the usage of the particular tool.
RESULTS
Singular value decomposition
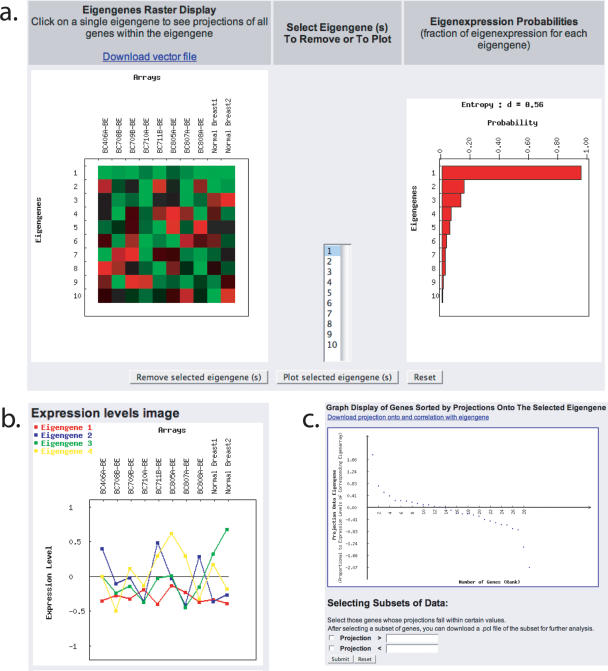
SVD is a mathematical approach that determines unique orthogonal gene and corresponding array expression patterns (i.e. ‘eigengenes’ and ‘eigenarrays’, respectively) in the data and this method has been successfully applied to microarray data analysis (10). Patterns uncovered by SVD can often be correlated with independent biological processes or with experimental artifacts, such as variations in the day of hybridization, array print or scanner calibration. Since the use of SVD (as well as KNNImpute, described below) results in relatively computationally intensive jobs, SMD provides SVD only from the Data Repository, thus restricting its use to registered users of the database. Since SVD cannot process data matrices with missing values, the user then has to decide whether to apply a simple algorithm to replace missing values by row (gene) averages or remove genes with missing values. Alternatively, the user can use KNNImpute (discussed below) to compute missing values before applying SVD to the dataset. The result of the SVD calculation is displayed as (i) a raster image of the eigengenes and arrays, and (ii) a bar graph of the probabilities of each eigengene vector, which indicate the fraction of overall expression information that they capture within the dataset (Figure 1). The user can plot individual eigengenes across the arrays or select an individual eigenvector on the raster image and look at the projection of all genes in the dataset to that eigengene. On subsequent pages projection values and correlations of genes in the dataset with the eigenvector can be downloaded. Expression values of genes whose projections on the eigengene are in some user-defined range can be exported/re-saved in the Data Repository in pcl file format for further analysis.
Figure 1.
Result of an SVD analysis in SMD. (a) Raster display of the eigengenes (left panel) and bar chart display of the probabilities of eigenexpression (right panel) of a sample dataset. (b) Plot showing the behavior of the top four eigengenes. (c) Projection of genes within an eigengene. This image shows how all genes in a dataset are projected onto a given eigengene. This is one way to determine those genes whose expression is contributed to by an eigengene.
KNNImpute
KNNImpute (11) is a tool that estimates missing values in data matrices. Data from k-nearest neighboring vectors are used to impute the missing values. Since many of the algorithms for microarray analysis work sub-optimally—or not at all—on data matrices that are missing data, rational methods of estimating missing values allow one to discard fewer sets of data. SMD provides a web-interface to the KNNImpute that is available from the Data Repository and can be applied to datasets in the pcl file format (http://smd.stanford.edu/help/formats.shtml#pcl). On the web-interface, the user has the option of specifying the number of nearest neighbors to use for calculating missing values. Since KNNImpute uses a computationally intensive algorithm, the process is placed in a job queue when it is launched and the job is executed when sufficient resources are available. When the job is complete, the user is alerted via Email and the resulting pcl file is available for further analysis in the user's Data Repository.
GO TermFinder
GO TermFinder uses annotations to Gene Ontology (GO) terms to determine whether a list of genes produced by any number of microarray analysis software has a significant enrichment of GO terms. SMD's implementation uses the open source GO::TermFinder package (12) made accessible via a web-interface to both registered and public SMD users.
Using GO TermFinder requires that SMD store GO and gene association data. Consequently, the database schema was extended with an ontology sub-schema to allow efficient storage and querying of ontology and gene association data. A mechanism was developed to perform weekly updates using the GO (from http://obo.sourceforge.net/) and a gene association file (from the Gene Ontology consortium's website) for each supported organism (human, mouse, yeast, etc.).
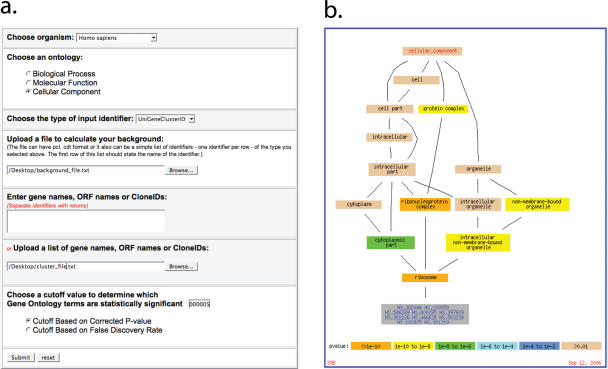
In SMD, GO TermFinder can be used with files uploaded by registered users or can be invoked directly from the graphical output of SMD's online clustering program (this feature is available for public as well as registered users). The user has to upload or select from the available options the reference set of genes from which the genes in the cluster to be analyzed was derived, select which component ontology to use for the analysis and set a threshold value for significance or false discovery rate. Significant results are presented in table and graphical format in the context of GO tree structure (Figure 2). Both the table and graphical output are downloadable for future reference.
Figure 2.
GO TermFinder analysis in SMD. (a) User interface to upload the required files (list of genes that make up an interesting cluster and background file from which the cluster was derived) and make selections of gene identifiers used in the uploaded files, the desired sub-ontology to use and significance value and parameter to use. (b) The graphical display of a positive result output from GO TermFinder. The graph shows the genes of the input cluster in the context of the relevant part of Gene Ontology. The GO terms that are found to have significant enrichment are colored according to the significance level.
FUTURE DIRECTIONS
SMD's current and future development focuses on two major paths. One of these builds on the above-discussed development of introducing ontologies in SMD. We are expanding on this theme and adding several new ontologies to allow accurate and searchable annotation of biological samples and experiments. We are developing dynamic tools that allow efficient and fast browsing and querying of these ontologies with the goal of using the ontology terms for easy and user-friendly annotation of experiments and samples. The other main development of SMD focuses on simplifying and generalizing our schema for the more efficient and flexible storage of biological data that annotate the genes represented on the microarrays. This will allow users to perform more powerful and flexible queries when retrieving their data, based on the gene annotations, as well as allow SMD to more easily accommodate annotation from additional organisms.
Acknowledgments
We thank the users of SMD for their constant feedback that helps us to improve SMD, and to the remote installers of SMD for detailed reports as to the ease of installing SMD that have helped us improve the process considerably. Funding for continued development of SMD is supported by NIH grant R01 HG003469 to G.S. and C.A.B. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ball C.A., Awad I.A., Demeter J., Gollub J., Hebert J.M., Hernandez-Boussard T., Jin H., Matese J.C., Nitzberg M., Wymore F., et al. The Stanford Microarray Database accommodates additional microarray platforms and data formats. Nucleic Acids Res. 2005;33:D580–D582. doi: 10.1093/nar/gki006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball C.A., Sherlock G., Parkinson H., Rocca-Sera P., Brooksbank C., Causton H.C., Cavalieri D., Gaasterland T., Hingamp P., Holstege F., et al. Standards for microarray data. Science. 2002;298:539. doi: 10.1126/science.298.5593.539b. [DOI] [PubMed] [Google Scholar]
- 3.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 4.Spellman P.T., Miller M., Stewart J., Troup C., Sarkans U., Chervitz S., Bernhart D., Sherlock G., Ball C., Lepage M., et al. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3:RESEARCH0046. doi: 10.1186/gb-2002-3-9-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazma A., Parkinson H., Sarkans U., Shojatalab M., Vilo J., Abeygunawardena N., Holloway E., Kapushesky M., Kemmeren P., Lara G.G., et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Killion P.J., Sherlock G., Iyer V.R. The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD) BMC Bioinformatics. 2003;4:32. doi: 10.1186/1471-2105-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gollub J., Ball C.A., Binkley G., Demeter J., Finkelstein D.B., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., et al. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 2003;31:94–96. doi: 10.1093/nar/gkg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alter O., Brown P.O., Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl Acad. Sci. USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., Altman R.B. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 12.Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]