Abstract
Protein Inter-atomic Distance Distributions (PIDD) is a dedicated database and structural bio-informatics system for distance based protein modeling. The database is developed to host and analyze the statistical data for protein inter-atomic distances based on their distributions in databases of known protein structures such as in the Protein Data Bank (PDB). PIDD is capable of generating, caching, and displaying the statistical distributions of the distances of various types and ranges. The collected information can be used to extract geometric restraints or mean-force potentials for protein structure determination including nuclear magnetic resonance structure determination and comparative model refinement. PIDD is supported with a friendly designed web interface so that users can easily specify the distance types and ranges, and retrieve, visualize or download the distributions of the distances as they desire. PIDD is freely accessible at http://www.math.iastate.edu/pidd
INTRODUCTION
The knowledge on inter-atomic distances in proteins is a valuable source of information for protein structural analysis and structure determination. The protein inter-atomic distances may be detected by using physical experiments such as nuclear magnetic resonance spectroscopy (NMR) (1), or estimated with the chemistry knowledge on various types of bond lengths and bond angles (2,3). However, in either case, only a small subset of all distances can be obtained due to various technical reasons (1,4). They can only be estimated approximately in certain ranges instead of exact values as well because of the inevitable estimation errors. Therefore, obtaining additional distance information beyond the current theoretical and experimental limitations is always important yet challenging for the further development of distance-based protein modeling.
In this paper, we introduce a computational approach of deriving distance data for proteins based on the distributions of the distances in the databases of known protein structures. In particular, we describe the development of a protein distance distribution database Protein Inter-atomic Distance Distributions (PIDD) for calculating and storing the distributions of the distances in databases of known protein structures and using the distribution data to derive distance constraints and mean-force potentials (5,6) for structural analysis and modeling.
The basic idea of our approach is that in order to estimate the distances for various pairs of atoms, we find all the information for how the distances for different pairs of atoms are distributed in known proteins or, more accurately, known protein structures. Then, for each distance, we assign a probability according to the distribution of the distances of the same kind. Such probability information can be very useful for evaluating estimated distances or building proper protein conformations. For example, in order to see if 5 Å is a proper distance between Cα in alanine and Cβ in tryptophan when the two residues are separated by a cysteine, we calculate all the distances of the same type in the known proteins in structural databases and then group the distances according to their lengths. We can then obtain the distribution of this type of distances within a given distance range, say in between 0 and 50 Å, where the probability for the distance to be 5 Å can be identified easily. Figure 1 shows more examples for protein inter-atomic distance distributions calculated from databases of known protein structures.
Figure 1.
Example distance distributions. The graph on the left is the distribution of the distances between Cα in Tyr and Cα in Tyr separated by Lys in sequence. The graph on the right is the distribution of the distances between Cα in Ser and Cα in Trp separated by Gly in sequence.
Indeed, based on our calculations on the distributions of the distances in the structures in Protein Data Bank (PDB) (7), we have found that (i) the majority of short to medium ranged distances are non-uniformly distributed, indicating that proteins do have preferences when forming these distances; (ii) as more and more protein structures are determined, good estimations on the distributions of the distances are possible, and they can be obtained with reasonable statistical significances; (iii) many distances in low-resolution structures have deviated from their average distributions by >2 SD, and in most cases, the deviations have been found in under-determined regions of proteins; and (iv) it follows that distance constraints or mean-force potentials can be derived from the distributions of the distances and be applied to ‘correct’ or ‘refine’ low-resolution structures (8,9).
Although the importance of the distance distribution data is easy to justify, the calculation of the data can be daunting, requiring a complete search for the distances in structural databases for each different distance type, and there can be millions of different distance types, defined in terms of the types of the two atoms related to the distance, the types of the two corresponding residues and the types of the residues separating them in the sequence. Even just storing and managing such an enormous amount of data can be quite challenging. For this reason, we have developed a database system for automatically generating, storing and analyzing all the distribution data for protein inter-atomic distances. The system consists of two coupled databases, one called the structural database for storing high-resolution structures downloaded from structural databases and another called the distance database for storing the distribution data for the distances. The data in the distance database are calculated and collected from the structural database. The distance database can be used by the users to store, query and analyze the distributions of any distances of interest. In any event, at the beginning, only the data for commonly used distance types are computed and stored, to avoid unnecessary space use. If the distributions for certain distances are requested, but not pre-calculated and pre-stored yet, they will be computed right away from the structural database and stored into the distance database afterwards. In this way, the database can eventually be developed to contain necessary distance distributions, yet does not have to keep all the overwhelming information. The database system is developed using MySQL. Currently, it has 2090 high-resolution structures downloaded from the PDB and up to 320 000 000 distance distribution records. The system is supported with a friendly designed web interface so that users can easily specify the distance types and ranges, and retrieve, visualize or download the distributions of the distances as they desire. It is accessible at http://www.math.iastate.edu/pidd freely.
SYSTEMS AND METHODS
Data source
When downloading the known protein structures from the PDB, we have considered only those containing the chains of amino acids rather than protein complexes such as protein–DNA, protein–RNA and protein–protein complexes. To obtain more accurate and reliable results, we only downloaded structures determined by X-ray crystallography with resolution >2.0 Å. In future, we will consider including NMR structures as well. To reduce the redundancy in homologous structures, only proteins with sequence similarities <70% were used. Based on these criteria, total 2090 qualified protein structures were selected from the PDB as on April 12, 2005.
Data structure
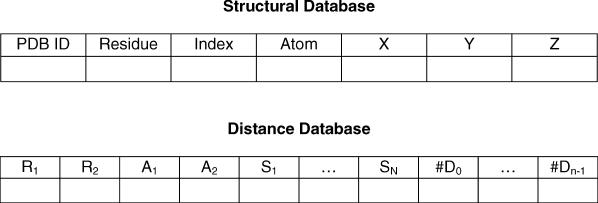
PIDD has two levels of databases, one called the structural database and another called the distance database. Both databases are implemented using MySQL. The structural database stores the sequence and structure information for a large set of high-resolution protein structures, with a similar data structure as the structural data represented in the PDB. Each record in the structural database is similar to an atom record in the PDB file, but contains a smaller number of fields. It has the PDB name of the protein, the residue name, the index for the atom, the atom name, and the x, y, z coordinates of the atom (see Figure 2). All the PDB files of the downloaded protein structures are converted into this format and stored in the structural database as MySQL database files. By using the MySQL database management system, the structure files can be processed much more efficiently and directly. No special scripts are required to parse the regular PDB text files. The distance database stores the distributions of the distances in known proteins calculated for every different type of distances. The calculations were based on the distributions of the distances in the downloaded structures in the structural database.
Figure 2.
Data structures of the databases. (Upper panel) The record of the atom in the structural database: PDB ID: ID of protein in the PDB; Residue: the name of the residue containing the atom; Index: the index for the atom; Atom: the name of the atom; X, Y, Z: x, y, z coordinates of the atom. (Lower panel) The record for the distribution of the distance, one for each different type: R1, R2: the two atoms; A1, A2: the two atoms; S1, …, SN: separating residues; #D0: the number of distances in [Di, Di+1], i = 0, …, n − 1.
In order to obtain the distribution data for the distances of various types and ranges, we specify the distances by using the types of the atoms it involves, the types of the residues containing the atoms and the types of the residues in between the two end residues in sequence. After calculating and collecting all the distances of each distance type from the structural database of PIDD, the statistical distribution of each distance type can be obtained. Note that although it is possible to include intra-residue distances, currently, we have considered only the inter-residue distances. Let D be the distance between two atoms, A1 and A2. Let R1 and R2 be the two residues where A1 and A2 are located, respectively. Let S1, … , SN be the residue sequence in between R1 and R2. Then, the distribution of the distance D between atoms A1 in R1 and A2 in R2 where R1 and R2 are separated by S1, … , SN can be represented by a distribution function P[A1, A2, R1, R2, S1, … , SN](D) and defined for any D in [Di, Di+1], where Di = 0.1 × i Å, i = 0, 1, … , n, to be the number of collected distances of this particular type in [Di, Di+1], normalized by the total number of collected distances of the same type in all [Di, Di+1], i = 0, 1, … , n.
Each record in the distance database therefore contains the distribution data for a particular type of distances, and it has the types of atoms, A1 and A2, the types of ending residues, R1 and R2, and the types of separating residues, S1, … , SN, that define the type of the distances followed by the number of distances of this type found in each of the distance intervals [Di, Di+1], i = 0, 1, … , n − 1.
System architecture
PIDD is implemented using MySQL. It consists of two databases, structural database and distance database, and three computational engines, such as search engine, distribution engine and visualization engine (Figure 3). In addition, there is a program written in Perl for automatically downloading the structures from the PDB and updating the structural database, and a web interface written in HTML for users to get online access to the system.
Figure 3.
PIDD system architecture. This automated system could generate and process the data dynamically. The system is implemented in MySQL and Perl. The user could access freely the database at http://www.math.iastate.edu/pidd. It requires specifying and inputting the distance type and then the user could choose to view the graph of distribution function as well as download the related results.
The structural database stores the sequence and structure information for a large set of high-resolution protein structures. The distance database stores the distribution data for the distances, with one record for one distance type. Since the distance type is defined in terms of the atom types, residues types and the separating residues, there can be a huge number of distance types and the amount of distribution data can be enormous. For example, if we assume that there are 10 different atoms types for A1 and A2, 20 different residue types for R1, R2, S1, … , SN, then even just for the distances with three separating residues (N = 3), there are already 320 million possible distance types. For this reason, we purposely design the system to have both structural and distance databases so that the distance database can actually be built dynamically from the structural database. More specifically, at the beginning, we only compute and store the distribution data for some commonly used distance types, which can certainly be queried or processed directly in the distance database. However, if the distributions for certain distances that are not pre-calculated and pre-stored are requested, they will be computed on fly from the structural database and stored into the distance database afterwards. In this way, the database can eventually be developed to contain all necessary distance distributions, yet does not have to be overwhelmed by the possible combinatorial growth of data, saving both storage space and search time.
The computational engines work together as follows. The search engine takes the query from a user and searches for the distribution of the specified type of distances in the distance database. If the requested distribution has been pre-calculated and pre-stored in the distance database, the search engine returns with it directly. Otherwise, the distances of the specified type will be computed and collected from the structural database and passed to the distribution engine. Based on the collected distances, the distribution engine calculates the distributions of the distances over discrete distance intervals and saves them in the distance database. The visualization engine is responsible for displaying the requested distribution function through a graphics interface. Figure 3 shows the architecture of PIDD graphically. Note that the structural database can be updated whenever new proteins are deposited into the PDB and the access to PIDD can be carried out conveniently through a well-designed web interface.
Features
A web user interface is designed so users can get access to PIDD anywhere online. It also provides various visualization tools and functions for researchers to display and analyze requested data. The users can obtain helps from the tutorial, references or related publications available at the website. The tutorial is well written and provides many examples.
The front page of the interface describes the PIDD system, its design purpose and the user guideline. More in-depth description about research on database-derived distance constraints and mean-force potentials and distance-based protein modeling is given in the research page. The links to tutorial, references and publications are also provided. Currently, the PIDD front page can be reached with its internet address (http://www.math.iastate.edu/pidd/).
Several pages are directed from the PIDD front page. One of them, as shown in Figure 4, allows the users to choose the distance type to be searched for via simple menu selections. Typically, the users follow three selection steps: (i) specify the two end residues and the number of separating residues; (ii) specify the types of the two atoms in the two end residues, respectively, and the types of all separating residues; and (iii) submit the query. The system returns with the distribution of the specified type of distances and displays it in a graph as shown in Figure 5. The current version of PIDD allows the users to specify up to three separating residues and handles one distance type per query, but it can be used simultaneously by multiple users.
Figure 4.
PIDD input selections. A user needs to specify the types of the two end residues (20 possibilities) and the number of separating residues first, and then choose the two atoms of the distance and the types of the separating residues.
Figure 5.
Graphics display. The distribution of the distances of the specified type is displayed in a graph. The distance range is up to 30 Å, and the length of each distance interval (bin) is either 0.1 or 0.2 Å.
Sample applications
The purpose for the development of PIDD is to provide an easy access to the information on how the inter-atomic distances are formed as revealed in their distributions in known proteins. Such information can be valuable for protein structural analysis, classification as well as modeling building. In particular, it can be used to extract geometric restraints or mean-force potentials for protein structure determination including NMR structure determination and comparative model refinement.
The distance distribution data have been used to analyze NMR determined structures as reported previously (8). The inter-atomic distances for 462 averaged and energy-minimized NMR structures downloaded from the PDB were examined and compared with their distribution functions (more specifically, for distances between atoms in two residues separated by zero or one residue). The results showed that many of these distances have deviations >2 SD. For example, the distribution of the distance between Cβ in Ala and the carbonyl C in Asp separated by one residue was found to have a mean ∼7.1 Å and SD 1.05 Å, whereas the distance between such a pair of atoms across the 20th and 22nd residues in the NMR structure 2GB1 was 4.6293 Å, which was 0.3707 Å smaller than the mean − 2 SD. More example cases of distance deviations in 2GB1 are given in Table 1. In fact, in each of the 462 NMR structures, similar deviations were found from 2 to 44%, or in an average of 21.98% of the residue pairs that are separated by one or zero residue along the protein backbone. The deviations were not only found among backbone atoms (N, O, C, Cα), but also between backbone (N, O, C, Cα) and side-chain atoms (Cβ). In most cases, the residues having such distance deviations were located on exposed parts of the proteins, which was consistent with the fact that the surface residues are usually of high mobility and more difficult to determine by using NMR.
Table 1.
Distance deviations in NMR determined structuresa
| Res. No. | Res. 1 | Atom 1 | Res. no. | Res. 2 | Atom 2 | Mean | 2 × SD | Distance |
|---|---|---|---|---|---|---|---|---|
| 19 | Glu | C | 20 | Ala | C | 3.1 | 0.4 | 3.62 |
| 20 | Ala | CB | 22 | Asp | C | 7.1 | 2.1 | 4.63 |
| 20 | Ala | CB | 22 | Asp | O | 7.8 | 2.5 | 3.53 |
| 21 | Val | N | 22 | Asp | O | 5.9 | 1.0 | 4.28 |
| 21 | Val | CB | 23 | Ala | N | 5.7 | 0.9 | 6.95 |
| 22 | Asp | CB | 23 | Ala | C | 5.4 | 0.6 | 4.69 |
aAtomic pairs (Atom 1 and Atom 2) across some of the residues (Res. 1 and Res. 2) in 2GB1 with distances deviated more than twice of their standard deviations (SD) from their average distributions (Mean) in known protein structures.
An important application of PIDD is structure determination or refinement. A set of distance constraints or mean-force potentials can be obtained by using the distribution data and applied to structure determination and refinement, e.g. to NMR structure determination and refinement. In general, a set of inter-proton distances of a protein can be obtained by using NMR spectroscopy. The protein structure can then be determined by solving a so-called distance geometry problem (10). However, many regions in NMR determined structures are often under-determined due to incomplete or inaccurate distances data. Overall, the quality and resolution of NMR determined structures are still not as high as X-ray crystallographic structures (11).
In order to increase the accuracy of the NMR determined structures, Cui et al. (8) and Wu et al. (9) used the distributions of the inter-atomic distances in known proteins as calculated in PIDD and derived a set of range constraints and mean-force potentials for the distances, and applied them to refining a set of NMR determined structures, along with original NMR experimental constraints. The results showed that with additional distance constraints or mean-force potentials, the structures were improved significantly in terms of standard measures, including the energies of the final structures, the Ramachandran plots, the RMSD values of the structures compared with X-ray reference structures, etc. For example, as shown in Figure 6, the percentage of the residues of the prion E200K in the most favorable region of the Ramachandran plot was increased from 85% (left) to 90% (right) after the protein was refined by using the database-derived distance constraints.
Figure 6.
Ramachandran plots for original and refined E200K. After employing additional distance constraints, the Ramachandran plot of NMR determined structures for prion E200K is improved significantly, with 85% of the residues in the most favorable region (left) increased to 90% of the residues in the most favorable region (right).
It is well-known that NMR determined structures are not as detailed as X-ray crystal structures. The discrepancies between the NMR and X-ray structures may be due to the flexibilities of the NMR structures in solution, whereas some of them may indeed be caused by the incorrectly formed regions in the NMR models. As indicated in the above applications, the distance distributions generated from PIDD can clearly be used to either find possible errors existing in NMR determined structures or generate additional distance constraints or potentials to refine the structures. There is also a great potential of using the same type of data for refining comparative models.
Future developments
The current version of PIDD has provided the basic functions for processing the data for protein distance distributions. More tools will be developed to facilitate various purposes of structural analysis including the tools for computing the distributions of the distances under more structural conditions, such as the distributions of the distances of certain types only when they are in alpha helices or beta sheets. Currently, we have only considered relatively short-range distances with maximal three separating residues in sequence. In future, we will also include all statistically significant long-range distance distributions. The reason that we have not considered the distances of all ranges is that many long-range distances either do not have clear distribution patterns or are difficult to sample and analyze. With the increasing number of high-resolution structures being determined, many structural properties, such as torsion angles, inter-atomic distances, residue volumes and side-chain orientations, can be analyzed from their statistical distributions in known proteins. Therefore, in future, we will extend our work on PIDD to the development of a general protein geometry database that includes the statistical distribution data for many other protein geometric properties besides the distances. Such a system will be able to provide more complete information on protein conformations and have even greater potentials as bioinformatics tools for protein structural analysis and structural modeling.
Acknowledgments
The authors would like to thank Peter Vedell, Ajith Gunaratne and Andrew Severin for helpful discussions, and the two anonymous referees for their valuable suggestions. The work is partially supported by the research funds provided by the Department of Mathematics, the Graduate Program on Bioinformatics and Computational Biology, and the Lawrence Baker Center for Bioinformatics and Biological Statistics at Iowa State University. Funding to pay the Open Access publication charges for this article was provided by the Lawrence Baker Center for Bioinformatics and Biological Statistics at Iowa State University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 2.Brooks C.L., Karplus M., Pettitt M. Proteins: A Theoretical Perspective of Dynamics, Structure, and Thermodynamics. John Wiley & Sons, NY; 1988. [Google Scholar]
- 3.Creighton T.E. Proteins: Structures and Molecular Properties, 2nd edn. Freeman and Company, NY; 1993. [Google Scholar]
- 4.Clore G.M., Gronenborn A.M. Determination of three-dimensional structures of proteins in solution by nuclear magnetic resonance spectroscopy. Protein Eng. 1987;1:275–288. doi: 10.1093/protein/1.4.275. [DOI] [PubMed] [Google Scholar]
- 5.Sippl M.J. Calculation of conformational ensembles from potentials of mean force. J. Mol. Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 6.Hendlich M., Lackner P., Weitckus S., Floeckner H., Froschauer R., Gottsbacher K., Casari G., Sippl M.J. Identification of native protein folds amongst a large number of incorrect models. The calculation of low energy conformations from potentials of mean force. J. Mol. Biol. 1990;216:167–180. doi: 10.1016/S0022-2836(05)80068-3. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F., Jr, Brice M.D., Rodgers, Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 8.Cui F., Jernigan R., Wu Z.J. Refinement of NMR-determined protein structures with database derived distance constraints. J. Bioinform. Comput. Biol. 2005;3:1315–1329. doi: 10.1142/s0219720005001582. [DOI] [PubMed] [Google Scholar]
- 9.Wu D., Jernigan R., Wu Z.J. Proteins. 2006. Refinement of NMR-determined protein structures with database derived mean force potentials. In press. [DOI] [PubMed] [Google Scholar]
- 10.Crippen G.M., Havel T.F. Distance Geometry and Molecular Conformation. UK: Research Studies Press; 1988. [Google Scholar]
- 11.Doreleijers J.F., Rullmann J.A., Kaptein R. Quality assessment of NMR structures: a statistical survey. J. Mol. Biol. 1988;281:149–164. doi: 10.1006/jmbi.1998.1808. [DOI] [PubMed] [Google Scholar]