Abstract
Here we present the Synthetic Gene Database (SGDB): a relational database that houses sequences and associated experimental information on synthetic (artificially engineered) genes from all peer-reviewed studies published to date. At present, the database comprises information from more than 200 published experiments. This resource not only provides reference material to guide experimentalists in designing new genes that improve protein expression, but also offers a dataset for analysis by bioinformaticians who seek to test ideas regarding the underlying factors that influence gene expression. The SGDB was built under MySQL database management system. We also offer an XML schema for standardized data description of synthetic genes. Users can access the database at http://www.evolvingcode.net/codon/sgdb/index.php, or batch downloads all information through XML files. Moreover, users may visually compare the coding sequences of a synthetic gene and its natural counterpart with an integrated web tool at http://www.evolvingcode.net/codon/sgdb/aligner.php, and discuss questions, findings and related information on an associated e-forum at http://www.evolvingcode.net/forum/viewforum.php?f=27.
INTRODUCTION
As the molecular biology revolution gains momentum, an increasing number and variety of ‘natural’ genes have been re-designed at the nucleotide level and synthesized in attempts to improve protein yields [reviewed in (1)]. Surprisingly, 60% of these synthetic genes do not have an entry in freely accessible nucleotide sequence databases, such as GenBank or EMBL. However, the molecular biology community could benefit from having easy access to a reference set of sequences of all such synthetic genes, as the systematic, quantitative rules for optimization remain obscure. For example, although many codon optimization experiments lead to an increase in protein yields, reports of negative results are not uncommon [e.g. (2,3)]. Indeed, given that the motivation for most genes re-designs is to improve protein yields, peer-reviewed publications are likely to be biased towards reports of success, such that the negative results we see underestimate the scale of the challenges that remain. In this context, the information contained in synthetic genes not only provides a valuable resource to guide biologists’ redesign of further genes but also serves a good training set of data for bioinformaticians to explore the underlying factors that actually affect protein expression. Given these considerations, we have constructed a relational database called the 'Synthetic Gene Database' (SGDB) to store the sequence of synthetic genes and associated information from all studies published to date.
DATABASE CONTENT AND STRUCTURE
Database content
We define a synthetic gene as an ‘a DNA molecule artificially constructed using a set of oligonucleotides without requiring a physical DNA template’. Thus synthetic genes are distinguishable from genes engineered by site-directed mutagenesis in the aspect of whether or not a physical DNA template was used during gene construction. By focusing on methodology, our definition leaves open the possibility that a synthetic gene contains one or more implicit amino acid substitutions relative to its natural counterpart. Indeed our database deliberately includes such synthetic genes (and they form a significant fraction of the synthetic genes reported in the peer-reviewed literature). Their relevance is that the most common purpose of gene synthesis is to optimize protein expression, and amino acid substitutions can create important effects here. As the major motivation for our database is to facilitate a deeper understanding of the relationship between gene sequence and gene expression, we wish to include all synthetic genes that contribute to a broad data foundation for researchers to explore such phenomena. Following the same motivation, to guide future studies of the relationship between gene design and protein expression, our database excludes synthetic gene sequences that lack associated peer-reviewed publications or experimental information.
In all we found more than 200 experiments that meet these criteria to date, and have thus been included in the database (the complete list of publications reporting synthetic genes may be found at http://www.evolvingcode.net/codon/sgdb/pub.php). Because each experiment may have reported more than one natural gene and multiple versions of the synthetic genes, our database actually contains more than 250 synthetic gene sequences.
In addition to the coding sequence of synthetic genes, SGDB also collects information on 5′-untranslated region (5′-UTR), 3′-UTR and various additional parameters (e.g. expression vectors, species, strains, assay methods, recoding methods, expression levels, etc.) that associate with gene expression.
Database structure
The SGDB contains three tables (entities) to minimize the redundancy of the information required in storage. These tables are ‘Literature’, ‘WTGene’ and ‘SyntheticGene’. The relationships between these three tables are illustrated in Figure 1.
Figure 1.
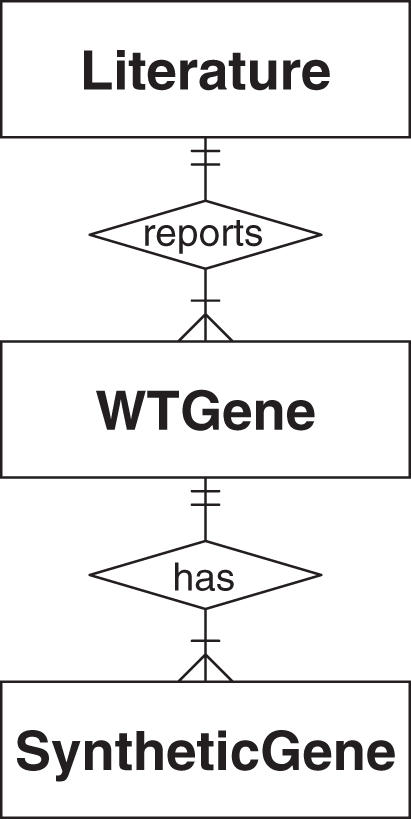
The Entity-Relationship model illustrates the logical structure of the SGDB. Each table (square) is an entity. The relationships between the three entities are: (i) each publication in the 'Literature' table reported one or many natural genes, conversely, each natural gene in the 'WTGene' table must have exactly one related publication; (ii) a natural gene may have one or many versions of synthetic genes in the 'SyntheticGene' table, while a synthetic gene must have exactly one natural counterpart.
Database implementation
The SGDB was implemented with MySQL database management system. To communicate with the database, a web interface has been developed in PHP and JavaScript at (http://www.evolvingcode.net/codon/sgdb/index.php). The sequence comparison tool was developed in Perl (source code available upon request). This database is maintained by the Apache web server of the UMBC EvolvingCode Research Group. The SGDB is freely available for browsing under all versions of Internet browsers.
RESULTS AND DISCUSSION
Data access
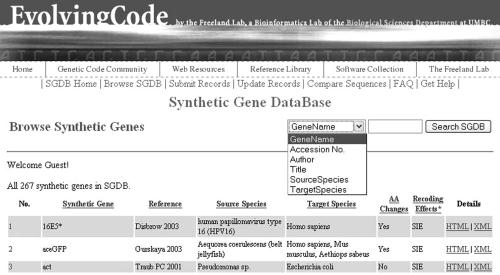
Users can browse all natural genes and synthetic genes in the SGDB at http://www.evolvingcode.net/codon/sgdb/wt.php and http://www.evolvingcode.net/codon/sgdb/browse.php (Figure 2), respectively. To find a synthetic gene of interest, users may query the SGDB at the homepage or any sub-pages via a search box according any of the following specified fields: gene name, GenBank accession no., author names, article title, source species or target species (Figure 2). In each case, a list of synthetic genes that satisfy the search terms will be returned.
Figure 2.
A screenshot of the SGDB. This browse page lists all synthetic genes in the SGDB. Each field underlined is sortable. A search box is present on every page of the SGDB, allowing users to search a synthetic gene according to various fields.
To facilitate data exchange for further analysis by interested users, we allow users to download an XML file for each publication (http://www.evolvingcode.net/codon/sgdb/tmp/). The XML schema can be found at http://www.evolvingcode.net/codon/sgdb/sgdb.xsd. We offer this schema as a new standard data description for synthetic genes; our focus on XML highlights our emphasis on future-flexibility as analysis and understanding here grows.
Current data trends
To date, the SGDB has collected 266 synthetic genes. Searching the database by date reveals that the number of published studies reporting one or more synthetic genes shows a dramatic increase after 1995 (http://www.evolvingcode.net/codon/sgdb/doc.php). This reflects the introduction of 'assembly PCR' (the classic methodology of gene synthesis) by Stemmer et al. (4) and suggests that the volume of data available for analysis is likely to increase significantly as further refinements of the synthesis technique continue to emerge [e.g. ‘Simplified Gene Synthesis', (5)].
Data submission
Although, the SGDB is a small database at this point of introduction, all indications predict continuing rapid growth in the number and diversity of synthetic genes reported in peer-reviewed literature. In this context, update of the SGDB will be most effective if undertaken by the broadest possible sub-section of the community that is creating new synthetic genes. Therefore, we created web forms for users to submit new records and update existing records in the SGDB (http://www.evolvingcode.net/codon/sgdb/submit.php and http://www.evolvingcode.net/codon/sgdb/update.php, respectively). As we continue to enter new data, we undertake to contact researchers to let them know that this database exists and that we encourage a community-wide, distributed development of the data resource.
Data analysis
A unique feature of the SGDB is that all information pertaining to a synthetic gene may be displayed side-by-side with its natural counterpart. To further help the comparison of each pair of genes, we developed an online sequence comparison tool that operates in four sequential steps: (i) translate the nucleotide sequence into amino acid sequence according to a specified genetic code (as the amino acid sequences are more similar than their nucleic acid counterparts such that the former are easy to align); (ii) use a dynamic programming algorithm to align the protein sequences of the synthetic gene and the natural gene (6); (iii) weight each codon at the aligned position according to a table listing quantitative estimates of 'fitness' (optimality) of the 64 codons (see below) and (iv) create a line plot using the alignment position as the x-axis and codon fitness values as the y-axis. The two differently-denoted lines afford users an easy visual comparison of estimated codon-translation-optimality at each aligned position. This line plot can be used to find rare codon clusters, which might dramatically affect protein translation [reviewed in (7)].
During protein translation, users can choose the standard genetic code or other non-standard genetic codes as described by NCBI (http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi).
To estimate the fitness of 64 codons, users can either choose to use the relative synonymous codon usage (RSCU) or w defined by Sharp and Li (8) downloaded from the CAI Calculator (a web tool offered by our group at http://www.evolvingcode.net/codon/cai/cai.php, unpublished data), or the fraction (in GCG format) defined by the CUTG (9). All measures reflect the significant body of research that links protein expression levels to background patterns of codon usage under the general idea that translationally optimal codons are those which match the most abundant tRNA species (10,11). At present, no more specific measure has been shown to clearly distinguish sequence features that contribute to differences in expression. In particular, the lack of standardized measures for protein expression renders direct quantitative comparisons extremely difficult at present. With such issues in mind, we have associated the SGDB with a forum so as to help stimulate discussion and standardization amongst synthetic gene researchers.
Discussion board
The design and synthesis of synthetic genes is a newly-emerging field that continues to undergo rapid theoretical and empirical development (1,12–17). During the design stage, the factor most often considered is that of codon usage. However, replacing a rare codon with an optimal codon requires that we understand what an 'optimal' codon is. The various methods that we offer to estimate codon fitness (above) merely represent some of the most popular and well supported generalizations, and it seems clear that much remains to be discovered. In terms of 'bottom up' thinking, we already know that specific codon replacements may change multiple properties of the mRNA beyond those of the codon itself, such as mRNA secondary structure (3,18,19) or 'codon context' (20–22). In terms of current knowledge, >20% of synthetic genes in our database did not increase protein yield even after supposed codon optimization. Against this background, it is striking to note that although a huge coding sequence space exists for any specific protein product (and average of just over 3n nucleic acid sequences for any given amino acid sequence of length n), very few studies have designed multiple versions of a synthetic gene to directly compare different algorithms for codon optimization. This omission significantly reduces the strength of current data interpretation. To address these frontiers of synthetic gene design, we created an e-forum (http://www.evolvingcode.net/forum/viewforum.php?f=27), which not only allows users to ask for help with our software, but also encourages users to participate in the discussion of each gene design study (each study corresponds to a thread in the e-forum). Our aim here is to reinforce the utility of the database with an online community of researchers who can share information and questions.
Future directions
As described in the introduction, we perceive two major directions for future development of the SGDB. On one hand, we plan to integrate the information that the SGDB contains about specific synthetic genes into existing software designed to facilitate gene design [e.g. the Synthetic Gene Designer (16)]. On the other hand, sequences of natural and synthetic genes can be co-analyzed, together with their associated expression data to improve our quantitative understanding of the rules of protein translation regulation. In particular, we would advocate for research that complement the current ‘broad and shallow’ data of one or a few synthetic genes for each of a wide diversity of proteins, with ‘narrow and deep’ data of multiple variations in coding strategy for a single protein product.
Acknowledgments
This project is supported by NSF award 0317349 (to S.J.F.) from DBI (Biological Databases and Informatics) Program. The authors thank Dr Alex Bateman, Dr Philip Farabaugh, Dr Janice Zengel, and two anonymous reviewers for insights and comments that have improved this database and manuscript. Funding to pay the Open Access publication charges for this article was provided by US National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gustafsson C., Govindarajan S., Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Kim C.H., Oh Y., Lee T.H. Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene. 1997;199:293–301. doi: 10.1016/s0378-1119(97)00384-3. [DOI] [PubMed] [Google Scholar]
- 3.Wu X., Jornvall H., Berndt K.D., Oppermann U. Codon optimization reveals critical factors for high level expression of two rare codon genes in Escherichia coli: RNA stability and secondary structure but not tRNA abundance. Biochem. Biophys. Res. Commun. 2004;313:89–96. doi: 10.1016/j.bbrc.2003.11.091. [DOI] [PubMed] [Google Scholar]
- 4.Stemmer W.P., Crameri A., Ha K.D., Brennan T.M., Heyneker H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 5.Wu G., Wolf J.B., Ibrahim A.F., Vadasz S., Gunasinghe M., Freeland S.J. Simplified gene synthesis: a one-step approach to PCR-based gene construction. J. Biotechnol. 2006;124:496–503. doi: 10.1016/j.jbiotec.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 7.Kane J.F. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 8.Sharp P.M., Li W.H. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura Y., Gojobori T., Ikemura T. Codon usage tabulated from the international DNA sequence databases; its status 1999. Nucleic Acids Res. 1999;27:292. doi: 10.1093/nar/27.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E.coli translational system. J. Mol. Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 11.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 12.Fuglsang A. Codon optimizer: a freeware tool for codon optimization. Protein Expr. Purif. 2003;31:247–249. doi: 10.1016/s1046-5928(03)00213-4. [DOI] [PubMed] [Google Scholar]
- 13.Gao W., Rzewski A., Sun H., Robbins P.D., Gambotto A. UpGene: application of a web-based DNA codon optimization algorithm. Biotechnol. Prog. 2004;20:443–448. doi: 10.1021/bp0300467. [DOI] [PubMed] [Google Scholar]
- 14.Jayaraj S., Reid R., Santi D.V. GeMS: an advanced software package for designing synthetic genes. Nucleic Acids Res. 2005;33:3011–3016. doi: 10.1093/nar/gki614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grote A., Hiller K., Scheer M., Munch R., Nortemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu G., Bashir-Bello N., Freeland S.J. The Synthetic Gene Designer: a flexible web platform to explore sequence manipulation for heterologous expression. Protein Expr. Purif. 2005;47:441–445. doi: 10.1016/j.pep.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Richardson S.M., Wheelan S.J., Yarrington R.M., Boeke J.D. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 2006;16:550–556. doi: 10.1101/gr.4431306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphreys D.P., Sehdev M., Chapman A.P., Ganesh R., Smith B.J., King L.M., Glover D.J., Reeks D.G., Stephens P.E. High-level periplasmic expression in Escherichia coli using a eukaryotic signal peptide: importance of codon usage at the 5′ end of the coding sequence. Protein Expr. Purif. 2000;20:252–264. doi: 10.1006/prep.2000.1286. [DOI] [PubMed] [Google Scholar]
- 19.Carlini D.B., Chen Y., Stephan W. The relationship between third-codon position nucleotide content, codon bias, mRNA secondary structure and gene expression in the drosophilid alcohol dehydrogenase genes Adh and Adhr. Genetics. 2001;159:623–633. doi: 10.1093/genetics/159.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buckingham R.H. Codon context and protein synthesis: enhancements of the genetic code. Biochimie. 1994;76:351–354. doi: 10.1016/0300-9084(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 21.Irwin B., Heck J.D., Hatfield G.W. Codon pair utilization biases influence translational elongation step times. J. Biol. Chem. 1995;270:22801–22806. doi: 10.1074/jbc.270.39.22801. [DOI] [PubMed] [Google Scholar]
- 22.Johnston J.C., Rochon D.M. Both codon context and leader length contribute to efficient expression of two overlapping open reading frames of a cucumber necrosis virus bifunctional subgenomic mRNA. Virology. 1996;221:232–239. doi: 10.1006/viro.1996.0370. [DOI] [PubMed] [Google Scholar]