Abstract
Variable number tandem repeat-PCR (VNTR-PCR) is a novel method developed for molecular typing of microorganisms. This method has proven useful in epidemiological studies in medical microbiology. Although hundreds of bacterial genomes have been sequenced, variable number tandem repeats (TRs) derived from comparative genome analyses are scarce. This may hamper their application to the surveillance of bacteria in molecular epidemiology. Here, we present a freely accessible variable number tandem repeat database (VNTRDB) that is intended to be a resource for helping in the discovery of putatively polymorphic tandem repeat loci and to aid with assay design by providing the flanking sequences that can be used in subsequent PCR primer design. In order to reveal possible polymorphism, each TR locus was obtained by comparing the sequences between different sets of bacterial genera, species or strains. Through this comparison, TRs which are unique to a genus can also be identified. Moreover, a visualization tool is provided to ensure that the copy number and locus length of repeats are correct. The VNTRDB is available at http://vntr.csie.ntu.edu.tw/.
INTRODUCTION
Many repetitive sequences exist in bacterial genomes. One family of repeats is tandem repeats (TRs). Some TRs are polymorphic due to variations in repeat copy number; such loci are often called variable number tandem repeats (VNTRs). Since the numbers of adjacent repeated units varies from individual to individual, VNTRs are inherently unstable units that undergo frequent variation in the number of copies through slippage strand misalignment during DNA synthesis (1) or double-strand break repair for instance (2). The variation can be found easily when bacteria contain TRs of different lengths in the same genomic locus (3,4). These phenomena imply that VNTR loci seem to be evolutionary hotspots. Due to their polymorphism, VNTRs have been used as DNA markers for molecular typing of several bacterial species, including Yersinia pestis (5,6), Francisella tularensis (7), Salmonella enterica (8,9), Mycobacterium tuberculosis (10,11), Xylella fastidiosa (12), Haemophilus influenzae (13), Staphylococcus aureus (14), Bacillus anthracis (15) and Neisseria meningitides (16,17) via an amplification-based method called PCR.
Nowadays, it is possible to search for TRs in a single bacterial genome via various TRs finding programs, such as REPuter (18), TRF (19), mreps (20) and ATRHunter (21). Furthermore, some databases, like TRDB (https://tandem.bu.edu/cgi-bin/trdb/trdb.exe), not only provide direct access to TRs but also provide analysis tools to assist the biologist in identifying polymorphic TRs. Nonetheless, recognizing polymorphism of TRs between different bacteria of the same genus or species is still not easily performed in a biological laboratory using these applications. This is due to the absence of the facility to perform multiple sequence comparisons with these programs in order to disclose the polymorphism of TRs. The GPMS database (http://minisatellites.u-psud.fr/)(22) uses this method. Specifically it combines TRF with pre-defined multiple genome comparisons to allow identification of VNTR loci that are polymorphic between the genomes compared.
In order to be recognized as a putative VNTR candidate, a TR should be mapped to its corresponding loci in other bacterial strains by its flanking sequences, and both locus length and copy number in each locus should be compared. Although this is the intuitive way to find putative VNTRs, when examining several phenomena in depth, we found that the copy number and locus length of reported TRs are sometimes uncertain. Variation in copy number may be due to the features of the algorithm used to determine copy number. Wraparound dynamic programming (WDP) is a technique commonly used to align potential TRs and determine copy number. For example, it is used by the TR finder algorithm (19) and consequently in other databases that use this algorithm (22). The alignment with the greatest score is usually chosen as the optimal alignment, but we have observed that there is the possibility to obtain two or more paths with the same maximal score in a local alignment using WDP. This would result in two alternative copy numbers being possible for one sequence and no facility to distinguish these based on the score alone. In this way, it is possible that the sequence from one strain could be listed as having two or more different number of copies depending on the alignment of the repeats (Figure 1). Ignoring this problem may lead to false polymorphisms being reported between different bacterial strains. Thus when the prediction of VNTR is based upon copy numbers alone, one may place false confidence in a result that may be flawed since it is not possible to be totally confident about the accuracy of the reported copy number. In addition, biologists often choose those TRs with higher copy number as candidates for molecular typing. The copy number may, however, vary depending on the parameters used in calculation. The more permissive the parameter set, the higher copy number reported. For example, a TR locus may have 20 copies of the repeat unit with a permissive parameter set but only four copies with a stricter one, since with the permissive set the repeats can be less conserved allowing the possibility of more divergent repeats to be included in the alignment. It suggests that the facility to dynamically set parameters is needed in order to best choose putative VNTR candidates, and the support of this function in Variable number tandom repeat database (VNTRDB) is convenient to users. Further more, although the variation in the locus length of the TR is normally due mostly to variation in the number of repeat copies between genomes, it may also be due in part to the deletion or insertion of nucleotides (Figure 2) or the transposition of a seemingly random nucleotide sequence within the TR locus. In addition to above situations, flanking sequence mutation (e.g. insertion or deletion) can also affect the lengths of PCR products unexpectedly (Figure 3). All these kinds of uncertainties may present false positive records to users. Together these observations, many of which may have been made before (22), emphasize that in order to determine the true reason for length variation at a putative VNTR locus, it is important to be able to visualize the sequence of a repeat and more crucially a multiple alignment of the locus in more than one strain. At present this latter facility does not seem to be readily accessible in publicly-available resources.
Figure 1.
An example of uncertain copy number. There are two different copy numbers (upper 12 and lower 3) reported here with the same alignment score and repeat pattern in this TR locus found in Mycobacterium leprae strain TN (from 300065 to 300138). The asterisk symbol denotes the mismatches between sequences.
Figure 2.
Visualization analyses of two possible VNTR loci. (a) A genuine VNTR locus in strains Ty2 and CT18 of S.enterica in yohM gene. The repeat pattern is ‘CACGAC’. (b) A ‘false positive’ VNTR locus of strains 9a5c and Temecula1 from X.fastidiosa which would probably not be suitable for a typing assay, Its repeat pattern is ‘ATTGACCTCATTCGT’. The asterisk symbol denotes the mismatches between sequences.
Figure 3.
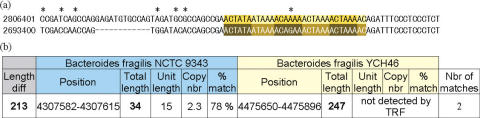
Examples of uncertain locus length. (a) Visualisation of the alignment of a possible VNTR loci. The upper sequence is from Bacteroides fragilis strain NCTC 9343, and the lower sequence is from B.fragilis strain YCH46. A 12 bp gap can be observed in this alignment. Because only a few of nucleotides exist between the gaps and the genuine TR, the primers must be designed to the 5′ of these gaps. Although the aligned repeats of both loci, which we call alignment length, are both exactly 31 bp, the PCR products will have a 12 bp difference. In VNTRDB, these 12 bp difference will affect the raw length but not the reported alignment length. (b) The loci were obtained directly from GPMS. Although total lengths of loci are different between these two Bacteroides strains, users cannot be sure whether this length difference is due to copy number difference alone or other sequence insertions or deletions. In VNTRDB, the corresponding locus in YCH46 has a locus length of 28 bp and raw length 244 bp suggesting that there are insertion/deletions resulting from changes other than repeat copy number differences. This combined with the fact that the visualization tool can only produce an alignment with very permissive parameters informs the user that the similarity of flanking sequences is very low and therefore the locus is probably not a genuine VNTR.
In order to solve the problems listed above and provide more accurate VNTR prediction to microbiologists, we have constructed a variable TR locus database, called VNTRDB (http://vntr.csie.ntu.edu.tw/). By comparing several bacterial genome sequences, the database records comprehensive information about a TR locus, including confidence in the level of polymorphism, two reported types of locus length and related gene information, some of which are not provided by related databases. To avoid assigning TRs to the wrong position, the flanking sequences of TRs were aligned and mapped to other genomes in order to identify the equivalent loci using BLAST with an Expect Value cut off of 0.01. Only those TRs with positive correlation between copy variance and difference in locus length are reported. Additionally, a visualization tool is provided to allow user-elimination of the potential problems resulting from copy number, locus length and flanking sequences of TRs, as explained previously. When visualizing the aligned results, users can further adjust parameters and reanalyze the alignment to confirm the robustness of the VNTR candidates derived from the algorithm.
In addition to providing comparative information between high-conserved sequences, the VNTRDB also provides some novel applications. One has the function of identifying potentially polymorphic TRs between distantly related sequences from several species or genera. This method is especially useful for those species where only one or two related strains have been sequenced. Another application finds putatively unique TRs which can be regarded as specific sequence markers for a particular microorganism. The implication is that these unique loci might be used in the rapid identification of bacteria that are difficult to isolate and culture, such as bacterial species of Mycobacterium, Mycoplasma, Clostridium, Chlamydia, Legionella, Listeria, Salmonella, etc. Most of them are also the important pathogens of diseases of human and animals. These applications are potentially very useful for clinical microbiologists.
CONSTRUCTION OF THE DATABASE
All possible VNTRs are identified by comparing all completely sequenced bacterial genomes. At the time of writing, 357 bacterial whole genomes from NCBI, Sanger Institute and TIGR are processed. To obtain the most comprehensive list of putative TRs, the widely used TR finding programs, TRF (19) and ATRHunter (21), were chosen to achieve optimal coverage of existing TRs. Subsequently, the flanking sequences of each TR were used to locate the corresponding loci between different bacterial strains through BLAST. The boundaries of corresponding loci, were adjusted by WDP (23,24) to obtain the new locus length and copy number of repeat. Here, two types of locus length were obtained. The first, called alignment length represents the theoretical locus length, which is the actual length of a TR. The other is the raw length of the corresponding locus before adjustment after alignment. The latter might affect the product length obtained in a PCR experiment and will be saved for later analysis of polymorphism. After the corresponding loci from each bacterial strain are determined, possible reporting problems are corrected and the confidence level for polymorphism within the corresponding loci is calculated. One possible reporting problem is a single locus being included twice in the database because the calculated positions of the overlaps between corresponding loci in two strains are different when each strain is used as the reference strain to search for the corresponding locus in the other strain. For example, if a locus in strain A is used to find the corresponding locus in strain B, the positions reported in strain A and strain B are 1000–1500 and 2000–2500 respectively. When strain B is used as the reference strain, then the same locus is reported as positions 2100–2600 overlapping positions 1100–1600 in strain A due to differences in sequences between the two strains and the effects of reversing the reference and query strains. Because different positions are recorded, these would be initially categorized as separate repeats but since the positions overlap, in this situation, the locus with the highest score will be picked as the representative of this repeat ‘cluster’. The confidence level of polymorphism intends to reflect the correlation between copy number and locus length. For a genuine VNTR or polymorphic TR, the variance in copy number between bacterial strains should show positive correlation with the lengths of the loci.
The web interface of VNTRDB contains three different browsing modes for choosing putative polymorphic TR, i.e. inter-genus, intra-genus and highly-conserved loci web pages. Since lateral transfer of genes is very common in bacteria, comparing TRs with bacteria that generally would show low levels of sequence conservation (i.e. belonging to different species or genera) may help users to discover potentially polymorphic TRs that they would not have otherwise found. It is possible if similar repeat loci with variable copy number are found in distantly-related bacterial sequences, that the same will also be polymorphic within a given bacterial species. After initial selection of a bacterial group in one of the three browsing modes, the system will direct the user to the main locus table in which the detailed information of polymorphic TRs is provided. This information consists of the location of repeat locus in each strain, locus length, copy number, repeat size, matched pattern, maximal score and the gene(s) involved (if the repeat overlaps a gene). This table also provides some convenient utilities. There is a region that allows selection of strains by the user and determines which strains or species are displayed in this table. The functions of ‘sort’ and ‘more criteria’ let users quickly pick TR loci by either default or user-selected parameter settings. The ‘locus location’ link shows the repeat and its flanking sequences from a given locus. This information would be required for primer design. Lastly, the ‘gene name’ link provides gene information to users, i.e. PID and its protein product. To rule out the problems we mentioned above, a visualization tool was developed and built into the locus display table so that the sequence alignment of two chosen loci can be examined by clicking the link in the ‘copy difference’ column. Because genuine VNTRs, whose polymorphism can be assessed by PCR amplicon length variation should have highly identical flanking sequences and a highly similar repeat pattern, the major difference between strains should be variability in copy number at each locus. This will be visualized as gaps within the alignment of the VNTR locus. For instance, the gaps of a genuine VNTR identified in gene yohM of S.enterica can be seen with the VNTRDB visualization tool (Figure 2a). This visualization tool can also detect false positive candidates with copy and length uncertainty. A false positive example from X.fastidiosa is shown in Figure 2b. Although the locus is reported as having one copy difference and 5 bp length variation, since there are no consecutive gaps visible in the alignment in one genome at least, the locus may not behaving as a true VNTR (variable number of TRs) since the variation is not a deletion or insertion of a whole repeat but sequence polymorphism is not related to the repeat sequence. In addition, users can observe the alignment result of the flanking sequences and choose the most conserved segments for primer design. In this way inappropriate primer design will be avoided. The visualization tool can also let users adjust parameters to produce the best alignment and in this way observe the change of copy number at the locus.
Although problematic records can be observed using this visualization tool, it may take too much time checking each repeat locus one by one. To take this into consideration, a confidence level of polymorphism corresponding to the correlation between copy number and locus length was provided to allow sorting of the VNTRs in order of priority with statistical significance. For a genuine VNTR, the variability of copy number between bacterial strains should positively correlate to their length of loci. If the positive correlation is >90% and flanking sequences are almost identical, these loci are good candidates of VNTR and will be marked as ‘H’ in confidence level of polymorphism. Symbol ‘L’ stands for lowest correlation and symbol ‘M’ for medium polymorphic level lying between level ‘L’ and ‘H’. Furthermore, features such as corresponding locus number, repeat pattern, length difference, copy variation and related gene information can be utilized to assist users to discriminate whether a given TR is polymorphic.
Additionally, users may deposit and manage their query results in freely registered space, called ‘My Query Result’ page. It permits collaborators to share and discuss their data privately, and may manage previously queried data when returning to the web site.
VALIDATION AND DISCUSSION
We evaluated our database by querying five bacterial species, i.e. M.tuberculosis, N.meningitides, S.aureus, S.enterica and Y.pestis. For each of these species, VNTR loci had been discovered in silico, tested experimentally, and used in DNA genotyping schemes (8–11,14,16,25). The evaluation showed that the entire set of published TRs of M.tuberculosis, N.meningitides, S.aureus and S.enterica can be successfully found by querying the VNTRDB, except for two loci, M02 and M06, in Y.pestis (Table 1). These two loci are filtered out in preprocessing step due to having very short locus length, (11 and 12 bp, respectively) and therefore not VNTR (also called minisatellites) but microsatellites. Microsatellites are reasonably rare in bacteria and often varies at very fast rate. For this reason and because PCR products containing small loci such as these will be difficult to size accurately to apply for bacterial typing. The results demonstrated that all of the useful TRs published in literature can be found by querying the VNTRDB. There are some VNTRs that have been discovered by laboratory experiments that exhibit no polymorphism in the presently published bacterial sequences. For example, typing Salmonella with VNTR-based method revealed that the locus SSTR2 is not polymorphic, and locus SSTR7 has two alleles with only one copy difference (9). This is exactly consistent with the query results of VNTRDB.
Table 1.
Known TR and VNTR candidates appearing in publications and VNTRDB
| Species | Mycobacterium tuberculosis | Neisseria meningitidis | Staphylococcus aureus | Salmonella enterica | Yersinia pestis |
|---|---|---|---|---|---|
| Strain used in VNTRDB | 4 | 3 | 9 | 7 | 4 |
| TRs in literature | 40 | 15 | 7 | 13 | 42 |
| TRs identified in VNTRDB | 40 | 15 | 7 | 13 | 40 |
| VNTRs in literature | 40 | 4 | 7 | 7 | 42 |
| VNTRs identified in VNTRDB | 36 | 4 | 7 | 7 | 36 |
| Percentage of published repeats in VNTRDB | 90% | 100% | 100% | 100% | 86% |
This table compares those TRs and VNTRs collected in literatures with those available from querying the VNTRDB. Two TRs of Y.pestis are filtered because the resulting PCR products are too short. All other TRs are recorded in VNTRDB. Although only between three and seven strains are used, nearly all of the TRs found in the database are described as being polymorphic as confirmed in the literatures. Those VNTRs described in the literature but not found in VNTRDB are absent because they are not polymorphic in those genome sequences that are currently available.
This VNTRDB has already assisted Chiou and his coworkers (17) to identify potential VNTR loci successfully in N.meningitidis for molecular subtyping and phylogenetic analysis. In this study, 23 potential VNTR loci were selected from VNTRDB based on the multiple comparisons of three genomes of N.meningitidis, strains Z2491, MC58 and FAM18. After evaluation of 10 genetically distinct N.meningitidis strains, 12 of the loci were chosen to use in genotyping, the remaining 11 were abandoned because of multiple or no PCR products. Four of the eleven loci are within opa genes, which exist in multiple copies with a range of repeat numbers in Neisseria species. None of the loci tested were monomorphic. This demonstrates that VNTRDB can provide microbiologists with a selection of polymorphic TRs for use in pilot studies and are likely to be polymorphic. Using this approach, the time taken to identify loci which are suitable for genotyping in bacteria will be greatly decreased.
Because of the improvements made in our algorithm to map loci, the VNTRDB can be used to search not only for the equivalent TR loci in highly-conserved genome sequences (e.g. between strains), but also in distantly-related bacteria. These distantly-related genome sequences may be from different species or even from different genera, e.g. Escherichia coli, Salmonella and Shigella flexneri. If the homologous TR loci found in bacteria from different genera or species are found to be polymorphic, this suggests that the same locus may also be polymorphic between different strains of the same species. Reasons why equivalent loci may be found in species from different genera include (i) the reclassification of bacteria (e.g. Pasteurella anatipestifer is now reassigned to a new genus, Rimerella anatipestifer, even though its genome sequence is very similar to other Pasteurella species); (ii) lateral gene transfer between bacteria (26); and (iii) bacteria of different genera having similar genome organization (e.g. Y.pestis versus Salmonella species). These scenarios imply that the more sequences that are compared, the more polymorphic TRs will be found. In genera, where the species are very closely related it may be possible to compare the closely related species and make predictions based on the VNTR observed in all species. For example, if only one S.felxneri strain has been sequenced by comparison with the very closely related E.coli strains predictions can be made about VNTRs that may be found in all Shigella strains.
CONCLUSION
Polymorphic TRs are very useful for the molecular typing of bacteria. The more information gathered from sequence data by biologists prior to starting lab experiments, the better the experimental results are likely to be. In order to help biologists find those TRs that are polymorphic, we constructed the VNTRDB to provide comprehensive information about bacterial VNTR loci in genome sequences. The accuracy of the database content was tested by comparing it with published TR loci. Additionally, a visualization tool is provided to allow problems resulting from copy number, locus length and flanking sequences to be resolved, which may make the experimental results unpredictable. Further more to enable the discovery of still more putative polymorphic TRs, we made comparisons between bacterial genomes that are not necessarily from the same species or with high levels of sequence identity. This is partly due to the fact that at the time of writing only 357 bacterial genomes have been made publicly available and for many species less than two strains have been sequenced.
Although a similar site (GPMS) has been reported by Denoeud and Vergnaud (22), what the database provides is mainly VNTRs discovered when comparing two highly-conserved strains of a single bacterial species. Although constructed using different strategies, these two databases are likely to serve the users in a complementary fashion.
Another potential function for the VNTRDB is the identification of unique TRs. Theoretically, any unique sequence can serve as DNA marker for the identification of bacteria, so a VNTR locus that is unique could be one such marker. In order to confirm the utility of these loci for identification, they would need to be tested experimentally.
ACCESSIBILITY
The VNTRDB can be accessed freely at http://vntr.csie.ntu.edu.tw. A mirror site to shorten the response time of web pages is also provided at http://www.hpa-bioinformatics.org.uk/VNTRUK/ in the United Kingdom.
Acknowledgments
This work was partially supported by grants NSC93-3112-B-002-022 and NSC92-2313-B-130-001 from the National Science Council, and grant COA95-13.3.2-BAPHIQ-B1 from Bureau of Animal and Plant Health Inspection and Quarantine. The authors also want to thank NCBI, TIGR and Sanger Institute for provide bacterial sequences for VNTR analysis. Funding to pay the Open Access publication charges for this article was provided by Institute for Information Industry (III), Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Strand M., Prolla T.A., Liskay R.M., Petes T.D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 2.Ozenberger B.A., Roeder G.S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weir B.S. Population genetics in the forensic DNA debate. Proc. Natl Acad. Sci. USA. 1992;89:11654–11659. doi: 10.1073/pnas.89.24.11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smouse P.E., Chevillon C. Analytical aspects of population-specific DNA fingerprinting for individuals. J. Hered. 1998;89:143–150. doi: 10.1093/jhered/89.2.143. [DOI] [PubMed] [Google Scholar]
- 5.Adair D.M., Worsham P.L., Hill K.K., Klevytska A.M., Jackson P.J., Friedlander A.M., Keim P. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 2000;38:1516–1519. doi: 10.1128/jcm.38.4.1516-1519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Fleche P., Hauck Y., Onteniente L., Prieur A., Denoeud F., Ramisse V., Sylvestre P., Benson G., Ramisse F., Vergnaud G. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 2001;1:2. doi: 10.1186/1471-2180-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farlow J., Smith K.L., Wong J., Abrams M., Lytle M., Keim P. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 2001;39:3186–3192. doi: 10.1128/JCM.39.9.3186-3192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Lee M.A., Ooi E.E., Mavis Y., Tan A.L., Quek H.H. Molecular typing of Salmonella enterica serovar typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 2003;41:4388–4394. doi: 10.1128/JCM.41.9.4388-4394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindstedt B.A., Heir E., Gjernes E., Kapperud G. DNA fingerprinting of Salmonella enterica subsp. enterica serovar Typhimurium with emphasis on phage type DT104 based on variable number of tandem repeat loci. J. Clin. Microbiol. 2003;41:1469–1479. doi: 10.1128/JCM.41.4.1469-1479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazars E., Lesjean S., Banuls A.L., Gilbert M., Vincent V., Gicquel B., Tibayrenc M., Locht C., Supply P. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 11.LeFleche P., Fabre M., Denoeud F., Koeck J.L., Vergnaud G. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2002;2:37. doi: 10.1186/1471-2180-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coletta-Filho H.D., Takita M.A., de Souza A.A., Aguilar-Vildoso C.I., Machado M.A. Differentiation of strains of Xylella fastidiosa by a variable number of tandem repeat analysis. Appl. Environ. Microbiol. 2001;67:4091–4095. doi: 10.1128/AEM.67.9.4091-4095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Belkum A., Melchers W.J., Ijsseldijk C., Nohlmans L., Verbrugh H., Meis J.F. Outbreak of amoxicillin-resistant Haemophilus influenzae type b: variable number of tandem repeats as novel molecular markers. J. Clin. Microbiol. 1997;35:1517–1520. doi: 10.1128/jcm.35.6.1517-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabat A., Krzyszton-Russjan J., Strzalka W., Filipek R., Kosowska K., Hryniewicz W., Travis J., Potempa J. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 2003;41:1801–1804. doi: 10.1128/JCM.41.4.1801-1804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keim P., Price L.B., Klevytska A.M., Smith K.L., Schupp J.M., Okinaka R., Jackson P.J., Hugh-Jones M.E. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yazdankhah S.P., Lindstedt B.A., Caugant D.A. Use of variable-number tandem repeats to examine genetic diversity of Neisseria meningitidis. J. Clin. Microbiol. 2005;43:1699–705. doi: 10.1128/JCM.43.4.1699-1705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao J.C., Li C.C., Chiou C.S. Use of a multilocus variable-number tandem repeat analysis method for molecular subtyping and phylogenetic analysis of Neisseria meningitidis isolates. BMC Microbiol. 2006;6:44. doi: 10.1186/1471-2180-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurtz S., Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 19.Benson G. Tandem repeats finder: a program to analyzed DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolpakov R., Bana G., Kucherov G. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res. 2003;31:3672–3678. doi: 10.1093/nar/gkg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wexler Y., Yakhini Z., Kashi Y., Geiger D. Finding approximate tandem repeats in genomic sequences. J. Comp. Biol. 2005;12:928–942. doi: 10.1089/cmb.2005.12.928. [DOI] [PubMed] [Google Scholar]
- 22.Denoeud F., Vergnaud G. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a Web-based resource. BMC Bioinformatics. 2004;5:4. doi: 10.1186/1471-2105-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischetti V., Landau G., Schmiedt J., Sellers P. Identifying periodic occurrences of a template with application to a portein structures. In: Apostolico A., Crochemore M., Galil Z., Manbers U., editors. Proceedings of the 3rd Annual Symposium on Combinatorial Pattern Matching. Springer-Verlag, Berlin 644: Lecture Notes in computers Science; 1992. pp. 111–120. [Google Scholar]
- 24.Miller W., Myers E. Approximate matching of regular expression. Bull. Math. Biol. 1989;51:5–37. doi: 10.1007/BF02458834. [DOI] [PubMed] [Google Scholar]
- 25.Klevytska A.M., Price L.B., Schupp J.M., Worsham P.L., Wong J., Keim P. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 2001;39:3179–3185. doi: 10.1128/JCM.39.9.3179-3185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesic B., Carniel E. Horizontal transfer of the high-pathogenicity island of Yersinia pseudotuberculosis. J. Bacteriol. 2005;187:3352–3358. doi: 10.1128/JB.187.10.3352-3358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]