Abstract
Replication of eukaryotic chromosomes initiates at multiple sites called replication origins. Replication origins are best understood in the budding yeast Saccharomyces cerevisiae, where several complementary studies have mapped their locations genome-wide. We have collated these datasets, taking account of the resolution of each study, to generate a single list of distinct origin sites. OriDB provides a web-based catalogue of these confirmed and predicted S.cerevisiae DNA replication origin sites. Each proposed or confirmed origin site appears as a record in OriDB, with each record comprising seven pages. These pages provide, in text and graphical formats, the following information: genomic location and chromosome context of the origin site; time of origin replication; DNA sequence of proposed or experimentally confirmed origin elements; free energy required to open the DNA duplex (stress-induced DNA duplex destabilization or SIDD); and phylogenetic conservation of sequence elements. In addition, OriDB encourages community submission of additional information for each origin site through a User Notes facility. Origin sites are linked to several external resources, including the Saccharomyces Genome Database (SGD) and relevant publications at PubMed. Finally, a Chromosome Viewer utility allows users to interactively generate graphical representations of DNA replication data genome-wide. OriDB is available at www.oridb.org.
INTRODUCTION
Genomic stability during cell proliferation demands accurate and precise DNA replication and segregation. DNA replication initiates at discrete sites, called replication origins. The size of eukaryotic chromosomes requires that they are replicated from multiple replication origins and these origins must be well spaced to ensure that no region is left unreplicated. Eukaryotic chromosome replication is primarily controlled at the level of replication origin activation. Understanding where replication origin sites are located and how these sites are specified is crucial to our understanding of DNA replication and genome integrity (1).
How eukaryotic replication origins are specified is best understood in the budding yeast Saccharomyces cerevisiae, where specific sequences that confer origin activity (called Autonomously Replicating Sequences or ARS elements) have been isolated. The ability of yeast ARS elements to support plasmid replication has facilitated the identification and analysis of many replication origins in the yeast genome (2–4). Furthermore, the chromosomal activity of replication origins can be analyzed by separating replication intermediates by either neutral–neutral two-dimensional (2D) gel electrophoresis (5–8) or by neutral–alkaline 2D gel electrophoresis (9,10). The neutral–alkaline method, and a modification of the neutral–neutral method (the ‘fork-direction gel’; 11) both permit determination of replication fork direction and consequently the proportion of cell cycles in which a particular chromosomal replication origin is activated (its observed ‘efficiency’).
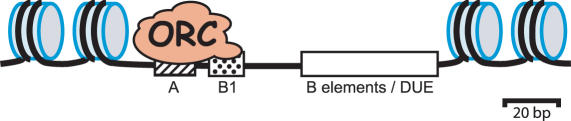
Combining plasmid assays for ARS activity with site-directed mutagenesis has revealed DNA elements required for replication initiation at origin sites. Budding yeast origins are almost exclusively intergenic (12) and consist of ∼200 bp sequences that can be divided into the A and B domains (Figure 1). The A domain contains an essential ARS consensus sequence (ACS), variously assigned as 11–17 bp in length that is the binding site for the Origin Recognition Complex (ORC) (13–15). ORC binding is required for the sequential recruitment of Cdc6, Cdt1 and MCM proteins during G1 to form a ‘pre-replication complex’ (pre-RC) that ‘licenses’ the origin for initiation in the subsequent S phase (16). A match to the ACS is essential but not sufficient for origin function, with only ∼500 of the ∼12 000 ACS matches in the yeast genome having replication origin function. Therefore additional surrounding sequences and/or chromatin states must be required to specify whether or not a particular ACS motif has replication origin function. The B region of an origin tends to be helically unstable (having a DUE or DNA unwinding element) and additionally contains a number of short sequence elements that contribute to origin activity, such as the B1, B2 and B3 elements of ARS1 (also known as ARSIV-463 or ARS416). The B1 element is thought to contribute to ORC recruitment (13–15); the B2 element resembles the ACS motif and is required for efficient loading of MCM proteins (17–19); the B3 element of ARS1 binds the transcription factor Abf1 (20) and excludes nucleosomes from the core origin sequences (19). Nucleosome exclusion is thought to be a general property of replication origins (12), however Abf1 does not bind at all origin sites and therefore other mechanisms must also be utilized to exclude nucleosomes.
Figure 1.
Cartoon of a Budding Yeast Replication Origin. Nucleosomes are excluded from the replication origin sequence elements which include the essential ACS (hatched box), the B1 (stippled box) and additional B elements (open box) that contribute to origin activity. The ACS and B1 elements are involved in ORC recruitment. Remaining B elements include a region of helical instability (the DUE or DNA Unwinding Element) as well as sequences that promote the loading of additional replication factors and the exclusion of nucleosomes.
The DUE, which overlaps the origin B elements, is presumed to facilitate origin unwinding (21,22). Sites where the duplex strands are most easily separated under the topological stresses that occur in vivo, called stress induced duplex destabilization (SIDD) sites, have been shown to collocate with replication origins (23). These SIDD sites are related to DUE sequences, and are similarly thought to facilitate localized DNA unwinding during the initiation of replication.
Four microarray-based studies mapped the approximate location of replication origins throughout the budding yeast genome. Two of these studies were based on measurement of the time at which each region of the yeast genome replicates, allowing approximate identification of origin sites as the earliest-replicating sequences in their locality (24,25). These studies identified origin sites that are active on the chromosome under the conditions analyzed. A third investigation used chromatin immunoprecipitation (ChIP) of origin-binding proteins (ORC and Mcm) to identify sites with potential origin activity, which were named proposed ARS (proARS) sites (26). Fourth, sites of replication initiation were mapped genome-wide by measuring the accumulation of single-stranded DNA (ssDNA) in cells challenged with the DNA replication inhibitor hydroxyurea (HU) (27). Each of these four studies produced separate lists of potential origin sites throughout the budding yeast genome, but none attempted to identify the individual ORC binding sequences (i.e. the ACS elements) [reviewed in (1)]. A recent investigation showed that the ACS elements of characterized replication origins tend to be phylogenetically conserved in closely related Saccharomyces species. Analysis of phylogenetic sequence conservation in likely origin regions allowed the genome-wide identification of ACS locations (termed proposed ACS or proACS) with base-pair resolution (12). In addition, this study confirmed origin activity associated with >200 of these proACS locations.
We have developed OriDB as a repository of information about S.cerevisiae DNA replication origins. The data used to compile OriDB has been collated from the genome-wide studies described above and from single origin studies, and OriDB thus brings together information that is currently difficult to access and compare because it has been presented disparately and spans the literature of the past quarter century. OriDB will provide a valuable resource for those working in the DNA replication field; moreover, by making the available datasets more accessible, the database will help researchers working in related fields analyze their results in relation to replication dynamics.
DATA COLLATION
Most of the information in the OriDB database is collated from four microarray-based studies, each of which produced a list of proposed origin sites (24–27), and a fifth study which produced a list of confirmed origin sites (12). We aimed to produce a single list of unique origin sites that assigns each origin location at the highest resolution available. First, it was necessary to develop criteria for OriDB to use in deciding whether closely spaced origin location assignments made by the various studies correspond to the same or distinct origins. We began by assessing the resolution of each microarray-based study by comparing its proposed origin locations with confirmed origin sites (12). Different source lists of origin location data were ranked, best first, based upon the estimated resolution of their origin location predictions. The six lists of origin sites used and the error values associated with each study are shown in Table 1.
Table 1.
Origin location datasets used by OriDB, ranked in order of estimated resolution of their origin assignments
| List | Data source | Origin location assignment | Estimated errors/bp |
|---|---|---|---|
| 1 | Cloned and assayed origins (12,26) | ARS-assayed fragment | +/− 0 |
| 2 | 2D gel-confirmed origins (24,27) | Analyzed restriction fragment | +/−0 |
| 3 | proARS sites identified by ChIP study (26) | proARS coordinate interval | +/−500 |
| 4 | Origins proposed by Copy Number Timing Study (25) | Peak location | +/−3500 |
| 5 | Origins proposed by ssDNA/HU study (27) | Peak location | +/−4000 |
| 6 | Origins proposed by Heavy:Light timing study (24) | Peak location | +/−7500 |
For each source of data the primary source reference used, the nature of the origin location information and the estimated errors are shown. Errors were calculated as described in (12). (For discussion of our treatment of ARS elements cloned and assayed on large fragments, see Supplementary Note 1).
These error values are taken into account by OriDB in deciding whether origins predicted by two different studies are the same. If origin location assignments overlap once appropriate errors are added, OriDB considers the assignments to represent a single origin. For example, origin sites identified by Raghuraman et al. (List 6) (24) and Yabuki et al. (List 4) (25) have estimated errors of 7.5 and 3.5 kb respectively. OriDB assigns origins identified by these two studies as the same if they lie <11 kb apart, but distinct if they lie >11 kb apart. (For further details of origin lists and resolution values see Supplementary Note 2.)
To amalgamate the data, the cohort of cloned origins (list 1) was first added to the database. To prevent duplicate entries, origins sites in the lower resolution datasets were labeled as ‘already included’ if the same site (assessed as described above) was added to the database from list 1. Next, those origins from list 2 not present on list 1 were added to the database. As before, lists 3–6 were then examined to ascertain whether they had identified each of the sites in list 2 and entries were labeled accordingly. Next, those origins on list 3 that had not yet been included in the database were added, again labeling lists 4–6 according to whether they had identified those sites. Outstanding assignments from the remaining lists, in order of study resolution, were then added in the same way. (For further details of this amalgamation process see Supplementary Note 3.)
Each proposed origin site is assigned a status by OriDB (Confirmed, Likely, or Dubious) that expresses our confidence that the site genuinely corresponds to an origin. ‘Confirmed’ origins are those have been cloned and tested by ARS assay and/or have been detected by 2D gel analysis. ‘Likely’ origin sites have been identified by two (or more) microarray studies but have not yet been confirmed. ‘Dubious’ origin sites are those identified by only one microarray study. In many cases, Dubious origin sites will correspond to ‘false positives’ arising due to the technical limitations associated with the experimental approach of the study concerned. The automated data-merge process has been developed to optimize the results produced, but will inevitably result in occasional inconsistencies or unexpected annotations. Occasionally, we have manually changed an automated Status assignment where there is evidence to suggest that it is incorrect; such alterations are marked with an asterisk (e.g. *Dubious ARS on Chromosome III at 85 kb) and explained in the User Notes tab for that Origin Site (see below). For all sites, we present as much information as possible to allow the user to make their own informed assessment.
Data from additional studies will be added to the database as they become available. This process may result in the criteria described above ‘evolving’. The status of predicted origins will certainly change as more origin sites are experimentally verified. At the time of writing OriDB contains 613 replication origin sites (279 Confirmed; 192 Likely; and 142 Dubious).
DATABASE ACCESS, INTERFACE AND CONTENT
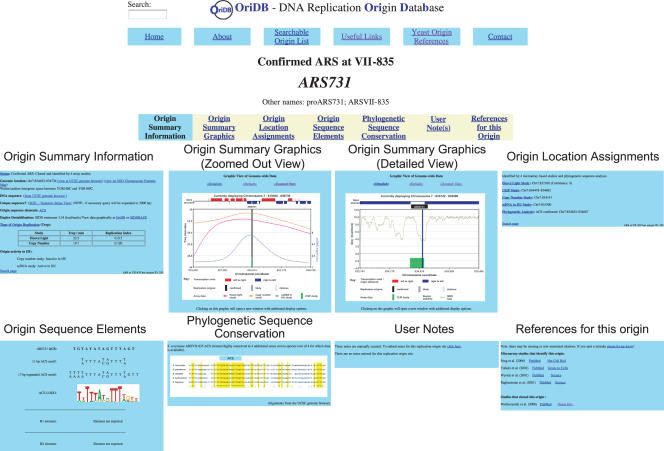
OriDB is freely available at http://www.oridb.org/. A user-friendly web-based interface invites visitors to explore the site and to examine the ‘Searchable Origin List’ that forms the heart of the database. The database can be searched using a search facility or by using the text box present on every web-page. Search results list the Name(s), Genomic location and assigned Status for each origin site that matches the entered search criteria, and provides a link to further details. These links take the user to the Origin Record pages. Each proposed or confirmed origin site appears as an Origin Record in OriDB, with each Record comprising seven dynamically generated pages presenting the collated information for that origin site. This information is displayed on each Origin Record under the following tab headings: Origin Summary Information; Origin Summary Graphics; Origin Location Assignments; Origin Sequence Elements; Phylogenetic Sequence Conservation; User Notes; and References for this Origin (see Figure 2). Brief descriptions of the information provided in each of these pages is given below; more complete descriptions are available on the OriDB About page (http://www.oridb.org/about.php#full).
Figure 2.
Screen Shots from the OriDB Origin Record for ARS731. Each origin site entry in OriDB comprises seven dynamically generated pages with a common Header (top). Example pages shown are: the Origin Summary Information; two (of the three available) Origin Summary Graphic views; Origin Location Assignments; Origin Sequence Elements; Phylogenetic Sequence Conservation; User Notes; and References for this Origin. Each page contains links to related pages within OriDB and to external sites. Where particular data is unavailable for an origin site this fact is explicitly stated.
Origin summary information
This tab presents a text summary of what is known about the origin site. The genome location (chromosome number and coordinate interval) assigned by the highest-resolution study to identify the origin, and the intergenic or genic location of the origin site are given. OriDB uses a static chromosome coordinate system that corresponds to that used by the UCSC genome browser (28) and the Oct 2003 release of the Saccharomyces Genome Database (SGD) (29). Links are provided to external databases (such as SGD and the UCSC Genome Browser) from which the DNA sequence of the origin site can be retrieved. Where appropriate some or all the following information may also be available: a summary of the origin substructure, including information about proposed and/or confirmed sequence elements and the stability of the DNA duplex at the origin site (‘Duplex Destabilization’—see below); time of replication during S phase; and the activity of the origin site when cells are treated with HU.
Origin summary graphics
This tab accesses a page allowing the display of three standardized graphic representations of DNA replication data for the origin site. The ‘Default’ and ‘Zoomed Out’ views of the origin locality show primary data from the four microarray-based studies (24–27), and graphically indicate the Status of the Origin site (Confirmed, Likely, or Dubious) and the gene structure for the chromosome window. The ‘Detailed’ view shows the free energy required to separate the DNA strands [superhelically induced duplex destabilization or SIDD data (23)] and the location of proposed or verified origin sequence elements. Replication origins are in general closely associated with SIDD minima, which are related to DUE sequences and are similarly thought to permit DNA unwinding during replication origin activation (23).
Clicking on any of these three standard graphics opens a more interactive graphic window that allows the user to specify display characteristics of dynamically generated plots (see discussion of the Chromosome Viewer below).
Origin location assignments
This tab displays the locations assigned for the origin site by the genome-wide studies and individual origin characterization studies, where available. The locations displayed on this page are as reported by the original studies—that is, they have not been expanded to include the errors described above for each study.
Origin sequence elements
The sequences of proposed or experimentally verified origin sequence elements are displayed on this tab. The essential ACS is shown in comparison to previously described A element consensi (12,30–32). Identified B elements are also shown.
Phylogenetic sequence conservation
Origin sequence elements are often phylogenetically conserved amongst the closely related sensu stricto Saccharomyces species (12). This tab indicates whether phylogenetic sequence conservation has been reported for any of the origin elements. ‘Highly conserved’ means that at least 12 out of 15 bp in the ACS are identical. Where conservation has been reported appropriate sequence alignments are presented (including sequences from S.cerevisiae, Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces kudriavzevii and Saccharomyces bayanus) (33,34). It should be noted that the sequence alignments used have not been manually adjusted for optimal origin conservation. Similarly, an origin could be incorrectly listed as ‘not conserved’ if genuine phylogenetic sequence conservation was not apparent from the alignments examined.
User notes
This tab presents any manually curated information about the origin site. OriDB users are encouraged to add further information about the origin using the link on this tab.
References for this origin
This tab lists references relevant to the origin site under various heading with links to external databases, such has PubMed (35). Only references that have been curated within OriDB are listed (71 at the time of writing). These references can be viewed by selecting the Yeast Origin References button present at the top of every page. Please contact the authors regarding corrections or additional references for inclusion.
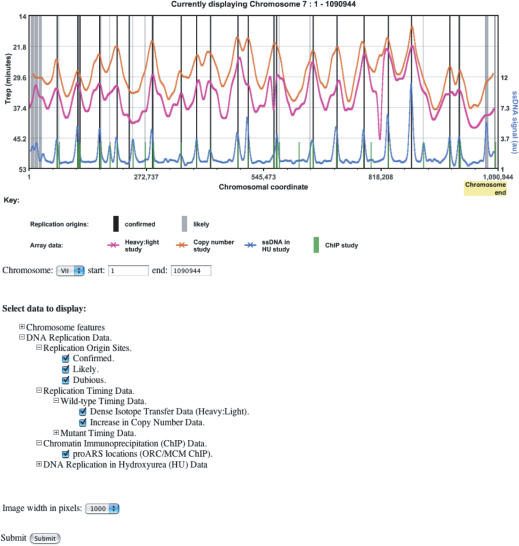
CHROMOSOME VIEWER
To complement the database of DNA replication origins, OriDB provides an interactive graphic interface for viewing chromosome data (Figure 3). Various entry points are available to this Chromosome Viewer utility: clicking on any of the graphic displays within an Origin Site Record opens a new window showing a Chromosome Viewer plot centered at that origin site; from the OriDB ‘Useful Links’ page, clicking on a chromosome number opens a new window with the Chromosome Viewer displaying available data for that entire chromosome. Various clickable options, available at the bottom of the Chromosome Viewer page, allow the user to specify display characteristics such as the chromosome and coordinate range to view and which datasets to display. Moving the mouse over the graphic gives further information about features on the plot and indicates available web links. These links include external web pages (e.g. SGD for further information on transcription units), OriDB pages (e.g. Origin Summary Pages) and the ability to navigate along the chromosome. Clicking on the chromosome scale bar (x-axis) recentres the display on the selected coordinate, whereas clicking within the graph window recenters and zooms in 2-fold on the selected coordinate. Buttons displayed above the graphic (not shown in Figure 3) allow further navigation. In summary, the Chromosome Viewer provides a powerful user interface to visualize a wide range of genomic data. This utility will facilitate better appreciation of the available data and understanding of the interplay between DNA replication and other chromosomal features.
Figure 3.
Example Screen Shot from the Interactive Chromosome Viewer. The OriDB Chromosome Viewer facility allows interactive graphic display of DNA replication data for any genomic location—in this case the whole of chromosome VII is shown. A control panel (below the plot) allows users to select datasets to display, the chromosome and coordinate range, and the size of the plot to be displayed. Users can also navigate using buttons above the plot (not shown).
DISCUSSION AND FUTURE DEVELOPMENTS
We present here a fully functional database that collates for the first time the available DNA replication origin datasets for budding yeast. This database will provide a valuable resource to those working in the field of DNA replication. By providing a consolidated dataset and powerful search facility, OriDB will also assist those working in related fields by making the existing datasets more accessible. Many areas of chromatin biology are impacted upon by DNA replication, and specifically by the location and nature of DNA replication origins. For example, it has been proposed the recombination hotspots co-localize with sites of DNA replication initiation (36), and the location of chromosome structural proteins (such as cohesion) may be influenced by the sites and activity of replication origins (37,38). Conversely, DNA replication and origin function are influenced by other chromosomal features, including transcription (39), chromatin structure (40) and chromosome context (41), and these relationships can be viewed using OriDB.
The structure of OriDB has been designed to allow for the incorporation of additional budding yeast datasets as they become available. Furthermore, we propose to develop OriDB to include parallel databases for other organisms as comparable DNA replication datasets are described.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
OriDB has been built on a large body of work only a fraction of which we have been able to mention here. We apologize to colleagues whose work has not been cited and remind readers that OriDB contains a more extensive and expanding list of cross-referenced citations. CAN is a Leverhulme Trust Early Career Fellow. ADD is Royal Society University Research Fellow. The contributions to this work by CJB and PAK were supported in part by Grant DBI-0416764 from the National Science Foundation. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.MacAlpine D.M., Bell S.P. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- 2.Stinchcomb D.T., Struhl K., Davis R.W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 3.Newlon C.S., Collins I., Dershowitz A., Deshpande A.M., Greenfeder S.A., Ong L.Y., Theis J.F. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- 4.Shirahige K., Iwasaki T., Rashid M.B., Ogasawara N., Yoshikawa H. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer B.J., Fangman W.L. The localization of replication origins on ARS plasmids in S.cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 6.Friedman K.L., Brewer B.J., Fangman W.L. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- 7.Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita M., Hori Y., Shinomiya T., Obuse C., Tsurimoto T., Yoshikawa H., Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–665. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]
- 9.Huberman J.A., Spotila L.D., Nawotka K.A., el-Assouli S.M., Davis L.R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J., Newlon C.S., Huberman J.A. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brewer B.J., Lockshon D., Fangman W.L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 12.Nieduszynski C.A., Knox Y., Donaldson A.D. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 2006;20:1874–1879. doi: 10.1101/gad.385306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowley A., Cocker J.H., Harwood J., Diffley J.F. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao H., Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl Acad. Sci. USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee D.G., Bell S.P. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diffley J.F. Regulation of early events in chromosome replication. Curr. Biol. 2004;14:R778–R786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Zou L., Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilmes G.M., Bell S.P. The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc. Natl Acad. Sci. USA. 2002;99:101–106. doi: 10.1073/pnas.012578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipford J.R., Bell S.P. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 20.Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 21.Umek R.M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988;52:559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 22.Natale D.A., Umek R.M., Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 1993;21:555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ak P., Benham C.J. Susceptibility to superhelically driven DNA duplex destabilization: a highly conserved property of yeast replication origins. PLoS Comput. Biol. 2005;1:e7. doi: 10.1371/journal.pcbi.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 25.Yabuki N., Terashima H., Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- 26.Wyrick J.J., Aparicio J.G., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M. Genome-wide distribution of ORC and MCM proteins in S.cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 27.Feng W., Collingwood D., Boeck M.E., Fox L.A., Alvino G.M., Fangman W.L., Raghuraman M.K., Brewer B.J. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell. Biol. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirschman J.E., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hong E.L., Livstone M.S., Nash R., et al. Genome Snapshot: a new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Houten J.V., Newlon C.S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol. Cell. Biol. 1990;10:3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theis J.F., Newlon C.S. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl Acad. Sci. USA. 1997;94:10786–10791. doi: 10.1073/pnas.94.20.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider T.D. Strong minor groove base conservation in sequence logos implies DNA distortion or base flipping during replication and transcription initiation. Nucleic Acids Res. 2001;29:4881–4891. doi: 10.1093/nar/29.23.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 34.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopes M., Cotta-Ramusino C., Liberi G., Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- 37.Suter B., Tong A., Chang M., Yu L., Brown G.W., Boone C., Rine J. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2004;167:579–591. doi: 10.1534/genetics.103.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glynn E.F., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieduszynski C.A., Blow J.J., Donaldson A.D. The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res. 2005;33:2410–2420. doi: 10.1093/nar/gki539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogelauer M., Rubbi L., Lucas I., Brewer B.J., Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 41.Cosgrove A.J., Nieduszynski C.A., Donaldson A.D. Ku complex controls the replication time of DNA in telomere regions. Genes Dev. 2002;16:2485–2490. doi: 10.1101/gad.231602. [DOI] [PMC free article] [PubMed] [Google Scholar]