Abstract
Comparasite is a database for comparative studies of transcriptomes of parasites. In this database, each data is defined by the full-length cDNAs from various apicomplexan parasites. It integrates seven individual databases, Full-Parasites, consisting of numerous full-length cDNA clones that we have produced and sequenced: 12 484 cDNA sequences from Plasmodium falciparum, 11 262 from Plasmodium yoelii, 9633 from Plasmodium vivax, 1518 from Plasmodium berghei, 7400 from Toxoplasma gondii, 5921 from Cryptosporidium parvum and 10 966 from the tapeworm Echinococcus multilocularis. Putatively counterpart gene groups are clustered and comparative analysis of any combination of six apicomplexa species is implemented, such as interspecies comparisons regarding protein motifs (InterPro), predicted subcellular localization signals (PSORT), transmembrane regions (SOSUI) or upstream promoter elements. By specifying keywords and other search conditions, Comparasite retrieves putative counterpart gene groups containing a given feature in common or in a species-specific manner. By enabling multi-faceted comparative analyses of genes of apicomplexa protozoa, monophyletic organisms that have evolved to diversify to parasitize various hosts by adopting complex life cycles, Comparasite should help elucidate the mechanism behind parasitism. Our full-length cDNA databases and Comparasite are accessible from http://fullmal.ims.u-tokyo.ac.jp.
INTRODUCTION
Malaria and other parasites are causes of worldwide health problems that need immediate actions based on scientific investigation. Thus, genome research has been enthusiastically pursued during the past decade and the entire genome sequences of various malarial species, such as Plasmodium falciparum, Plasmodium vivax and Plasmodium yoelii, have been determined (1,2). We have also constructed full-length cDNA libraries and collected full-length cDNAs in malarial species. The obtained cDNA information together with physical cDNA clones were made publicly available from our database Full-malaria (http://fullmal.ims.u-tokyo.ac.jp) (3).
In these years, we have expanded our full-length cDNA database to various kinds of additional malarial species and other apicomplexan parasites, including Toxoplasma, Cryptosporidium and Echinococcus. We determined the 5′ end one-pass sequences of numerous clones. The sequences were mapped onto the genome sequences and compiled in a database for every species [collectively called Full-Parasites hereafter: Plasmodium species (http://fullmal.hgc.jp/pf/); Toxoplasma species (http://fullmal.hgc.jp/tg/); Cryptosporidium species (http://fullmal.hgc.jp/cp/); Echinococcus species (http://fullmal.hgc.jp/em/)]. Furthermore, we have developed a new database, Comparasite (http://fullmal.hgc.jp/comp_index.html), to integrate these individual databases, enabling trans-species comparative searches between putative counterpart genes. This was performed to improve the annotations currently attached to malarial parasites as well as to provide physical and informational resources for a wider set of parasite species. The additional information should be extremely useful for understanding the biologies of the parasites as well as developing anti-parasitic drugs or vaccines (4).
Moreover, by further enhancing the data contents of individual databases as well as enhancing functionalities of Comparasite, we attempt to create a foundation for elucidating mechanisms underlying parasitism: how apicomplexa protozoa, monophyletic organisms, have evolved to diversify to parasitize various hosts by adopting complex life cycles. To this end, apicomplexa can be regarded as the most successful obligatory parasitic protozoa that had evolved from the common ancestor to adapt to various hosts having various life cycles; thus, they provide ideal models for comparative genomics. We also expect that intensive comparative studies of parasite genes would eventually give insights into how a life has accommodated itself to occasionally drastic environmental changes. Using 3000–6000 genes, they parasitize various organs of host organisms. Although their basic ultra-structures are well conserved, the genomes have diversified enormously in size, which vary from 9 Mb of Cryptosporidium parvum to 65 Mb of Toxoplasma gondii and G+C contents, which vary from 19% of P.falciparum to 55% of T.gondii [(1,2,5); http://www.toxodb.org/toxo-release4-0/home.jsp]. Here, we introduce the expansion of our full-length cDNA databases to six apicomplexa parasites as well as a brand new integrated database, Comparasite, for comparative studies of transcriptomes of parasites defined by full-length cDNAs. Interspecies comparisons of full-length cDNA sequences of mutually putative counterpart genes will help not only to refine gene annotations but also to elucidate the process of evolution and the molecular mechanisms behind parasitism.
DATA PRODUCTION
Data resources (Full-Parasites)
We constructed full-length cDNA libraries using the oligo-capping and V-capping methods (Table 1) (6,7). In each library, we calculated the frequency of putative full-lengthness to be ∼80%. For the details of the procedures of the construction and the evaluation of the cDNA library, see the reference and our website (http://fullmal.hgc.jp/comparas/Experimental.htm). Using the constructed cDNA libraries, we determined the 5′ end one-pass sequences of P.falciparum (12 484 cDNAs), P.vivax (9633), P.yoelii (11 262), Plasmodium berhgei (1518), T.gondii (7400 + 1018 full sequences) and C.parvum (5921) and Echinococcus multilocularis (10 966).
Table 1.
Statistics of the data contents for each of the individual full-length cDNA databases
| Species | Host | Stage | Library method | Number of cDNA sequenced | Number of RefFull | DB (URL) |
|---|---|---|---|---|---|---|
| Plasmodium falciparum | Human | Erythrocytic, gametocyte | Oligo-capping | 12 484 | 1465 | Fullmal (http://fullmal.hgc.jp) |
| Plasmodium vivax | Human | Erythrocytic, gametocyte | Oligo-capping | 11 262 | 1566 | Fullmal (http://fullmal.hgc.jp) |
| Plasmodium yoelii | Mouse | Erythrocytic, gametocyte | Oligo-capping | 9633 | 1206 | Fullmal (http://fullmal.hgc.jp) |
| Plasmodium berghei | Mouse | Erythrocytic, gametocyte | Oligo-capping | 1518 | 416 | Fullmal (http://fullmal.hgc.jp) |
| Toxoplasma gondii | Mammals | Tachyzoite | Oligo-capping | 7400 | 762 | FullToxo (http://fullmal.hgc.jp/tg/) |
| Cryptosporidium parvum | Human/cow | Sporozoite | Oligo-capping | 5921 | 682 | FullCrypto (http://fullmal.hgc.jp/cp/) |
| Echinococcus multilocularis | Dog/fox | Larva | V-capping | 10 966 | ND | FullEchino (http://fullmal.hgc.jp/em/) |
Data process (Full-Parasites)
After trimming the vector sequence and ambiguously sequenced parts, we mapped the cDNAs sequences onto the corresponding genomic sequences using Blat (http://genome.ucsc.edu/cgi-bin/hgBlat), except for those of E.multilocularis (its genome is not available). The relative positions compared with the so-called annotated genes (as of attached to genome sequences) were thereby calculated and the cDNAs overlapping the annotated gene were defined as representing the annotated gene. Except for T.gondii and in particular P.falciparum, for which intensive efforts have been made to genome annotations, annotations attached to the genomes might be still inadequate, seeming still mainly based on mere computational predictions. Possibly reflecting this fact, in many cases (including some cases, even in T.gondii and P.falciparum genes), significant parts of the annotated genic regions were inconsistent with cDNAs, including 5′-untranslated regions (5′-UTRs) [Note: usually a gene prediction program does not predict non-coding regions, and therefore, it is impossible to predict exact transcriptional start sites and the following UTRs (1,2,5)]. Therefore, the cDNA sequences were merged with the annotated genes to complement the missing or possibly incorrect annotated parts. We defined the resultant virtual putative full-length cDNAs as ‘RefFulls’. We generated RefFull sequences for P.falciparum (1465 RefFulls), P.vivax (1566), P.yoelii (1206), P.berhgei (416), T.gondii (762 + 1018 full cDNAs) and C.parvum (682).
The RefFull sequences obtained were used for determining ORFs. Basically, the longest ORFs were depicted. However, all the ORFs larger than 20 amino acids in length are stored in the database, so that analysis of shorter ORFs is also supported by advanced search options. Subsequently, amino acid sequences deduced from the ORFs were subjected to functional annotations. We included annotations resulting from homology search [BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/)], protein motif search [InterPro (http://www.ebi.ac.uk/interpro/) and Pfam (http://www.sanger.ac.uk/Software/Pfam/)], hydropathy plot [using the standard protocol (3)] and predictions of subcellular localization signals [PSORT (http://psort.hgc.jp/)] and transmembrane domains [SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html)].
Transcription start site (TSS) and promoter identification (Full-Parasites)
Another useful feature extracted from full-length cDNAs is the precise positional information of the TSSs of the mRNAs. Since in many cases, the important cis-regulatory elements are embedded in the proximal regions of the TSSs), it should be useful to analyze these upstream regions as putative transcriptional regulatory regions (8). As for the RefFull cDNAs, we extracted the genomic sequences corresponding to −500 to +100 (TSS is designated as 0) and annotated the presence of representative promoter elements, which are TATA box, GC-rich stretch [GC box or CpG islands in the case of higher mammals: in some parasites they are reported to be this kind of promoter motif; (9)]. For the search for TATA box, position weight matrix search V$TATA_01 as of TRANSFAC and MATCH (http://www.gene-regulation.com/pub/databases.html#transfac) was used. Since sequence composition of the TATA box in malarial parasites was reported to be deviated from those in other species we also used TATA box matrix calculated from malarial genes (10). The sexual-stage-specific element of P.falciparum [SSSP (11)] is also searched.
Comparative studies (Comparasite)
For comparative studies, putative counterpart gene groups were defined as follows. Similarly with the approach taken by comparative genomics studies between P.falciparum and P.yoelii and between other cases (1,2), the protein sequences of the annotated genes of P.falciparum, of which genome sequence is most complete, were used as queries to search homologous regions of the genome sequences of the other malarial parasites using TBLASTN program (http://www.ncbi.nlm.nih.gov/BLAST/) and only the mutually best hit regions were selected. Annotated genes that overlap with the homologous region were designated as corresponding homologues. Based on these homologies, the contig sequences of P.vivax, P.yoelii, P.berghei, T.gondii and C.parvum were aligned with the genome sequences of P.falciparum (Table 2).
Table 2.
Statistics of the number of RefFulls corresponded to the putative counterpart genes in each species
| Species | 5 | 4 | 3 | 2 | 1 | 0 | Total |
|---|---|---|---|---|---|---|---|
| Plasmodium falciparum | 1 | 18 | 44 | 140 | 444 | 818 | 1465 |
| Plasmodium vivax | 1 | 20 | 49 | 150 | 363 | 983 | 1566 |
| Plasmodium yoelii | 1 | 19 | 49 | 155 | 329 | 653 | 1206 |
| Plasmodium berghei | 1 | 18 | 39 | 89 | 74 | 195 | 416 |
| Toxoplasma gondii | 1 | 17 | 30 | 41 | 85 | 588 | 762 |
| Cryptosporidium parvum | 1 | 18 | 33 | 49 | 101 | 480 | 682 |
Number of RefFulls corresponded to putative counterpart genes with indicated number of species.
DATABASE DESCRIPTIONS
Full-Parasites: Full-malaria and six additional full-length cDNA databases
Overview (Full-Parasites)
Our databases started in 2000 with Full-malaria (12), which is a database of full-length cDNAs of the human malarial parasite, P.falciparum. After several rounds of updates, our malarial databases now collectively contain P.falciparum, P.vivax and murine malarial parasites, P.yoelii and P.berghei, which consist of 12 484, 9633, 11 262 and 1518 full-length cDNAs, respectively. In addition, we have expanded the database to cover cDNAs of tachyzoites of T.gondii, sporozoites of C.parvum and larval stage parasites of tapeworm, E.multilocularis, which consist of 7400, 5921 and 10 966 full-length cDNAs, respectively [URLs of the top pages are as follows: Plasmodium species (http://fullmal.hgc.jp/pf/); Toxoplasma (http://fullmal.hgc.jp/tg/); Cryptosporidium (http://fullmal.hgc.jp/cp/); Echinococcus (http://fullmal.hgc.jp/em/)]. Each entry is connected to representative genome databases, such as PlasmoDB (http://www.plasmodb.org/plasmo/home.jsp), CryptoDB (http://cryptodb.org/cryptodb/) and ToxoDB (http://www.toxodb.org/toxo-release4-0/home.jsp)].
In each species, cDNAs were clustered into RefFulls (Pf: 1465 RefFulls; Pv: 1566; Py: 1206; Pb: 416; Tg: 762; Cp: 682) and were subjected to functional annotations. Currently attached functional annotations include (i) the protein motif and GO terms, identified by InterProScan and Pfam; (ii) the subcellular localization signals, predicted by PSORT; (iii) hyrdopathy plot; (iv) transmembrane domain predicted by SOSUI. Sequences of the promoter region of the RefFulls were also analyzed in terms of promoter elements using TRANSFAC and other position weight matrices for the TATA box, the CpG-like element and SSSP (see above) and attached annotations are presented. For further details in functional annotation procedures, cut-offs and other parameters/criteria, see our website (http://fullmal.hgc.jp/comparas/Glossary.htm).
Interface of database(s) (Full-Parasites)
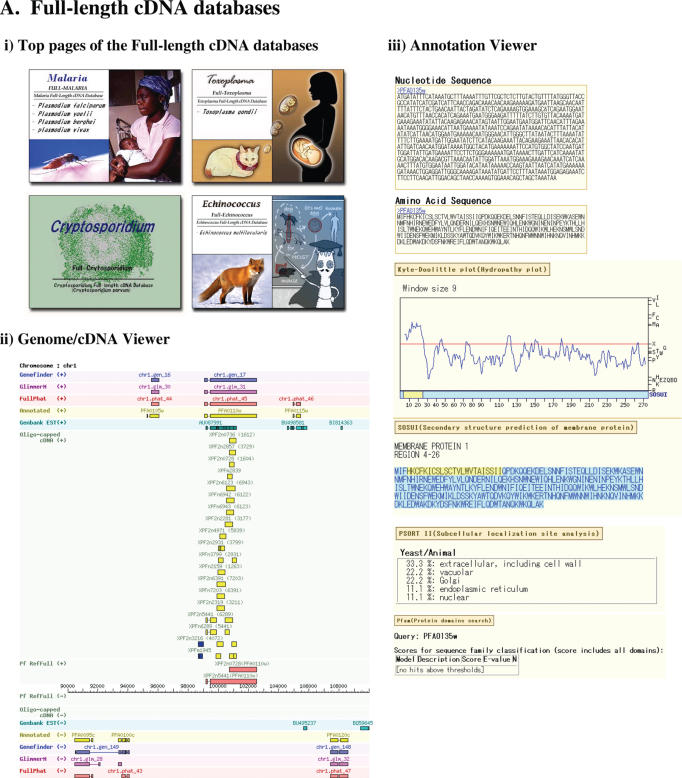
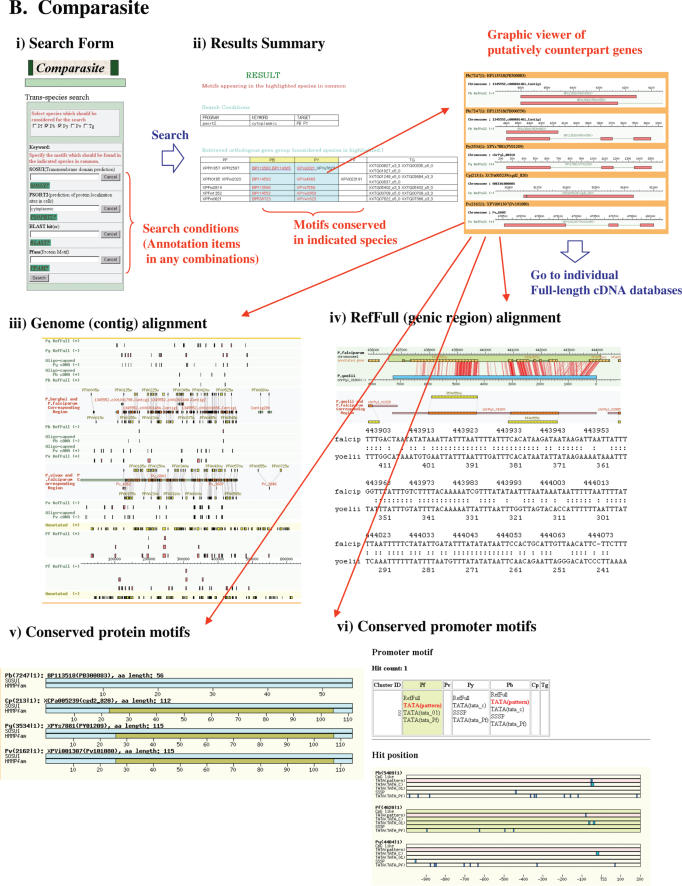
From the top page of each of the databases, the user can search the respective databases by inputting keywords (cDNA/annotated gene ID), genomic positions, presence or absence of the various kinds of annotation features attached to RefFulls or BLAST search. The main part of the databases in each species consists of a user-friendly dynamic interface, supporting seamless zooming in/out from the genomic level. From the interface, the user can easily find necessary information about the retrieved gene, regarding its genomic locations, basal individual physical cDNAs and their consistency of the annotated genes. By following the link to functional annotation page, the user finds the ORF structures and aforementioned attached annotations (Figure 1A). For further details of the functionality, see the reference. We also included the links to the comparative viewer from the annotation page, providing the user with a means to evaluate the annotations of individual species from the comparative point of view with other species (also see Figure 1B)
Figure 1.
Screen shots of the individual databases of the full-length cDNAs (A) and Comparasite (B).
Comparasite: an integrated database for comparative studies
Overview (Comparasite)
Comparasite is an integrated database consisting of the aforementioned six individual full-length cDNA databases (http://fullmal.hgc.jp/comp_index.html). This database is constructed in the expectation that meticulous comparisons of each homologous RefFull, including gene structures, protein features and promoter elements should reveal common or unique features underlying the parasitism in each of the species. For the comparative studies, annotations attached to RefFulls, which are virtual hybrids of our full-length cDNAs and annotated genes (see Data process), are subjected to the search to determine whether annotated feature(s) appear in common or in a species-specific manner. When the user attempts to avoid the false-positively annotated information, only the hits appearing in multiple species in common should be considered. On the contrary, when the user is interested in species-specific features of the parasitism, unique hits should be considered (Figure 2). It is also intriguing to examine whether the putative counterpart genes have similarities or differences in the annotated cis-regulatory elements. It has long been supposed that many characteristic and distinct features in different organisms might be caused by the changes in the regulations of genes, rather than the changes in the function of genes themselves (13). Actually the TATA-binding protein of malarial parasites has unique amino acid sequence compared with other organisms, possibly accommodating the transcription machinery properly at the TSSs in the extremely AT-rich genomes (14). It is also worth to mention that various phenotypes of apicomplexa parasites, with different host ranges, organ specificities and life cycles provide opportunities to produce libraries representing the stages, which are difficult in other species. Before the completion of cataloging, all the expressed genes from every life-cycle stage of all the apicomplexa parasites, we can analyze making full use of bioinformatics, handling both protein features of expressed genes and respective nucleotide features of transcription control elements.
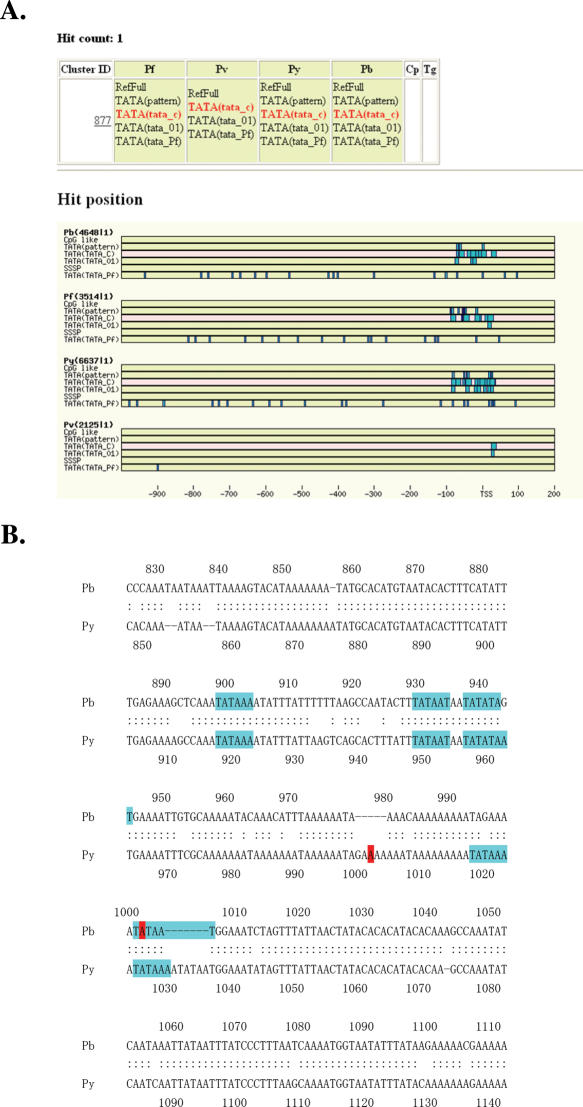
Figure 2.
Example of comparison of putative promoter elements. (A) Positions of conserved promoter elements. The predicted positions of the indicated promoter elements in the promoters of putative orhthlogous gene group, XPF2n2934 (Pf), XPVi011046 (Pv), XPYw0792 (Py), BP114799 (Pb) are shown by blue boxes. (B) Sequence alignment of the surrounding regions of TSSs in Py and Pb genes. Predicted TATA boxes and identified TSSs are shown by blue and red boxes, respectively.
Interface of database (Comparasite)
In the top page, Comparasite supports versatile searches, designed for multifaceted comparative analyses of protozoan parasite genes (Figure 1B). In any combination of the species, the user can specify which particular annotated features should be contained or not within mutually putative counterpart genes groups. For the search, any combination of the annotation items (SOSUI, PSORT, BLAST, Pfam and TRANSFAC; also see the above section) could be used. For example, the user can search for the gene groups with the features; gene group should be an ‘antigen’ with predicted ‘transmembrane domain’ and have ‘protein kinase’ motif; these features should commonly appear in at least Plasmodium species and Toxoplama.
The results page is a graphical view of the transcriptomes mapped onto the genome sequences. For each entry, the data are connected to the database of individual species with close links to further detailed information of the gene in the corresponding species. The sequence alignments were also implemented, so that the user can empirically understand which part of the transcripts/promoters/ORFs should be conserved and in which part the commonly/uniquely annotated features are located.
Current statistics
Full-Parasites
Current statistics of the databases are summarized in Table 1. Table 2 shows number of RefFulls corresponded to putative counterpart genes with indicated number of species.
Comparasite
Tables 3 and 4 show the statistics focusing on functional annotations. Table 3 shows number of annotation terms identified in each species. Table 4 shows the number of putative counterpart gene groups containing corresponding functional annotation terms common in indicated number of species.
Table 3.
Number of annotation terms identified from the RefFulls in each species
| Annotations for RefFull (category) | Pf | Pv | Py | Pb | Tg | Cp |
|---|---|---|---|---|---|---|
| ‘Antigen’ (keyword) | 107 | 41 | 0 | 11 | 3 | 12 |
| ‘Transcription’ (keyword) | 42 | 16 | 0 | 3 | 2 | 9 |
| ‘Kinase’ (Pfam) | 116 | 38 | 41 | 0 | 0 | 0 |
| ‘Mitochondria’ (PSORT) | 81 | 117 | 49 | 31 | 67 | 19 |
| ‘Transmembrane domain’ (SOSUI) | 508 | 250 | 166 | 62 | 79 | 43 |
| ‘Transporter’ (GO term) | 131 | 92 | 60 | 27 | 30 | 20 |
| ‘TATA box’ (promoter) | 1157 | 601 | 530 | 227 | 42 | 233 |
| ‘TATA box; Pf’ (promoter) | 1328 | 1007 | 741 | 273 | 158 | 299 |
Table 4.
Number of putatively counterpart genes containing corresponding annotation terms in indicated number of species in common
| Annotations for RefFull (category) | 6 | 5 | 4 | 3 | 2 |
|---|---|---|---|---|---|
| ‘Antigen’ (keyword) | 0 | 0 | 4 | 14 | 56 |
| ‘Transcription’ (keyword) | 0 | 0 | 1 | 6 | 31 |
| ‘Kinase’ (Pfam) | 0 | 1 | 3 | 4 | 29 |
| ‘Mitochondria’ (PSORT) | 0 | 0 | 3 | 5 | 25 |
| ‘Transmembrane domain’ (SOSUI) | 0 | 4 | 3 | 21 | 108 |
| ‘Transporter’ (GO term) | 0 | 5 | 1 | 18 | 5 |
| ‘TATA box’ (promoter) | 0 | 2 | 14 | 87 | 443 |
| ‘TATA box; Pf’ (promoter) | 0 | 17 | 46 | 192 | 646 |
Search example
Comparasite
For an example of the search using Comparasite, follow the link as follows: Comparasite top (http://fullmal.hgc.jp/comp_index.html; cookie is obtained here); select the species, Pf, Py, Pv and Pb and specify the search by: ‘Ribosomal’ in Pfam section; from the search results, select the cluster ID 4525 (ribosomal protein S4; http://fullmal.hgc.jp/cgi-bin/comp_main_view.cgi?UID=2&SEE=1,2&MXX=750&VPLN=sp005|scale|cl06|sp003|&CLSID=4525); further links can be followed by clicking Pf RefFull to, for example, genome alignment viewer (http://fullmal.hgc.jp/cgi-bin/fulmal_contig_aln.cgi?UID=2&SEE=1,2&CMI=1408) and functional annotation viewer (http://fullmal.hgc.jp/cgi-bin/fulmal_gen_detail.cgi?UID=2&SEE=1,2<P=0&SPI=0304&HID=11&BSS=233731&BSE=233734&STD=0&STP=0&BAS=232861&BAE=235331) and so on.
Annotation definitions, glossary, clone and data repository
cDNA clones registered in the database are freely available and should serve as indispensable resources to explore functions of genes to combat the relevant diseases. All the database services as well as the used raw data are publicly and freely accessible to anonymous users without any restrictions from our download sites (they can be followed from the top page of each of the full-length cDNA databases and from Comparasite).
Full-Parasites
For further details and functionality of each of the Full-Parasite databases, refer to our previous documentations (3,12). Download site and help pages can be followed from the top page.
Comparasite
Especially for the new part of our series of databases, Comparasite, we arranged detailed user manual and used technical terms, definitions and parameters for the annotations are precisely described in Glossary and Experimental Procedure sections in our websites (http://fullmal.hgc.jp/comparas/Glossary.htm; http://fullmal.hgc.jp/comparas/Experimental.htm).
CONCLUSIONS AND FUTURE PERSPECTIVES
Comparasite, consisting of a series of full-length cDNA databases of apicomplexa parasites, provides bases not only for analyzing gene functions of individual species in a concrete and versatile ways but also for understandings the biological concepts of parasitism in a more abstract way. Furthermore, in future detailed experimental validation of gene functions in each species, our full-length cDNA databases should serve as an important interface for looking into cDNA clone resources, as each of the database entries represents physical full-length cDNAs, which should serve as indispensable reagents for any kinds of experimental purposes.
Our cDNA project, especially regarding Comparasite, is at the early stage. As shown in Table 1, the data coverage is quite incomplete. However, we believe that this is still largest (unique) among the apicomplexan cDNA databases. Also, we believe our database will become more useful as our cDNA collection is enlarged. Determinations of the entire sequences of RefFull clones are also underway to replace RefFull sequences with physically confirmed cDNA sequences, which will revise existing annotated gene structures. Currently, because of unavailability of the genome sequence, Echincoccus cDNAs could not be fully utilized for comparative studies. However, on release of the genomic information, this part should be immediately incorporated into the current cDNA databases in full. The new libraries from other apicomplexan parasites, including Eimeria, Theileria and Babesia, which are all parasitic species representing an additional three genera, will also be produced to expand Comparasite, which will make our database further unique from other parasite database, such as PlasmoDB, CryptoDB and ToxoDB.
Alternative computational methods for the annotations should also be considered. Especially, for defining putative counterpart (putative orthologous) genes, results obtained from gene-based methods, such as OrthoMCL, should also be considered (15). Eventually, refinements of the annotations, including manual scrutiny of the putatively defined mutually counterpart genes, will further improve the fidelity of the database contents. For this, we sincerely welcome feedbacks from users. We will compile comments from users on discrepancies of the annotations and on future directions. Whenever they reach some consensus, we will also send the comments to the other databases of this community. With further enhanced functionalities as well as improved fidelity of the individual data, our series of apicomplexan full-length cDNA databases, wrapped up by Comparasite, will allow us to reveal the detail of both conservation and specification that have resulted from the process of evolution of parasitism.
Acknowledgments
We are grateful to Keiko Toya for excellent programming work. Full-length cDNA libraries were produced in collaboration with Dr Josef Tuda of Sam Ratulangi University and Mihoko Imada of Keio University (P.vivax), Dr Akiko Shibui of Tokyo Science University and Prof Sadao Nogami of Nihon University (P.berghei), Prof Xuenan Xuan of Obihiro Unieversity of Agriculture and Veterinary Medicine and Prof Chihiro Sugimoto of Hokkaido University (T.gondii), Dr Isao Kimata of Osaka City University (C.parvum), Drs Yuzaburo Oku, Noriaki Nonaka, Jun Matsumoto of Hokkaido University and Prof Masao Kamiya of Rakuno Gakuen University (E.multilocularis). Large-scale sequencing was performed by Dr Atsushi Toyoda of Riken. This database has been constructed and maintained by a Grant-in-Aid for Publication of Scientific Research Results from Japan Society for Promotion of Science. Funding to pay the Open Access publication charges for this article was provided by a Grant-in-Aid for Publication of Scientific Research Results from Japan Society for Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlton J.M., Angiuoli S.V., Suh B.B., Kooij T.W., Pertea M., Silva J.C., Ermolaeva M.D., Allen J.E., Selengut J.D., Koo H.L., et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe J., Suzuki Y., Sasaki M., Sugano S. Full-malaria 2004: an enlarged database for comparative studies of full-length cDNAs of malaria parasites, Plasmodium species. Nucleic Acids Res. 2004;32:D334–D338. doi: 10.1093/nar/gkh115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibui A., Shiibashi T., Nogami S., Sugano S., Watanabe J. A novel method for development of malaria vaccines using full-length cDNA libraries. Vaccine. 2005;23:4359–4366. doi: 10.1016/j.vaccine.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 5.Abrahamsen M., Templeton T., Enomoto S., Abrahante J., Zhu G., Lancto C., Deng M., Liu C., Widmer G., Tzipori S., et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y., Sugano S. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 2003;221:73–91. doi: 10.1385/1-59259-359-3:73. [DOI] [PubMed] [Google Scholar]
- 7.Kato S., Ohtoko K., Ohtake H., Kimura T. Vector-capping: a simple method for preparing a high-quality full-length cDNA library. DNA Res. 2005;12:53–62. doi: 10.1093/dnares/12.1.53. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita R., Suzuki Y., Wakaguri H., Tsuritani K., Nakai K., Sugano S. DBTSS: database of human transcription start sites, progress report 2006. Nucleic Acids Res. 2006;34:D86–D89. doi: 10.1093/nar/gkj129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollack Y., Kogan N., Golenser J. Plasmodium falciparum: evidence for a DNA methylation pattern. Exp. Parasitol. 1991;72:339–344. doi: 10.1016/0014-4894(91)90079-c. [DOI] [PubMed] [Google Scholar]
- 10.Ruvalcaba-Salazar O.K., del Carmen Ramirez-Estudillo M., Montiel-Condado D., Recillas-Targa F., Vargas M., Hernandez-Rivas R. Recombinant and native Plasmodium falciparum TATA-binding-protein binds to a specific TATA box element in promoter regions. Mol. Biochem. Parasitol. 2005;140:183–196. doi: 10.1016/j.molbiopara.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Dechering K.J., Kaan A.M., Mbacham W., Wirth D.F., Eling W., Konings R.N., Stunnenberg H.G. Isolation and functional characterization of two distinct sexual-stage-specific promoters of the human malaria parasite Plasmodium falciparum. Mol. Cell. Biol. 1999;19:967–978. doi: 10.1128/mcb.19.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe J., Sasaki M., Suzuki Y., Sugano S. FULL-malaria: a database for a full-length enriched cDNA library from human malaria parasite, Plasmodium falciparum. Nucleic Acids Res. 2001;29:70–71. doi: 10.1093/nar/29.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King M.C., Wilson A.C. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 14.McAndrew M.B., Read M., Sims P.F., Hyde J.E. Characterization of the gene encoding an unusually divergent TATA-binding protein (TBP) from the extremely A+T-rich human malaria parasite Plasmodium falciparum. Gene. 1993;124:165–171. doi: 10.1016/0378-1119(93)90390-o. [DOI] [PubMed] [Google Scholar]
- 15.Li L., Stoeckert C.J., Jr, Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]