Abstract
The MITOMAP (http://www.mitomap.org) data system for the human mitochondrial genome has been greatly enhanced by the addition of a navigable mutational mitochondrial DNA (mtDNA) phylogenetic tree of ∼3000 mtDNA coding region sequences plus expanded pathogenic mutation tables and a nuclear-mtDNA pseudogene (NUMT) data base. The phylogeny reconstructs the entire mutational history of the human mtDNA, thus defining the mtDNA haplogroups and differentiating ancient from recent mtDNA mutations. Pathogenic mutations are classified by both genotype and phenotype, and the NUMT sequences permits detection of spurious inclusion of pseudogene variants during mutation analysis. These additions position MITOMAP for the implementation of our automated mtDNA sequence analysis system, Mitomaster.
INTRODUCTION
MITOMAP is a comprehensive database of human mtDNA variation and its relation to human evolution and disease. The need for a curated and thus reliable database of mitochondrial gene sequence variation with easily used web-based query tools is becoming increasingly essential as the importance of mtDNA variation in the common age-related metabolic and neurodegenerative diseases, aging and cancer is elucidated. The mtDNA encompasses the 13 core polypeptide genes that define the efficiency of the mitochondrial energy generating system oxidative phosphorylation (OXPHOS), the tRNAs and rRNAs for their expression, and a 1121 np control region that regulates mtDNA transcription and replication. The mtDNA also has a unique genetics, being maternally inherited, present in thousands of copies per cell, and capable of encompassing various percentages of mutant and normal molecules (or heteroplasmy) (1). Hence, the mtDNA imparts a quantitative genetics to the vital, systemic function of energy production (1,2).
The 16 569 nt pairs (np) of the closed circular mtDNA are each numbered sequentially and the ‘standard’ base at each position (3) is specified in the ‘revised Cambridge Reference Sequence’ (rCRS), GenBank J01415.2. This permits any new mtDNA sequence to be unambiguously defined by specifying only the deviant positions and bases relative to the rCRS. Thus, all of the information in MITOMAP can be digitally interrelated.
The human mtDNA has a very high mutation rate, and this has generated three classes of sequence variants: pathogenic, adaptive and neutral. Pathogenic mutations damage conserved gene functions and as the deleterious mutation increases in its percentage heteroplasmy, it erodes systemic cellular energy output. This deficiency is acted on by natural selection and the mutation is soon eliminated from the population as hereditary disease. Hence, all deleterious mutations in the population are relatively recent. Adaptive mtDNA mutations also alter conserved functions but these alterations are advantageous in certain environments. Consequently, they become established in the individual (or homoplasmic) and increase in the population that occupies the compatible environment. Neutral mutations accumulate by chance, but those linked to advantageous mutations can become enriched in particular populations by hitchhiking.
Mutations in the mtDNA not only arise in the female germline, but also accumulate in the post-mitotic cells of the body with age. These somatic mtDNA mutations now appear to be the aging clock and may also be important in the etiology of certain cancers (1,2).
Pathogenic mtDNA mutations can be divided into those that alter more than one gene (inter-gene or contiguous gene defects) and those that affect only one gene (intra-gene or single gene defects). All contiguous gene defects involve rearrangement (insertion-deletion) mutations. Intra-gene mutations include small insertion-deletion mutations or base substitution mutations. The intra-gene mutations can either alter polypeptide coding genes or protein synthesis (rRNA and tRNA) genes. The functional severity of pathogenic nucleotide substitutions can be estimated, in part, through the inter-species conservation index (CI) and for rRNA or tRNA genes by the effect of the base change on the RNA secondary structure. Since the clinical effect of a mutation is the product of both the severity of the mutation and its percentage of heteroplasmy, the clinical phenotypes of heteroplasmic mtDNA mutations can be highly variable among the maternal relatives (1,2). Since mtDNA mutations can arise in both the germline and somatic tissues, they have been associated with both degenerative diseases and cancers (4–6).
Adaptive mutations alter functions and thus are primarily single nucleotide substitution mutations. Moreover, since they are retained in the population by selection, they are frequently ancient and present at polymorphic frequencies. About 25% of the polypeptide amino acid substitution (missense) mutations present in the human mtDNA phylogenetic tree are adaptive (7,8), ∼10–20% of tRNA mutations appear to be adaptive, as do various rRNA variants (9). Many of these same mutations have also been reported as somatic mutations in cancer cells suggesting that tumors adapt to their changing energetic environment through the evolution of mtDNA variation (10). A surprisingly high proportion of cancer control region somatic mutations are also population polymorphisms (10), suggesting that modulation of mtDNA replication and transcription is an important adaptive strategy.
Since mtDNA mutations accumulate sequentially along maternal mtDNA lineages, the mtDNAs of region-specific branches of the mtDNA tree are founded by a common mtDNA haplotype and thus share a set of common polymorphisms. From this founder, additional mutations are added sequentially over time forming groups of related haplotypes is called a haplogroup. Since selection may have caused the haplogroup to flourish in the new environment, different haplogroups may be functionally different (8,9). Evidence that haplogroups can be functionally different comes from the fact that haplogroups are associated with predilection to or protection from various clinical phenotypes. For instance, haplogroup J increases the risk of developing blindness in association with the milder Leber Hereditary Optic Neuropathy (LHON) mutations (11–13) and haplogroup T increases risk for depression (14). However, other haplogroups are protective against Alzheimer Disease (15–17) and Parkinson Disease (17–19) and are associated with increased longevity (8,20–24).
Throughout the evolution of the symbiosis between the nuclear-cytosol and the mitochondrial organisms, sequences from the mtDNA have been transferred to the nDNA with the result that the vast majority of mitochondrial genes are now located in the nucleus. The transfer of mtDNA sequences to the nDNA continues today, but differences in the mtDNA genetic code that arose prior to the origin of the fungal-animal mtDNA lineage render the mtDNA sequences unintelligible to the nuclear-cytosol system (2). Consequently, all recent mtDNA transfers into the human nDNA have resulted in nuclear-mtDNA pseudogenes (NUMTs) (25). NUMTs are potentially problematic since they can be inadvertently amplified using the polymerase chain reaction (PCR), sequenced, and the pseudogene variants mistaken for pathogenic mtDNA mutations (26,27).
While the mtDNA retains the 13 central OXPHOS proton and electron carries for the OXPHOS circuitry, all of the structural genes necessary to assemble the mitochondrial OXPHOS complexes as well as all of the genes needed to create a mitochondrion are now encoded by the nucleus, a total of ∼1500 mitochondrial genes. Therefore, many mutations that affect mitochondrial function are the result of nDNA mutations (1,2).
A mtDNA SEQUENTIAL MUTATION PHYLOGENETIC TREE
A number of factors must be considered when evaluating the nucleotide variants observed in a new mtDNA sequence relative to the rCRS. These include the association between different variants on particular branches of the mtDNA tree (the haplogroup), the time in human mtDNA radiation that the variant appeared and the position of the variant in the protein or RNA gene. All of these considerations were utilized in developing our sequential mutational phylogenetic tree of the human mtDNA.
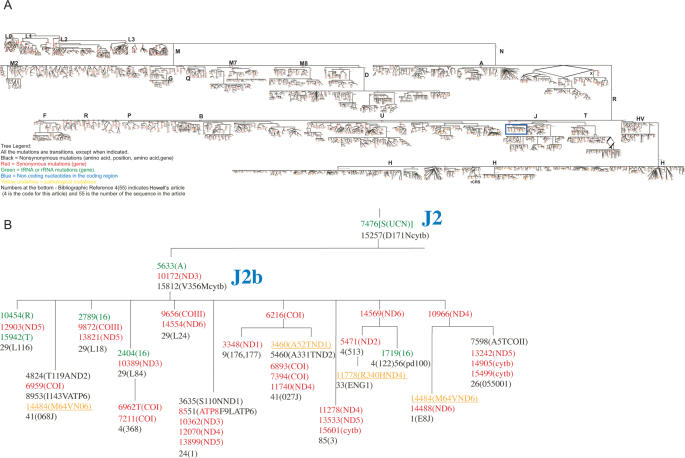
Our mtDNA tree encompasses the information in 2959 mtDNA coding region sequences, using the rCRS (GenBank J01415.2) as the reference. While our tree was assembled primarily using mtDNA coding region single nucleotide polymorphisms (SNPs), informative control region SNPs are also being incorporated when available. However, this dataset is incomplete since the control region sequences have not been reported for many of the published mtDNA sequences used in building the tree. Each sequence was analyzed for possible sequencing errors, identified by base changes inconsistent with the most likely haplogroup. Sequences with multiple potential errors were not included in the dataset. Our tree is now available in MITOMAP, http://www.mitomap.org. Partial representations of the tree are presented in Figure 1.
Figure 1.
(A) Sequential mutational phylogenetic tree of 2959 mtDNA coding region sequences. The full tree is available in PDF format at http://www.mitomap.org/mitomap-phylogeny.pdf. All mutations are transitions except where indicated. Black polymorphisms represent nonsynonymous mutations (amino acid, position, amino acid, gene), red synonymous mutations (gene), green tRNA or rRNA mutations (gene), blue indicates non-coding nucleotides in the coding region and underlined yellow represent pathological mutations. Large blue letters indicate a haplogroup designation and large black letters allow for navigation of the tree at low magnification. Numbers in black below the final polymorphism are the bibliographic references and sequence identifiers. For example, 4(55) indicates Howell's article (4 is the code for this article) and 55 is the number of the sequence in the article). The bibliographic references are available on the mitomap website. (B) Magnification of blue box in Figure1A showing details of the tree for subhaplogroup J2b.
The core topology of the tree was generated by the neighbor-joining program from MEGA (28) using 1060 human mtDNA sequences. The branching order used is highly robust as demonstrated by its consistency within 1000 bootstrap iterations. Separate neighbor-joining trees were also built for each major haplogroup, again with the branching order being robust based in 1000 bootstrap iterations. Then using sequence alignment and sequence comparisons, the positions of the individual nucleotide changes were incorporated into the various branches and nodes of the tree. Since the core tree encompasses all of the major mtDNA haplogroups, new mtDNA sequences can be continually incorporated into the existing tree manually without necessity for further computer analysis. Atypical variants such as insertion-deletions are also added manually and unsolved parallel mutations are resolved by network analysis.
The overall branching order of the human mtDNA tree has been clear for many years (29–32). However, the presence of identical mutations in multiple different parts of the phylogeny was initially puzzling. This was resolved when we concluded that many of these recurrent mutations were due to convergent evolution resulting from adaptive selection acting on specific functional variants (8,9).
The major branches of the tree are labeled with alphabetical letters. The letters were assigned as the mtDNA lineages became apparent. Because Native Americans were found to have a limited number of mtDNA lineages (33), the four major Native American haplogroups were designated A, B, C and D (34,35). Subsequent analysis added haplogroups in Europe (H, I, J, T, U, Uk, V, W and X) (36,37), Asia (many derivatives of macro-haplogroups M and N which radiated out-of-Africa) (38,39) and Africa (haplogroups L0, L1, L2 and L3) (7,40,41). A fifth Native American haplogroup was also found to be related to Eurasian mtDNA haplogroup X (42). The sub-branching order of the haplogroups has been further refined by incorporating subsequent nomenclatural proposals (43) and application of the information derived from complete mtDNA sequences (7,44,45). Since the African mtDNAs lie at the root of the mtDNA phylogeny (29–31), the African haplogroups are presented at the beginning (top) of the phylogenetic tree (Figure 1).
The complete phylogeny with all of the ordered SNPs requires 126 pages to print. To make the tree more accessible, it was integrated and transferred into the Canvas X (ACD Systems, Inc.) program. The PDF file was then exported to http://www.mitomap.org/. In this form, the entire tree can be viewed at any resolution desired, from ultra-low resolution, where all of the branches can be observed but no detail can be perceived, to ultra-high resolution, where the branching order is lost but the individual nucleotide changes can be seen (Figure 1).
The nucleotide position of each change is located in the tree in the order that they arose, and the nature of the mutation, transition versus transversion, given. If the nucleotide change alters a tRNA or an rRNA gene, the mutation is presented in green with the gene designated. If the mutation alters a polypeptide gene, but is synonymous, then it is presented in red with the gene identified. However, if the mutation alters the amino acid sequence of a polypeptide gene, the mutation is presented in black with the affected gene identified and the codon position and amino acid substitution reported. Non-coding variants are shown in blue, and few representative pathogenic mutations are included in the tree in yellow. In this way, all of the common mtDNA sequence changes, their relationship to each other and their potential significance can easily be observed.
Because of how our tree was developed, it is invaluable for analyzing the nature of the nucleotide differences found in any new mtDNA sequence relative to the rCRS. By tracing the nucleotide variants in the new mtDNA sequence along the branching order of our tree, the new sequence's haplogroup is defined, the time that each variant arose (ancient versus recent) is determined and the effect of the sequence variant on the polypeptide or RNA gene revealed.
Ancient functional variants associated with a new sequence may be adaptive variants and may have also arisen independently in other branches of the phylogeny. Many of these have already been identified and discussed (8,9). Variants that do not appear in our phylogenetic tree are likely to be relatively recent mutations. If the mtDNA sequence is from a potentially maternally inherited disease patient and the new mutation is potentially functionally significant, the mutation becomes a candidate for a pathogenic mutation. Neutral variants can be either ancient or recent, but do not alter an important mitochondrial function.
With these rules, a mtDNA sequence from a patient with a potentially maternally inherited disease would be analyzed by first comparing the sequence to the rCRS, and all differences will be listed. These variants would then be used to trace down the various branches of the phylogenetic tree until you arrive at the end of a terminal branch. All sequence variants that have been used thus far are haplogroup-associated polymorphisms. Any variants that are left over must be recent and require additional analysis. If a residual nucleotide change alters an amino acid in a mtDNA polypeptide or the sequence of a tRNA or rRNA, then it is a candidate for a pathogenic mutation. For polypeptide mutations, calculation of the CI of the variant amino acid can give further insight into pathogenicity (8). Similarly, for a tRNA or rRNA sequence change, if the variant alters a conserved nucleotide and/or affects the secondary structure of the RNA then it becomes a candidate for a pathogenic mutation (9). Additional criteria suggesting pathogenicity could include the presence of heteroplasmy and a correlation between the severity of the pathogenic mutation and the percentage of mutant mtDNAs present in various family members along a maternal lineage. Another criterion could be the similarity between the pathological presentation of the mutation and that of a similar type of pathogenic mutation that presents with a similar phenotype.
In certain cases, multiple patients from unrelated families may be found that have closely related mtDNA haplotypes. In these situations, it is possible that the background mtDNA haplogroup is also contributing to the pathogenicity of the deleterious mutation. This type of phenomenon has been documented for the milder pathogenic mutations at nt G11778A, T14484C, and T10663C which cause LHON, which are preferentially associated with mtDNA haplogroups J and T (11–13).
PATHOGENIC MTDNA MUTATION TABLES
One strong criterion for identifying a pathogenic mtDNA mutation would be if the same mutation had been identified in other patients with similar clinical presentation. To facilitate this type of comparison, the MITOMAP mutational tables have been completely updated and upgraded. The current searchable tables are divided into three major classes: (i) mutations associated with LHON, (ii) mutations that affect polypeptide sequences but present with other phenotypes and (iii) mutations that alter tRNAs and rRNAs and present with a various multi-system clinical problems. These tables and the associated clinical diseases are explained in depth in our various clinical reviews (2,46,47). As an example, the LHON disease mutations have been sub-classified into two main categories. The first category includes well characterized and causative mutations designated the ‘top 10’ primary LHON mutations. This group includes the three most common LHON mutations (G3460A, G11778A and T14484C) with account for ∼95% of LHON patients. The second mutation category includes mutations that have been reported in a single family and thus require additional independent reports to confirm their causal association with LHON.
If in the process of analyzing an mtDNA sequence from a patient, one of the recent sequence differences is also present in one of the pathogenic mutational tables, then this mutation becomes a good likely candidate for the disease mutation. However, it must be remembered that the mtDNA haplogroup background can be a contributing factor in some diseases such as LHON. Hence, it is important that the role of the mutation be considered in the larger context of what we know about human mitochondrial genetics (2).
MITOCHONDRIAL DISEASES OF NUCLEAR GENETIC ORIGIN
While the mtDNA encodes 37 critical genes for OXPHOS (13 polypeptides, 22 tRNAs and 2 rRNAs) the nDNA encodes in the order of 1500 additional mitochondrial proteins and mutations in these genes can also cause mitochondrial disease. Hence, many of the more severe mitochondrial diseases can be caused by mutations in nDNA encoded genes for the structural or assembly proteins of the OXPHOS complexes and a spectrum of disorders can result from nDNA mutations that affect mtDNA replication (48). To alert MITOMAP users to these important classes of mitochondrial disease, MITOMAP now includes summary tables of the known nDNA-encoded mitochondrial diseases (2).
NUMT PSEUDOGENE DATABASE
The nDNA also encodes 247 fragments of the mtDNA of sufficiently recent origin to be easily mistaken for legitimate mtDNA sequences. Inadvertent PCR amplification of one of these sequences during a clinical study can result in the erroneous conclusion that the sequence variants within the NUMT pseudogene are the cause of the disease (26,27).
To address this problem, we have developed a MITOMAP human NUMT database (25). By scanning the NUMT sequences that encompass the same region as that of clinical interest, the potential nDNA pseudogene variants can be identified.
REFERENCE DATABASE
The MITOMAP database currently encompasses over 5100 references. These pertain to the mtDNA sequence, sequence variation and mitochondrial role in a wide variety of diseases. This reference database is searchable by author and subject word.
MITOMAP COMPUTATIONAL SYSTEM AND DATABASE MAINTENANCE
All of the data for MITOMAP is now managed in PostgreSQL (http://www.postgresql.org/), an open-source database management system (i.e. DBMS) with object-relational modeling capabilities. All programs are written in the Perl programming language.
Data for MITOMAP are initially extracted by the database curators and stored, formatted and screened in Excel spreadsheets. Updated versions of the curated data are then periodically uploaded to the server, where Perl programs implementing the ParseExcel and DBI modules are used to parse the values from the spreadsheets and populate the data structures implemented in PostgreSQL. Error screening is performed both by the curator and by the population programs.
The MITOMAP servers are located at the University of California, Irvine, on a dual-processor server running RedHat Linux as its operating system. This system is accessed through a dynamic web interface using an Apache webserver.
DISCUSSION
By adding the mtDNA mutational tree, updating the depth and breadth of the mtDNA disease mutation tables and creating a NUMT pseudogene sequence database, we have now generated the informational infrastructure that will permit MITOMAP to evolve from a simple factual database to an intelligent analytical system. The sequential mutation mtDNA phylogenetic tree places all of the factual information about mtDNA sequence variation into an evolutionary and functional context. This single integrated entity thus automatically sorts mtDNA variants into ancient versus recent mutations, defines the gene affected and provides preliminary information on the potential functional implications of the mutation.
From this foundation, MITOMAP is now poised to implement our much more powerful data integration and analysis system, MITOMASTER. The MITOMAP-MITOMASTER system will permit the user to up-load a new mtDNA sequence, and the system will automatically identify the sequence variants relative to the rCRS, determine the mtDNA haplogroup, indicate the potentially functional variants, check for NUMT pseudogene variants and provide a list of potential disease causing mutations. Hence MITOMAP-MITOMASTER will soon be an important bioinformatics tool in the analysis of human mtDNA sequence variation.
Acknowledgments
This research has been supported by NIH grants NS213L8, HL64017, AG13154 and an Ellison Foundation Senior Investigator Grant awarded to Douglas C. Wallace; NIH Biomedical Informatics Training Grant T15 LM007443, NSF grant EIA-0321390 and a UCI Laurel Wilknening Faculty Innovation Award to Pierre Baldi; and Spanish Fondo de Investigacion Sanitaria grant # PI050647, Diputacion General de Aragon Grupos consolidados B33 and by Ciber Enfermedades raras (CB06/07/0043). Funding to pay the Open Access publication charges for this article was provided by AG24373.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wallace D.C. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace D.C., Lott M.T., Procaccio V. Mitochondrial genes in degenerative diseases, cancer and aging (Chapter 13). In: Rimoin D.L., Connor J.M., Pyeritz R.E., Korf B.R., editors. Emery and Rimoin's Principles and Practice of Medical Genetics. 5th Edition. London: Churchill Livingstone; pp. 194–298. [Google Scholar]
- 3.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 4.Petros J.A., Baumann A.K., Ruiz-Pesini E., Amin M.B., Sun C.Q., Hall J., Lim S., Issa M.M., Flanders W.D., Hosseini S.H., et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl Acad. Sci. USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace D.C. Mitochondria and Cancer: Warburg Address. Cold Spring Harb. Symp. Quant. Biol. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 6.Brandon M., Baldi P., Wallace D.C. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 7.Mishmar D., Ruiz-Pesini E.E., Golik P., Macaulay V., Clark A.G., Hosseini S., Brandon M., Easley K., Chen E., Brown M.D., et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Pesini E., Mishmar D., Brandon M., Procaccio V., Wallace D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Pesini E., Wallace D.C. Evidence for adaptive selection acting on the tRNA and rRNA genes of the human mitochondrial DNA. Hum. Mutat. 2006;27:1072–1081. doi: 10.1002/humu.20378. [DOI] [PubMed] [Google Scholar]
- 10.Brandon M.C., Baldi P., Wallace D.C. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 11.Brown M.D., Starikovskaya E., Derbeneva O., Hosseini S., Allen J.C., Mikhailovskaya I.E., Sukernik R.I., Wallace D.C. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup. J. Hum. Genet. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- 12.Brown M.D., Sun F., Wallace D.C. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am. J. Hum. Genet. 1997;60:381–387. [PMC free article] [PubMed] [Google Scholar]
- 13.Torroni A., Petrozzi M., D'Urbano L., Sellitto D., Zeviani M., Carrara F., Carducci C., Leuzzi V., Carelli V., Barboni P., et al. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am. J. Hum. Genet. 1997;60:1107–11021. [PMC free article] [PubMed] [Google Scholar]
- 14.McMahon F.J., Chen Y.S., Patel S., Kokoszka J., Brown M.D., Torroni A., DePaulo J.R., Wallace D.C. Mitochondrial DNA sequence diversity in bipolar affective disorder. Am. J. Psychiatry. 2000;157:1058–1064. doi: 10.1176/appi.ajp.157.7.1058. [DOI] [PubMed] [Google Scholar]
- 15.Chagnon P., Gee M., Filion M., Robitaille Y., Belouchi M., Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am. J. Med. Genet. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 16.Carrieri G., Bonafe M., De Luca M., Rose G., Varcasia O., Bruni A., Maletta R., Nacmias B., Sorbi S., Corsonello F., et al. Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer's disease. Hum. Genet. 2001;108:194–198. doi: 10.1007/s004390100463. [DOI] [PubMed] [Google Scholar]
- 17.van der Walt J.M., Dementieva Y.A., Martin E.R., Scott W.K., Nicodemus K.K., Kroner C.C., Welsh-Bohmer K.A., Saunders A.M., Roses A.D., Small G.W., et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci. Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 18.van der Walt J.M., Nicodemus K.K, Martin E.R., Scott W.K., Nance M.A., Watts R.L., Hubble J.P., Haines J.L., Koller W.C., Lyons K., et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson Disease. Am. J. Hum. Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghezzi D., Marelli C., Achilli A., Goldwurm S., Pezzoli G., Barone P., Pellecchia M.T., Stanzione P., Brusa L., Bentivoglio A.R., et al. Mitochondrial DNA haplogroup K is associated with a lower risk of parkinson's disease in Italians. Eur. J. Hum. Genet. 2005;13:748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- 20.De Benedictis G., Rose G., Carrieri G., De Luca M., Falcone E., Passarino G., Bonafe M., Monti D., Baggio G., Bertolini S., et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 21.Coskun P.E., Ruiz-Pesini E.E., Wallace D.C. Control region mtDNA variants: longevity, climatic adaptation and a forensic conundrum. Proc. Natl Acad. Sci. USA. 2003;100:2174–2176. doi: 10.1073/pnas.0630589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanova R., Astrinidis A., Lepage V., Kouvatsi A., Djoulah S., Hors J., Charron D. Mitochondrial DNA polymorphism in the French population. Biomed. Pharmacother. 1999;53:207–212. doi: 10.1016/S0753-3322(99)80089-4. [DOI] [PubMed] [Google Scholar]
- 23.Ross O.A., McCormack R., Curran M.D., Duguid R.A., Barnett Y.A., Rea I.M., Middleton D. Mitochondrial DNA polymorphism: its role in longevity of the Irish population. Exp. Gerontol. 2001;36:1161–1178. doi: 10.1016/s0531-5565(01)00094-8. [DOI] [PubMed] [Google Scholar]
- 24.Niemi A.K., Hervonen A., Hurme M., Karhunen P.J., Jylha M., Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 25.Mishmar D., Ruiz-Pesini E., Brandon M., Wallace D.C. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum. Mutat. 2004;23:125–133. doi: 10.1002/humu.10304. [DOI] [PubMed] [Google Scholar]
- 26.Davis R.E., Miller S., Herrnstadt C., Ghosh S.S., Fahy E., Shinobu L.A., Galasko D., Thal L.J., Beal M.F., Howell N., et al. Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc. Natl Acad. Sci. USA. 1997;94:4526–4531. doi: 10.1073/pnas.94.9.4526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Wallace D.C., Stugard C., Murdock D., Schurr T., Brown M.D. Ancient mtDNA sequences in the human nuclear genome: a potential source of errors in identifying pathogenic mutations. Proc. Natl Acad. Sci. USA. 1997;94:14900–14905. doi: 10.1073/pnas.94.26.14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 29.Johnson M.J., Wallace D.C., Ferris S.D., Rattazzi M.C., Cavalli-Sforza L.L. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J. Mol. Evol. 1983;19:255–271. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- 30.Cann R.L., Stoneking M., Wilson A.C. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 31.Merriwether D.A., Clark A.G., Ballinger S.W., Schurr T.G., Soodyall H., Jenkins T., Sherry S.T., Wallace D.C. The structure of human mitochondrial DNA variation. J. Mol. Evol. 1991;33:543–555. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- 32.Wallace D.C. 1994 William Allan Award Address. Mitochondrial DNA variation in human evolution, degenerative disease, and aging. Am. J. Hum. Genet. 1995;57:201–223. [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace D.C., Garrison K., Knowler W.C. Dramatic founder effects in Amerindian mitochondrial DNAs. Am. J. Phys. Anthropol. 1985;68:149–155. doi: 10.1002/ajpa.1330680202. [DOI] [PubMed] [Google Scholar]
- 34.Schurr T.G., Ballinger S.W., Gan Y.Y., Hodge J.A., Merriwether D.A., Lawrence D.N., Knowler W.C., Weiss K.M., Wallace D.C. Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am. J. Hum. Genet. 1990;46:613–623. [PMC free article] [PubMed] [Google Scholar]
- 35.Torroni A., Schurr T.G., Yang C.C., Szathmary E.J., Williams R.C., Schanfield M.S., Troup G.A., Knowler W.C., Lawrence D.N., Weiss K.M., et al. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992;130:153–162. doi: 10.1093/genetics/130.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torroni A., Lott M.T., Cabell M.F., Chen Y., Laverge L., Wallace D.C. MtDNA and the origin of Caucasians. identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am. J. Hum. Genet. 1994;55:760–776. [PMC free article] [PubMed] [Google Scholar]
- 37.Torroni A., Huoponen K., Francalacci P., Petrozzi M., Morelli L., Scozzari R., Obinu D., Savontaus M.L., Wallace D.C. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballinger S.W., Schurr T.G., Torroni A., Gan Y.Y., Hodge J.A., Hassan K., Chen K.H., Wallace D.C. Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations [published erratum appears in Genetics 1992 Apr;130(4):957] Genetics. 1992;130:139–152. doi: 10.1093/genetics/130.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torroni A., Miller J.A., Moore L.G., Zamudio S., Zhuang J., Droma R., Wallace D.C. Mitochondrial DNA analysis in Tibet implications for the origin of the Tibetan population and its adaptation to high altitude. Am. J. Phys. Anthropol. 1994;93:189–199. doi: 10.1002/ajpa.1330930204. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y.S., Torroni A., Excoffier L., Santachiara-Benerecetti A.S., Wallace D.C. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. Am. J. Hum. Genet. 1995;57:133–149. [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y.S., Olckers A., Schurr T.G., Kogelnik A.M., Huoponen K., Wallace D.C. mtDNA variation in the South African Kung and Khwe and their genetic relationships to other African populations. Am. J. Hum. Genet. 2000;66:1362–1383. doi: 10.1086/302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown M.D., Hosseini S.H., Torroni A., Bandelt H.J., Allen J.C., Schurr T.G., Scozzari R., Cruciani F., Wallace D.C. mtDNA Haplogroup X: an ancient link between Europe/Western Asia and North America? Am. J. Hum. Genet. 1998;63:1852–1861. doi: 10.1086/302155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macaulay V., Richards M., Hickey E., Vega E., Cruciani F., Guida V., Scozzari R., Bonne-Tamir B., Sykes B., Torroni A. The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am. J. Hum. Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace D.C., Brown M.D., Schurr T.G., Chen E., Chen Y.-S., Starikovskaya Y.B., Sukernik R.I. Global mitochordrial DNA variation and the origin of Native Americans. In: Aloisi M., Battaglia B., Carafoli E., Danieli G.A., editors. The Origin of Humankind. Conference Proceedings of the International Symposium; 14–15 May 1998; Venice. Venice: IOS Press; 2000. pp. 9–11. [Google Scholar]
- 45.Ingman M., Kaessmann H., Paabo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 46.Wallace D.C., Lott M.T. Mitochondrial genes in degenerative diseases, cancer and aging. In: Rimoin D.L., Connor J.M., Pyeritz R.E., Korf B.R., editors. Emery and Rimoin's Principles and Practice of Medical Genetics. Fourth ed. London: Churchill Livingstone; 2002. pp. 299–409. [Google Scholar]
- 47.Wallace D.C., Lott M.T., Brown M.D., Kerstann K. Mitochondria and Neuro-opthalmological disease. In: Scriver C.R., Beaudet A.L., Sly W.S., Valle D., editors. The Metabolic and Molecular Basis of Inherited Disease. Eighth ed. Vol. II. New York: McGraw-Hill; 2001. pp. 2425–2512. [Google Scholar]
- 48.Rubio-Gozalbo M.E., Dijkman K.P., van den Heuvel L.P., Sengers R.C., Wendel U., Smeitink J.A. Clinical differences in patients with mitochondriocytopathies due to nuclear versus mitochondrial DNA mutations. Hum. Mutat. 2000;15:522–532. doi: 10.1002/1098-1004(200006)15:6<522::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]