Abstract
With the advent of automated and high-throughput techniques, the number of patent applications containing biological sequences has been increasing rapidly. However, they have attracted relatively little attention compared to other sequence resources. We have built a database server called Patome, which contains biological sequence data disclosed in patents and published applications, as well as their analysis information. The analysis is divided into two steps. The first is an annotation step in which the disclosed sequences were annotated with RefSeq database. The second is an association step where the sequences were linked to Entrez Gene, OMIM and GO databases, and their results were saved as a gene–patent table. From the analysis, we found that 55% of human genes were associated with patenting. The gene–patent table can be used to identify whether a particular gene or disease is related to patenting. Patome is available at http://www.patome.org/; the information is updated bimonthly.
INTRODUCTION
Recent advances in high-throughput sequencing technologies have enabled us to determine many genomic sequences quickly and cheaply. The use of biological sequence information has greatly facilitated the R&D process in the pharmaceutical, agricultural, medical and chemical industries (1). Filing a patent application is the key step to protect sequence intellectual property. Sequences patent owners can gain tremendous value and control over the exploitation of the sequences (2). Over the past several decades, the number of patent applications containing nucleic acid or amino acid sequences has been increasing rapidly.
For patent-related biological sequences, there are both public and commercial databases that provide patent information (3). Public efforts are represented by GenBank (4), the European Molecular Biology Laboratory (EMBL) (5) and the DNA Database of Japan (DDBJ) (6). GenBank obtains patent sequences from the patents issued by the US Patent and Trademark Office (USPTO). EMBL and DDBJ also obtain sequences from their respective patent offices. The websites of these three public databases offer patent sequence data downloading, simple keyword searching and alignment searching. In addition, PatGen (7), an integrated database containing sequence data from public resources, provides access to non-redundant sequence information, as well as an abstract service. The commercial sector is led by Chemical Abstract Service (http://www.cas.org/) and Derwent (http://www.derwent.com/) with their corresponding databases: CAS Registry and Derwent GENESEQ.
Although these patent databases provide good information on patent-related sequences, they have little biological information on the sequences, except for their organism information. The mapping of the sequences and biological resources, such as gene and disease, will provide an opportunity for understanding of bio-resources' patentable targets (8). Especially, patent-related gene information can be used to identify whether a particular gene has been patented or published and to reveal if a patent has been infringed upon (9). Recently, Jensen and Murray (10) reported that ∼20% of human genes have been claimed as US intellectual property from the analysis of the sequences in US patents with bioinformatical methods. However, they used only the sequences at GenBank, and their analysis was focused on human genes. According to their knowledge, no attempt has been made to annotate the biological sequences in patents or applications thoroughly and to analyze them from a biological perspective.
In this report, we describe Patome, a database server containing the patent-related biological sequences and their analysis data. To build Patome, we downloaded sequence data from publicly available databases and created a non-redundant sequence set. The non-redundant sequences were annotated with RefSeq (11) and associated with Entrez Gene (12), the Online Mendelian Inheritance in Man (OMIM) (13) and Gene Ontology (GO) (14). The annotation results and the analysis data were integrated into a relational database and served via web-based user interfaces.
DATABASE CONTENTS AND ANALYSIS PIPELINE
Dataset
We downloaded the sequences from the patent divisions of GenBank (ftp://ftp.ncbi.nih.gov/genbank), EMBL (ftp://ftp.ebi.ac.uk/pub/databases/embl/patent), and DDBJ (ftp://ftp.ddbj.nig.ac.jp/database/ddbj). Sequence data were also obtained from the World Intellectual Property Organization (WIPO) (ftp://ftp.wipo.int/pub/published_pct_sequences) and the Publication Site for Issued and Published Sequences (PSIPS) database (http://seqdata.uspto.gov/) maintained by USPTO. As PSIPS does not provide a conventional ftp service, we downloaded sequence listings one-by-one from the PSIPS search interface. All the sequences obtained were derived from the sequence listings disclosed in the issued patents or published patent applications mainly in USA, Japan, Europe and by WIPO.
We created a non-redundant sequence set by removing the redundancy in the five databases. If the sequences had the same publication number and the same SEQID, they were considered duplicates. As on June 1, 2006, the number of non-redundant sequences were 38 151 115, including 34 762 740 nucleic acid sequences and 3 388 375 amino acid sequences.
In the non-redundant sequences, there is a large number of short fragments, which are mostly primers or synthetic constructs. These fragments are very short and not directly related to gene functions. Therefore, we excluded the nucleic acids (<100 bases) and the amino acids (<10 residues) in a similarity-based annotation. The sequence data also had many spelling errors and informal names in their organism fields. They were corrected either by using NCBI taxonomy database or manually for use in the next process.
Annotating the disclosed sequences
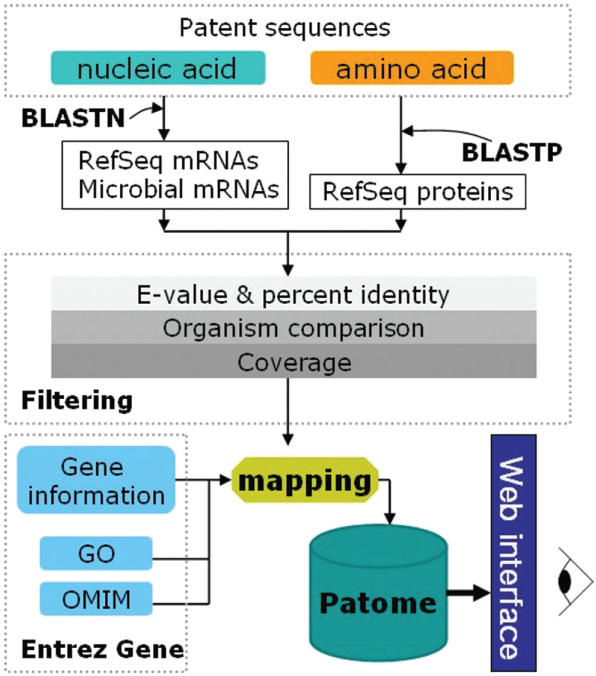
We developed an automated pipeline to identify the genes from which the disclosed sequences were derived (Figure 1). First, the nucleic acid sequences were compared with both the RefSeq mRNA database and complete sequenced microbial mRNA by using BALSTN (15). The amino acid sequences were compared with RefSeq proteins by using BALSTP. In the patent field, BLAST search is a general method that is used to identify functions of unknown sequences and to determine whether sequences are novel (16). The BLAST results were filtered by 98% identity and an E-value of 1 × 10−20.
Figure 1.
Overview of the analysis flow in Patome. The downloaded sequences are divided into nucleic acids and amino acids. Nucleic acids are annotated with both RefSeq mRNAs and microbial mRNAs. Amino acids are annotated with RefSeq proteins. The BLAST results are filtered with three filtering methods: E-value & percent identity, organism comparison and coverage. From the mapping between the annotated data and biological database information (Gene, GO, OMIM), a gene–patent association table is built. The Patome database is constructed to store annotation and analysis data and to make this information available to users via web interfaces.
In general, the BLAST program was widely used to identify cDNAs or proteins similar to a query sequence by retrieving putative homologs with performing local alignments (17). Thus, their results could have orthologous sequences of query sequences (e.g. human query sequence matches with mouse sequences of RefSeq). In the study, the main purpose of BLAST searching is to find gene sequences of the query sequences. Therefore, we applied two additional filtering methods to the BLAST results, in order to remove orthologs and find the exact gene in the same organism as that of a query sequence.
First, we utilized the organism name of a query sequence, called an organism comparison method, in which an organism of the best hit RefSeq should be the same as that of the query sequence. By applying this method, spurious matches to orthologous genes could be filtered out. However, this method could not be applied if a query's organism was unknown.
Second, we applied a coverage method. If a sequence was derived from a gene sequence, it should either contain most of the gene sequence or be a part of it. Hence, we divided the BLAST results into two types according to the alignment coverage against RefSeq sequences. They are full- and partial-length type. The full-length type is defined if an alignment length is >80% of that of the matched RefSeq sequence. The partial-length type is defined if an alignment length is >95% of that of the query sequence, except full-length types. BLAST alignments should be either full-length or partial-length type to go through the filtering.
From the filtered BLAST results, the sequence function was assigned with the best RefSeq hit. Of the 8 471 504 nucleic acid sequences, 1 075 764 (13%) were annotated with RefSeq mRNAs or microbial mRNAs. Of the 2 470 671 amino acid sequences, 1 486 625 (50%) were annotated with RefSeq proteins. The filtered BLAST results were saved as an annotation table that consists of the patent information (publication and SEQID) and the annotation information (RefSeq number, alignment length, alignment coverage and type, and E-value).
Building a gene–patent association table
RefSeq numbers of the annotation table served as a bridge to link the disclosed sequences and gene information, whose association can be represented as a gene–patent table that contains genes and their related patent sequence information. To build the gene–patent table, we extracted linking information between the sequences and the corresponding RefSeq number from the annotation table, and their relationship was translated to a gene–patent table by using a gene2refseq file from the Entrez Gene database. Entrez Gene also has cross-link information to other biological databases represented by gene2GO and mim2gene files. These cross-links were added into the gene–patent table.
For example, if two sequences from a ‘SEQID 39020 of WO0171042’ and a ‘SEQID 20352 of US6703491’ are assigned a RefSeq number, ‘NM_143536’, in the annotation table, it means that the RefSeq number, ‘NM_143536’, was linked to the two sequences. As the RefSeq number, ‘NM_143536’ was cross-linked with a GeneID ‘43614’ in the gene2refseq, we can conclude that the GeneID, ‘43614’ was disclosed twice in issued patents or patent applications.
From the analysis of the sequences, we found that 55% of the human genes were associated with patents or patent applications, the highest percentage among organisms. Oryza sativa (rice) ranked second with 25% (12 045 genes). The summary of patent-related genes in the major organisms is shown in Table 1. For the gene–patent analysis, it is necessary to distinguish sequences of granted patents from those of patent applications (18). Only granted patents are given intellectual property rights. In this study, the sequences were derived from either issued patents or published patent applications. Accordingly, it cannot be guaranteed that the genes appearing in the gene–patent table are patented.
Table 1.
Summary of patent-related genes in the major organisms
| Organism | No. of genesa | No. of patent-related genes | Patent-related genes (in %) |
|---|---|---|---|
| Homo sapiens | 39 216 | 21 478 | 55 |
| Oryza sativa | 47 275 | 12 045 | 25 |
| Drosophila melanogaster | 21 146 | 8 503 | 40 |
| Arabidopsis thaliana | 31 386 | 5 262 | 17 |
| Mus musculus | 60 714 | 4 479 | 7 |
| Rattus norvegicus | 23 973 | 3 542 | 15 |
aThe number of genes is from NCBI Entrez Gene.
SEARCHING AND DOWNLOADING THE PATOME DATABASE
Patome database server is composed of a web interface and an MySQL database management system. The web interface is implemented in static HTML pages, Java Server Pages (JSP) (http://java.sun.com/products/jsp/) and servlet (http://java.sun.com/products/servlet/) programs for database searching. MySQL is used to store the disclosed sequence information and their annotation and analysis data.
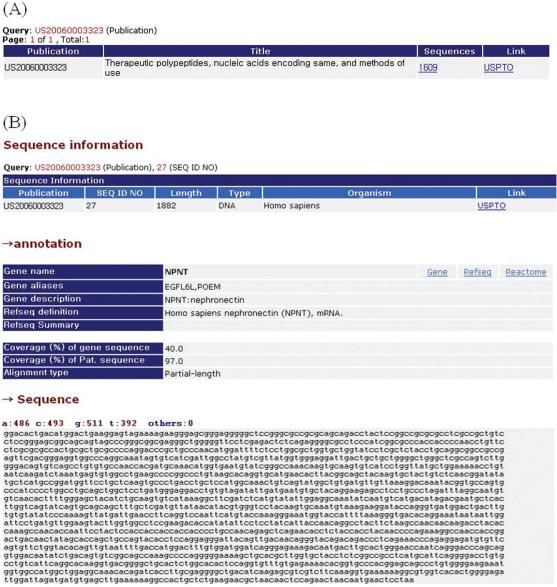
Patome can be accessed through web interface for querying (Figure 2). The querying interface allows the user to search against the patent sequence data and their analysis data. The patent sequence data can be searched by patent (or application) number, GenBank accession number and title. The search results consist of sequence information (length, type, organism, sequence), annotation information (Refseq, gene, GO, OMIM, coverage) if it exists and the same sequences information as a query. The analysis data search interface allows the user to search for the gene name (or symbol), Entrez Gene ID and RefSeq number. In addition, the logical operators, OR (∣) and AND (&), can be used to combine search words in the title and gene searches.
Figure 2.
Output of the Patome search. (A) Output of only patent (or application) number search. (B) Output of both patent (or application) number and SEQID search.
The display of search results also contains outgoing links to external databases for sequences and patents. For example, patent (or application) numbers are linked to the USPTO database (http://www.uspto.gov/patft/index.html) for US patents or applications, and the esp@cenet patent database server (http://ep.espacenet.com/) at the European Patent Office for the others. Gene names in the search output are linked to Entrez Gene and Reactome (19). The entire content of Patome annotation data is available for download from our website. We provide a zip-compressed annotation file. The data are presented in simple tab-delimited text file (for easy parsing of the data).
Detailed statistics on patent sequences are provided in the ‘statistics’ link on the Patome homepage. This includes the national (including WIPO), length and organism distributions of the sequences. These distributions are represented as tables. We also present statistics on gene-associated patents of major organisms in the homepage.
Acknowledgments
B.L. thanks Dr YoungGyun Cho at the Korean Intellectual Property Office (KIPO) for helpful discussion. We especially thank Maryana Bhak for editing this manuscript. This work was supported by the Korean Ministry of Science and Technology (MOST) under grant number M10407010001-04N0701-00110. DL was supported by the National Research Laboratory Grant (2005-01450) and the Korean Systems Biology Research Grant (2005-00343) from the Ministry of Science and Technology. KHL was supported by the Ministry of Science and Technology/Korea Science and Engineering Foundation (KOSEF) through the Advanced Information Technology Research Center (AITrc). Funding to pay the Open Access publication charges for this article was provided by the Ministry of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yoo H., Ramanathan C., Barcelon-Yang C. Intellectual property management of biosequence information from a patent searching perspective. World Pat. Inform. 2005;27:203–211. [Google Scholar]
- 2.Jones R. Errors in patent application sequence listings. Nat. Biotechnol. 2003;21:1239–1240. doi: 10.1038/nbt1003-1239. [DOI] [PubMed] [Google Scholar]
- 3.Xu G.G., Webster A., Doran E. Patent sequence databases. World Pat. Inform. 2002;24:95–101. [Google Scholar]
- 4.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cochrane G., Aldebert P., Althorpe N., Andersson M., Baker W., Baldwin A., Bates K., Bhattacharyya S., Browne P., van den Broek A., et al. EMBL Nucleotide Sequence Database: developments in 2005. Nucleic Acids Res. 2006;34:D10–D15. doi: 10.1093/nar/gkj130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okubo K., Sugawara H., Gojobori T., Tateno Y. DDBJ in preparation for overview of research activities behind data submissions. Nucleic Acids Res. 2006;34:D6–D9. doi: 10.1093/nar/gkj111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouse R.J., Castagnetto J., Niedner R.H. PatGen—a consolidated resource for searching genetic patent sequences. Bioinformatics. 2005;21:1707–1708. doi: 10.1093/bioinformatics/bti202. [DOI] [PubMed] [Google Scholar]
- 8.Farnley S., Morey-Nase P., Sternfeld D. Biotechnology—a challenge to the patent system. Curr. Opin. Biotechnol. 2004;15:254–257. doi: 10.1016/j.copbio.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Crespi R.S. Patents on genes: clarifying the issues. Nat. Biotechnol. 2000;18:683–684. doi: 10.1038/76548. [DOI] [PubMed] [Google Scholar]
- 10.Jensen K., Murray F. Intellectual property. Enhanced: intellectual property landscape of the human genome. Science. 2005;310:239–240. doi: 10.1126/science.1120014. [DOI] [PubMed] [Google Scholar]
- 11.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Groot A.S., Bosma A., Chinai N., Frost J., Jesdale B.M., Gonzalez M.A., Martin W., Saint-Aubin C. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine. 2001;19:4385–4395. doi: 10.1016/s0264-410x(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 17.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stott M., Valentine J. Impact of gene patenting on R&D and commerce. Nat. Biotechnol. 2003;21:729–731. doi: 10.1038/nbt0703-729. author reply 731. [DOI] [PubMed] [Google Scholar]
- 19.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]