Abstract
EPConDB (http://www.cbil.upenn.edu/EPConDB) is a public web site that supports research in diabetes, pancreatic development and beta-cell function by providing information about genes expressed in cells of the pancreas. EPConDB displays expression profiles for individual genes and information about transcripts, promoter elements and transcription factor binding sites. Gene expression results are obtained from studies examining tissue expression, pancreatic development and growth, differentiation of insulin-producing cells, islet or beta-cell injury, and genetic models of impaired beta-cell function. The expression datasets are derived using different microarray platforms, including the BCBC PancChips and Affymetrix gene expression arrays. Other datasets include semi-quantitative RT–PCR and MPSS expression studies. For selected microarray studies, lists of differentially expressed genes, derived from PaGE analysis, are displayed on the site. EPConDB provides database queries and tools to examine the relationship between a gene, its transcriptional regulation, protein function and expression in pancreatic tissues.
INTRODUCTION
Diabetes mellitus is a significant health problem affecting ∼177 million people worldwide with ∼4 million deaths per year attributed to the presence of the disease (http://www.who.int/diabetes/facts/en/). Diabetes, resulting from either the immuno-destruction or inadequate function of insulin-secreting beta cells located in pancreatic islets, occurs when glucose levels in the blood are not maintained within a normal range. Over time, high blood glucose levels can cause damage to almost every organ of the body.
EPConDB, the Endocrine Pancreas Consortium Database, is a public web site that was developed in order to serve the bioinformatics needs of the diabetes research community. The web site provides information about genes expressed in the pancreas. EPConDB, a resource of the Beta Cell Biology Consortium (BCBC, http://www.betacell.org), collects information and data from public sources, BCBC members and diabetes researchers and presents it in a useful manner. Users can easily query EPConDB for annotations and expression results. EPConDB also contains information about reagents publicly available through the BCBC, including the human and mouse PancChips (1,2) and the mouse PromoterChips (3,4). A major research focus of many BCBC members is the analysis of the transcriptional networks guiding the development and differentiation of the endocrine pancreas. EPConDB provides information to assist in the identification of transcription factor binding sites or cis-regulatory modules controlling the transcriptional program regulating development and function of the endocrine pancreas.
CONTENT
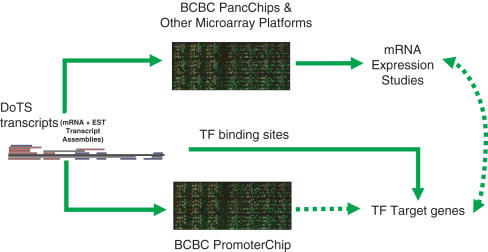
EPConDB is transcript-based and uses DoTS (Database Of Transcribed Sequences, http://www.allgenes.org), a human and mouse transcript index, which was created from all available mRNA and EST sequences from GenBank and dbEST. Currently, EPConDB uses release 9 of human DoTS and release 10 of mouse DoTS. DoTS transcripts are consensus sequences created from clustering and assembling the sequences (Figure 1) and are assigned identifiers, e.g. DT.443798. Computational annotation is applied to the human and mouse DoTS transcripts (5,6), including Entrez Gene (7) assignment and coding potential, and the annotations are included in EPConDB. DoTS annotation was instrumental in the discovery of novel mouse transcripts, expressed in the pancreas (6). DoTS transcripts are assembled to preserve alternative transcripts, so it is possible to have more than one DoTS transcript associated with a particular gene. For example, human PAX4 has two alternatively spliced forms (DT.40127947 and DT. 91748890). Associations between DoTS transcripts and elements on microarray platforms provide a link to the expression results for Entrez Genes and binding sites regulating their expression (Figure 1).
Figure 1.
Concepts and data used in the EPConDB web site. DoTS Transcripts, which are consensus sequences generated by the assembly of ESTs and mRNAs, are associated with elements on microarray platforms based upon their GenBank accession numbers and RefSeqs. Using the arrays, gene expression is determined under multiple experimental conditions. For the PromoterChip, the tiles are assigned DoTS transcripts based upon Entrez Genes. Transcription factor binding site predictions are made in regions conserved between human and mouse that are in or near a PromoterChip tile. Using the PromoterChip, binding sites can be determined experimentally and employed to validate binding site predictions. Overall, the relationship between a gene, its transcriptional regulation and expression under various conditions can be examined. The colored lines represent sequences used to create a DoTS transcript, black lines represent mRNAs or RefSeqs, grey lines ESTs, red lines represent annotated 5′ ESTs and blue lines annotated 3′ ESTs.
EPConDB presents expression results obtained using the BCBC microarrays, e.g. mouse PancChip 6 and human PancChip 1 (1,2); other microarray platforms, e.g. Affymetrix gene expression arrays (8) (Figure 1); and additional expression platforms, e.g. MPSS [massively parallel signature sequencing (9)] and RT–PCR (10). The expression studies are fully annotated according to the MIAME (11) standards including biomaterials and experimental protocols. Links to RAD (RNA Abundance Database) (12) from EPConDB provide additional study information. Currently, there are 24 studies from 18 different laboratories that can be downloaded from the EPConDB Experiments page. Annotation files and protocols, associated with each of the BCBC arrays are available from the EPConDB Downloads page.
For the BCBC Mouse PromoterChips (5A and 5B), EPConDB supplies the association between DoTS transcripts and the promoter elements (tiles) along with predicted transcription factor binding sites (Figure 1). The mouse PromoterChip 5A has tiles covering proximal and distal promoters while the 5B array has tiles covering proximal and distal promoters, enhancers, microRNA genes and highly conserved regions (13). The binding site predictions are made by considering conserved putative sites in or near a tile. The sites were predicted using positional weight matrices from TRANSFAC (14) and JASPAR (15) and a scoring threshold set to control the false positive and true positive prediction rate. Transcription factor binding data from ‘ChIP-on-chip’ experiments using BCBC mouse PromoterChips (3,4) detects transcription factor target genes and can also be used to validate the predicted binding sites (Figure 1).
EPConDB provides lists of differentially expressed genes, derived from the PaGE (Patterns from Gene Expression) (16) analysis of multiple pancreatic microarray studies. The gene lists are accessed from the Experiments Page and can also be generated using the queries for Up or Down regulated Expression (Table 1). Using these queries, a user can set the fold change parameter to find transcripts differentially expressed within a specified range (Figure 2).
Table 1.
Functionalities of the EPConDB web site
| Expression |
| Gene expression profiles |
| Lists of differentially expressed genes |
| Database queries |
| Find a transcript or gene using |
| Gene name or symbol, accession or RefSeq, Entrez Gene id, MGIa Gene id, IMAGEb clone id |
| Find transcripts or genes expressed in tissues |
| Transcription factor expression |
| Q-value tissue expression |
| Up-regulated expression, down-regulated expression, EST expression profile |
| Find genes by transcriptional regulation |
| Predicted transcription factor target genes |
| Find transcripts or genes associated with the BCBC PancChips/PromoterChips using |
| RefSeq, Entrez Gene id, Array Element id |
| Find transcripts by chromosomal location |
| BLAT alignment, diabetes candidate region |
| Find transcripts by annotation |
| GO functions, Gene trap insertions, signal peptide, transmembrane domains |
| Query history/my transcripts |
| Displays the queries performed by the user and the results |
| User can combine query results |
| Analysis and tools |
| BLAST |
| Genome Browser |
| T1DBase GBrowse |
| Binding site collection query |
| Locate specific arrangements of transcription factor binding sites in promoter regions |
| Reportmaker |
| Create and download a file containing query results |
| Downloads |
| BCBC Chip annotations and protocols |
| Expression data from studies |
| List of novel cDNA clones |
| EPConDB tutorial |
aMouse Genome Informatics (http://www.informatics.jax.org/).
bIntegrated Molecular Analysis of Genomes and their Expression (http://image.llnl.gov/).
Figure 2.
(A) The results page from a query for transcripts with Down regulated Expression in total pancreas as compared with islet tissue having a fold change in expression between 8-fold and 10-fold and a PaGE confidence of 0.9. Twenty-nine transcripts, including a transcript for the gene, Isl1, ISL1 transcription factor, LIM/homeodomain (islet 1), were returned from the query. (B) The transcript page for DT.489821, a DoTS transcript associated with the gene, Isl1. On the transcript page, annotations are displayed including a link to the T1DBase/EPConDB Gene Page and links to the T1DBase Genome Browser. Links to other views for the transcript such as Protein features, EST anatomy and Expression Profile are also available. (C) The Expression Profile link displays gene expression profiles derived from expression studies. Clicking on an individual profile displays the graph and additional information about the study. (D) The T1DBase GBrowse view shows the transcript aligned to the genome. Other views include the BCBC PromoterChip elements and predicted transcription factor binding sites.
QUERIES AND DATA-MINING TOOLS
Queries of EPConDB allow investigators to obtain information about their genes of interest and to find lists of genes with common features, e.g. genes regulated by a particular transcription factor and expressed in a particular tissue. Queries are listed on the EPConDB Query Transcripts page, accessible from the blue navigation bar, and are listed in Table 1. The queries are organized according to finding transcripts by an identifier, expression in a tissue, regulation, presence on a microarray, chromosomal location and annotation, e.g. function. EPConDB provides appropriate forms for each query, allowing users to specify search parameters (e.g. human or mouse). Example queries for expression in a tissue include a novel measure of tissue specificity, termed Q-value (17), to identify transcripts expressed in the pancreas, and the Transcription Factor Expression query to find transcription factors expressed in fetal or adult tissues (10). An example query for regulation includes using the Predicted Transcription Factor Target Genes query to find genes regulated by a specific transcription factor based upon the presence of a predicted binding site. An example transcript that resulted from a query for transcripts with Down regulated Expression in pancreas as compared with purified islets is shown in Figure 2A.
DoTS transcript annotation is displayed on the EPConDB transcript page (Figure 2B). Links to other annotations or views, available for the transcript, are included on the page. Expression profiles are accessed using the Expression Profile link and are displayed as graphs (Figure 2C). The transcript page also provides links to BCBC web pages describing publicly available antibody reagents. As part of a collaboration between EPConDB and T1DBase (www.t1dbase.org) (18), a type 1 diabetes-specific bioinformatics resource, EPConDB has integrated the T1DBase genome browser (GBrowse) (19), as shown in Figure 2D. The genome browser includes tracks for the BCBC promoter chip tiles, predicted transcription factor binding sites and DoTS transcripts.
As a user queries EPConDB, the web site keeps track of the results using the query history mechanism accessible from the ‘My Transcripts’ button located on each page. The results from queries can be further combined to narrow lists of transcripts. Using the Reportmaker tool, the resulting list of transcripts, are formatted and assembled into a report that can be downloaded. An online tutorial, available from the Help link on the navigation bar, supplies additional query examples. Queries for finding DoTS transcripts, Entrez Genes and accession numbers, associated with elements on the BCBC arrays, are readily available from the EPConDB Home page. A user can also use BLAST (20) to perform similarity searches for their sequence of interest against the human and mouse DoTS transcripts.
A Binding Site Collection Query available from the Analysis link allows users to search for arrangements of binding sites in promoters or introns. The search is performed on genomic sequence using DoTS transcripts aligned to the sequence to guide the search. Users select the distance constraint relative to the putative start of transcription, one or more binding sites to include in the search, how close to each other the binding sites must be and whether or not the order of the binding sites is important. A list of binding sites for pancreas-related factors is provided or the user can define their own sites.
IMPLEMENTATION
The EPConDB web interface (version 3.42) is currently implemented as a Java Servlet. Plans are underway to convert the web site to Java Server Pages, using a web development kit actively maintained (http://www.gusdb.org/wdk/). The underlying database accessed by EPConDB is a relational database utilizing the Genomics Unified Schema (GUS, http://www.gusdb.org) (21) and Oracle 10g. The expression studies are contained in RAD (http://www.cbil.upenn.edu/RAD), (12) a portion of the GUS schema reserved for storing expression data, analysis and annotations.
DISCUSSION AND FUTURE PLANS
Expression studies are continuously added to EPConDB, and contributions from all sources for relevant studies are welcome. The gene expression profiles are updated to include results from these studies. Additional studies are analyzed for differentially expressed genes and the resulting Gene Lists are added to the Up or Down regulated Expression Queries. New binding site predictions for other pancreatic transcription factors (e.g. Nkx2.2) are included in the Predicted Transcription Factor Target Genes Query. New versions of human and mouse DoTS transcripts are under development and will be incorporated into EPConDB.
Current plans for EPConDB include integrating data from transcription factor binding studies including ChIP-on-chip and SACO (serial analysis of chromatin occupancy) (22). The data can provide evidence for the validation of computationally predicted binding sites, thus providing high confidence target genes for transcription factors. As the number of expression studies in EPConDB increases, gene lists, created from joint analysis of multiple studies reflecting genes differentially expressed in the same context (e.g. differentiation of pancreatic cells) will be presented on EPConDB. There is also interest in incorporating data into EPConDB from protein expression studies, including sequences influenced by microRNAs (23) regulating their translation. It is the goal of EPConDB to provide insight into the cis-regulatory modules operating in gene expression networks crucial for the development and differentiation of the endocrine pancreas through integration and analysis of these different data sources.
Acknowledgments
EPConDB is supported by NIDDK grant, UO1 56947. Funding to pay the Open Access publication charges for this article was provided by the NIDDK.
Conflict of interest statement. None declared.
REFERENCES
- 1.Scearce L.M., Brestelli J.E., McWeeney S.K., Lee C.S., Mazzarelli J., Pinney D.F., Pizarro A., Stoeckert C.J., Jr, Clifton S., Permutt M.A., et al. Functional genomics of the endocrine pancreas: The Pancreas Clone Set and PancChip, New Resources for Diabetes Research. Diabetes. 2002;51:1997–2004. doi: 10.2337/diabetes.51.7.1997. [DOI] [PubMed] [Google Scholar]
- 2.Kaestner K.H., Lee C.S., Scearce L.M., Brestelli J.E., Arsenlis A., Le P.P., Lantz K.A., Crabtree J., Pizarro A., Mazzarelli J., et al. Transcriptional Program of the endocrine pancreas in mice and humans. Diabetes. 2003;52:1604–1610. doi: 10.2337/diabetes.52.7.1604. [DOI] [PubMed] [Google Scholar]
- 3.Rubins N.E., Friedman J.R., Le P.P., Zhang L., Brestelli J., Kaestner K.H. Transcriptional networks in the liver: hepatocyte nuclear factor 6 function is largely independent of Foxa2. Mol. Cell Biol. 2005;16:7069–7077. doi: 10.1128/MCB.25.16.7069-7077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le P.P., Friedman J.R., Schug J., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y., King B.L., Parvizi B., Brunk B.P., Stoeckert C.J., Jr, Quackenbush J., Richardson J., Bult C.J. Integrating computationally assembled mouse transcript sequences with the Mouse Genome Informatics (MGI) database. Genome Biol. 2003;4:R16. doi: 10.1186/gb-2003-4-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazzarelli J.M., White P., Gorski R., Brestelli J., Pinney D.F., Arsenlis A., Katokhin A., Belova O., Bogdanova V., Elisafenko E., et al. Novel genes identified by manual annotation and microarray analysis in the pancreas. Genomics. 2006 doi: 10.1016/j.ygeno.2006.04.005. 10.1016/j.ygeno.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockhart D.J., Dong H., Byrne M.C., Follettie M.T., Gallo M.V., Chee M.S., Mittmann M., Wang C., Kobayashi M., Horton H., et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 9.Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 10.Kong Y.M., MacDonald R.J., Wen X., Yang P., Barbera V.M., Swift G.H. A comprehensive survey of DNA-binding transcription factor gene expression in human fetal and adult organs. Gene Expr. Patterns. 2006 doi: 10.1016/j.modgep.2006.01.002. 10.1016/j.modgep.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., et al. Minimum Information About a Microarray Experiment (MIAME): toward standards for microarray data. Nature Genetics. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 12.Manduchi E., Grant G.R., He H., Liu J., Mailman M.D., Pizarro A.D., Whetzel P.L., Stoeckert C.J., Jr RAD and the RAD Study-Annotator: an approach to collection, organization and exchange of all relevant information for high-throughput gene expression studies. Bioinformatics. 2004;20:452–459. doi: 10.1093/bioinformatics/btg428. [DOI] [PubMed] [Google Scholar]
- 13.Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knuppel R., Dietze P., Lehnberg W., Frech K., Wingender E.J. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol. 1994;1:191–198. doi: 10.1089/cmb.1994.1.191. [DOI] [PubMed] [Google Scholar]
- 15.Sandelin A., Alkema W., Engström P., Wasserman W., Lenhard B. JASPAR: an open access database or eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant G.R., Liu J., Stoeckert C.J., Jr A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;11:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
- 17.Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smink L.J., Helton E.M., Healy B.C., Cavnor C.C., Lam A.C., Flamez D., Burren O.S., Wang Y., Dolman G.E., Burdick D.B., et al. T1DBase, a community web-based resource for type 1 diabetes research. Nucleic Acids Res. 2005;33:D544–D549. doi: 10.1093/nar/gki095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Davidson S., Crabtree J., Brunk B.P., Schug J., Tannen V., Overton G.C., Stoeckert C.J., Jr K2/Klesli and GUS: experiments in integrated access to genomic data sources. IBM Systems J. 2001;40:512–531. [Google Scholar]
- 22.Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Cuellar T.L., McManus M.T. MicroRNAs and endocrine biology. J. Endocrinol. 2005;187:327–332. doi: 10.1677/joe.1.06426. [DOI] [PubMed] [Google Scholar]