Abstract
The RNA Editing Database (REDIdb) is an interactive, web-based database created and designed with the aim to allocate RNA editing events such as substitutions, insertions and deletions occurring in a wide range of organisms. The database contains both fully and partially sequenced DNA molecules for which editing information is available either by experimental inspection (in vitro) or by computational detection (in silico). Each record of REDIdb is organized in a specific flat-file containing a description of the main characteristics of the entry, a feature table with the editing events and related details and a sequence zone with both the genomic sequence and the corresponding edited transcript. REDIdb is a relational database in which the browsing and identification of editing sites has been simplified by means of two facilities to either graphically display genomic or cDNA sequences or to show the corresponding alignment. In both cases, all editing sites are highlighted in colour and their relative positions are detailed by mousing over. New editing positions can be directly submitted to REDIdb after a user-specific registration to obtain authorized secure access. This first version of REDIdb database stores 9964 editing events and can be freely queried at http://biologia.unical.it/py_script/search.html.
INTRODUCTION
RNA editing is a post-transcriptional process whereby a genetic message is modified from the corresponding DNA template by means of substitutions, insertions and deletions (1,2). It is widely dispersed between distant lines of evolution such as unicellular organisms, viruses and various species of eukaryotes, including animals and plants (1,2). According to the definition of Price and Gray (3), RNA editing describes only those processes of nucleotide alterations which result in different or additional nucleotides in the RNA. It is thus distinguished from the classical post-transcriptional RNA modification in which single RNA nucleosides are chemically changed, for example, by the addition of a methyl group or by other site-specific alterations (4,5).
RNA editing has a major impact on genes and gene expression in mitochondria and chloroplasts (1,2,6,7). The translation of mitochondrial mRNAs in kinetoplastids is meaningless without massive RNA editing of the transcripts by insertion and deletion of uridines (U) (6,7). In some kinetoplastid mRNAs, editing creates >90% of the amino acid codons. The corresponding genomic DNA sequences are barely recognizable by their residual sequence similarity and are appropriately called ‘cryptogenes’ (6,7).
The impact of RNA editing can also be crucial in cases where the physical extent of RNA editing is less dramatic and comparatively small (1,2,7). In plant mitochondrial and chloroplast transcripts, for instance, the replacement of a limited number of cytidines (C) by uridines (U) results in the translation of functionally competent and evolutionarily conserved polypeptides (8). RNA editing is thus an essential post-transcriptional process which ensures functional expression in a wide range of organisms (1,2,6,7).
Current primary databases do not always include editing information and, most frequently, they contain simple exception notes indicating the existence of RNA editing modifications, but omitting details about the editing type or the nucleotide positions affected. Moreover, present day primary databases do not accommodate an appropriate space to unambiguously store the post-transcriptional changes induced by RNA editing.
Nonetheless, some editing information can be retrieved from other secondary databases such as ChloroplastDB and the organelle genome database GOBASE, while neither is specialized to store RNA editing events (9,10). ChloroplastDB is a web-based database for fully sequenced plastid genomes, whereby allocated editing events are of course limited to only those occurring in chloroplasts. The GOBASE database is generated by an accurate parsing of GenBank flat-files and, thus, mainly includes the annotation details provided in primary databases. A compilation of specific edited mitochondrial sequences can be found at the U-insertion/deletion edited sequence database (11); however, the allocated editing information is restricted to post-transcriptional events in the mitochondria of kinetoplastid protozoa.
In any case, the retrieval of RNA editing events tends to be tedious and time consuming, especially when editing positions and related details are not included in current databases, but can be identified only in the pertinent primary literature.
In order to fill this gap and to take into account the physiological importance of RNA editing in organellar as well as nuclear and viral gene expression in a wide range of organisms, we started this initiative to collect and store all editing alterations such as insertions, deletions and substitutions in a novel and original specialized database called REDIdb (RNA EDIting database). This database now facilitates a number of diverse queries concerning information within and around the process of RNA editing. The new possibilities include, for example, the retrieval of editing sites not currently annotated in other databases and so far only accessible as scattered data reported in specific publications. REDIdb simplifies the recovery of edited sequences from user-specified species to perform comparative analyses or to generate training sets of sequences to test computational methods for editing site detection (12,13). Moreover, REDIdb allows to store editing information related to specific biochemical, physiological, pathological and/or mutant situations as, for example, specific instances of cytoplasmic male sterility (CMS) of plants and their respective associations with changes in the RNA editing patterns (14).
The REDIdb database can be freely queried at http://biologia.unical.it/py_script/search.html. This initial version is restricted to extra-nuclear cell organelles as targets, but REDIdb is ready to be extended to also cover nuclear and viral editing events.
DATABASE CONSTRUCTION
Present day GenBank records do not contain a standard feature to store RNA editing information. Editing sites are currently annotated in the feature table as ‘misc-feature’, which is reserved to parameters of biological interest that cannot be described by any other feature key. With the aim to collect editing events, all primary organellar sequence records which show changes due to RNA editing were downloaded in the flat-file format from GenBank. The procedure of extracting and storing editing sites was carried out as follows:
For each selected GenBank record, the feature table was parsed by ad hoc Python scripts utilizing modules from the Biopython project (15).
Editing positions and related nucleotide sequences were extracted from the parsed feature table of a GenBank entry.
Each editing site was assigned to the corresponding nucleotide sequence based on the genomic coordinates for that editing site.
Editing positions and nucleotide sequences were used to generate in silico complementary DNAs (cDNAs).
Before completing the annotation, editing sites and the corresponding genomic and cDNA sequences were manually checked to ensure data consistency.
All parsed editing information was stored in the database.
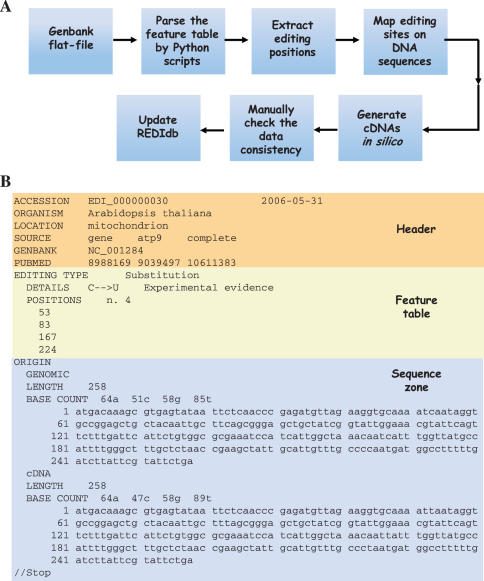
Following the above procedure (Flow chart depicted in Figure 1A), a large number of editing sites were included in REDIdb, even though sometimes several inconsistent GenBank annotations were found. Such divergent annotations were usually the results of human errors and were corrected by hand to the best of our knowledge by referring to the pertinent primary literature before inclusion in the database. In cases where editing sites were not reported in GenBank flat-files or only exception notes were found, the relevant primary literature sources were used to retrieve additional RNA editing events. When plant mitochondrial genes were taken into account, data consistency was assessed by comparing the parsed annotations with a highly reliable compilation of RNA editing sites in plant mitochondrial genes (kindly provided by J.P. Mower), which was designed to and is currently used to efficiently predict post-transcriptional RNA editing events in plant mitochondrial transcripts (12). On the other hand, inconsistent and conflicting GenBank annotations in kinetoplast genes were improved by comparing the annotations with those stored in the U-insertion/deletion edited sequence database (11).
Figure 1.
(A) Flow diagram of the various steps used during database construction and (B) dissection of the REDIdb flat-file structure.
The editing information accommodated in REDIdb is organized in specific flat-files, in which it is possible to distinguish a header containing the main features of the record (accession number, organism, intracellular location, gene name, GenBank and PubMed cross-links), a feature table with all editing events and details such as positions, editing type(s) and detection, and a sequence zone with both the genomic sequence and the corresponding edited transcript. An example of a REDIdb flat-file is given in Figure 1B for the atp9 gene in mitochondria of Arabidopsis thaliana (more details about REDIdb flat-file structure are available at http://biologia.unical.it/py_script/structure.html).
DATABASE CONTENT
The RNA editing database presently stores 9964 editing events distributed over 706 different nucleotide sequences. Sixty-seven per cent of these editing alterations are due to substitutions, 30% to insertions and 3% to deletions. With respect to sequence location, 486 sequences come from mitochondria, the remaining 220 sequences are from chloroplasts.
REDIdb is currently in the process of expansion to also include editing events in nuclear and viral sequences. The statistics page of the REDIdb website (http://biologia.unical.it/py_script/statistics.html) can be consulted for further details about the continuously updated current content of the database.
DATABASE WEB INTERFACE
REDIdb is a relational database developed using the MySQL program. It can be accessed through an easy CGI/Python-based web interface at http://biologia.unical.it/py_script/search.html.
Each REDIdb web page provides help and links to simplify browsing through the facilities of the website.
Individual entries of interest can be searched in REDIdb by appropriate query strings containing alternatively or in any combination: the gene name, the intracellular location, the type of molecule (such as tRNA, rRNA, intron) or the organism. Each record of the REDIdb database can also be retrieved by its specific accession number (more help on REDIdb searching is available at http://biologia.unical.it/py_script/help.html).
Browsing of each REDIdb entry and the identification of editing sites is facilitated by two ad hoc tools: one graphically displays genomic and cDNA sequences, the other shows the corresponding alignment and the functional protein sequence deduced from the edited cDNA. In both views editing sites are highlighted in colour and their relative positions are accessed in detail by mousing over.
Similar to other biological databases, all REDIdb entries can be downloaded in the flat-file format to quick-search by appropriate user-defined scripts. Moreover, REDIdb allows the user to directly submit new edited sequences. This submission is limited to registered users and submitted sequences need to have been previously annotated in primary databases. Username and password for uploading RNA editing data can be easily obtained from the REDIdb staff by the completion of a generic form (see the http://biologia.unical.it/py_script/registration.html page).
An overview of the REDIdb web interface is shown in Figure 2.
Figure 2.
An overview of the REDIdb web interface. (A) The main search page. (B) The ‘result’ page for the query string ‘atp9 arabidopsis’. (C) Graphic display of genomic and cDNA sequences in which editing sites are highlighted in colour. (D) The alignment of genomic and cDNA sequences with the editing sites displayed in colour. (E) Fasta format of genomic and cDNA sequences.
IMPLEMENTATION
REDIdb database is implemented in MySQL version 4.0 while all facilities to store and interact with the database were written in v2.3.5 of the Python scripting language making use of two non-standard libraries such as MySQL-python v1.2.0 and Biopython v1.41 (14). All procedures are executed on a Windows 2003 server and the database will be maintained at the Area Informatica e Telematica of the Università della Calabria (Italy).
FUTURE PROSPECTS
The addition of new RNA editing events to REDIdb will continue with the inclusion of post-transcriptional editing alterations from nuclear and viral nucleotide sequences. However, the authors solicit comments concerning existing entries, errors or omissions, and suggestions for improvements (see the http://biologia.unical.it/contact.html page).
Further planned developments include the addition of similarity searching capabilities by means of BLAST programs and an increased portability of database entries using a well-structured XML file format.
Acknowledgments
The authors are grateful to J.P. Mower for providing a compilation of edited plant mitochondrial genes and for helpful comments. The authors thank the Centro di Eccellenza per il Calcolo ad Alte Prestazioni for making available computing facilities and Dr F. Di Maio from the Area Informatica e Telematica of the Università della Calabria for providing technical support about the MySQL server. They also acknowledge the colleagues of the Molekulare Botanik Laboratory at the Universität Ulm for valuable and useful suggestions on the REDIdb web interface. Funding to pay the Open Access publication charges for this article was provided by grants from Dipartimento di Biologia Cellulare, Università della Calabria and the Italian Ministero dell'Istruzione, Università e Ricerca (MIUR).
Conflict of interest statement. None declared.
REFERENCES
- 1.Brennicke A., Marchfelder A., Binder S. RNA editing. FEMS Microbiol. Rev. 1999;23:297–316. doi: 10.1111/j.1574-6976.1999.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 2.Gray M.W. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life. 2003;55:227–233. doi: 10.1080/1521654031000119425. [DOI] [PubMed] [Google Scholar]
- 3.Price D.H., Gray M.W. Editing of tRNA. In: Grosjean H., Benne R., editors. Modification and Editing of RNA. Washington, DC: ASM Press; 1998. pp. 289–306. [Google Scholar]
- 4.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rozenski J., Crain P.F., McCloskey J.A. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton T.L., Landweber L.F. Rewriting the information in DNA: RNA editing in kinetoplastids and myxomycetes. Curr. Opin. Microbiol. 2002;5:620–626. doi: 10.1016/s1369-5274(02)00379-x. [DOI] [PubMed] [Google Scholar]
- 7.Maier R.M., Zeltz P., Kössel H., Bonnard G., Gualberto J.M., Grienenberger J.M. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 8.Regina T.M., Lopez L., Picardi E., Quagliariello C. Striking differences in RNA editing requirements to express the rps4 gene in magnolia and sunflower mitochondria. Gene. 2002;286:33–41. doi: 10.1016/s0378-1119(01)00802-2. [DOI] [PubMed] [Google Scholar]
- 9.Cui L., Veeraraghavan N., Richter A., Wall K., Jansen R.K., Leebens-Mack J., Makalowska I., de Pamphilis C.W. ChloroplastDB: the chloroplast genome database. Nucleic Acids Res. 2006;34:D692–D696. doi: 10.1093/nar/gkj055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien E.A., Zhang Y., Yang L., Wang E., Marie V., Lang B.F., Burger G. GOBASE—a database of organelle and bacterial genome information. Nucleic Acids Res. 2006;34:D697–D699. doi: 10.1093/nar/gkj098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson L., Wang S.H., Thiemann O.H., Alfonzo J.D., Maslov D.A., Avila H.A. U-insertion/deletion edited sequence database. Nucleic Acids Res. 1998;26:170–176. doi: 10.1093/nar/26.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mower J.P. PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinformatics. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picardi E., Quagliariello C. EdiPy: a resource to simulate the evolution of plant mitochondrial genes under the RNA editing. Comput. Biol. Chem. 2006;30:77–80. doi: 10.1016/j.compbiolchem.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Hernould M., Suharsono S., Zabaleta E., Carde J.P., Litvak S., Araya A., Mouras A. Impairment of tapetum and mitochondria in engineered male-sterile tobacco plants. Plant Mol. Biol. 1998;36:499–508. doi: 10.1023/a:1005946104983. [DOI] [PubMed] [Google Scholar]
- 15.Mangalam H. The Bio* toolkits—a brief overview. Brief Bioinformatics. 2002;3:296–302. doi: 10.1093/bib/3.3.296. [DOI] [PubMed] [Google Scholar]