Abstract
The serum response factor (SRF) is essential for embryonic development and maintenance of muscle cells and neurons. The mechanism by which this factor controls these divergent pathways is unclear. Here we present a genome-wide view of occupancy of SRF at its binding sites with a focus on those that vary with cell type. We used chromatin immunoprecipitation (ChIP) in combination with human promoter microarrays to identify 216 putative SRF binding sites in the human genome. We performed independent quantitative PCR validation at over half of these sites that resulted in 146 sites we assert to be true binding sites at over 90% confidence. Nearly half of the sites are bound by SRF in only one of the three cell types we tested, providing strong evidence for the diverse roles for SRF in different cell types. We also explore possible mechanisms controlling differential binding of SRF in these cell types by assaying cofactor binding, DNA methylation, histone methylation, and histone acetylation at a subset of sites bound preferentially in smooth muscle cells. Although we did not see a strong correlation between SRF binding and epigenetics modifications, at these sites, we propose that SRF cofactors may play an important role in determining cell-dependent SRF binding sites. ELK4 (previously known as SAP-1 [SRF-associated protein-1]) is ubiquitously expressed. Therefore, we expected it to occupy sites where SRF binding is common in all cell types. Indeed, 90% of SRF sites also bound by ELK4 were common to all three cell types. Together, our data provide a more complete understanding of the regulatory network controlled by SRF.
The serum response factor (SRF) is an essential gene, expressed throughout the human body and throughout development (Gauthier-Rouviere et al. 1991; Belaguli et al. 1997). It plays important roles in human disease and development, specifically in muscle cells and neurons. Because of the essential roles of SRF throughout the body, identifying the genes it regulates provides key information to understanding its functions.
SRF is a MADS-box transcription factor first identified for its role in the response of tissue culture cells to serum (Treisman et al. 1992). Early work demonstrated that genes controlling cell proliferation such as Fos, Junb, Fosb, and Egr1 were regulated by SRF (Treisman 1986; Qureshi et al. 1991; Lazo et al. 1992; Perez-Albuerne et al. 1993). SRF has also been linked to human diseases including cancer and heart disease (Iyer et al. 1999; Chang et al. 2003; Nelson et al. 2005; Parlakian et al. 2005). A common set of genes activated in response to serum are expressed similarly in tumor samples and serum-induced tissue culture cells, demonstrating a link between this regulatory program and tumor cells (Chang et al. 2004, 2005).
Given its well-described role in cell proliferation, the link between SRF and cancer is not surprising; however, recent studies have shown that it is also essential for development and maintenance of smooth muscle cells and neurons. SRF is necessary for mesoderm formation (Arsenian et al. 1998), cardiac development and maintenance (Miano et al. 2004; Parlakian et al. 2005), neuronal migration and circuit assembly (Alberti et al. 2005; Knoll et al. 2006), and learning (Etkin et al. 2006). The requirement for SRF in these processes may result from its role in regulating cytoskeletal proteins and cytoskeleton remodeling proteins, such as actin and cofilin.
The distinct roles for SRF in cell proliferation and cell-type specific maintenance suggest the genes it regulates may differ between cell types and environments. Studies to date have focused on identifying SRF binding sites in a single tissue or analyzing the gross phenotypes of SRF knockouts without identifying the downstream effectors. SRF is highly expressed in muscle but detectable throughout the body, leaving the unanswered question of how SRF effects its changes in divergent pathways (Treisman 1986; Belaguli et al. 1997). Many groups have hypothesized that SRF binding is determined by the presence of cofactors, which can be tissue specific. SRF interacts with many different cofactors. These include ubiquitously expressed cofactors of the Ets transcription factor family—ELK1, ELK4 (previously known as SAP-1 SRF-associated protein-1]), and NET1 (for review, see Buchwalter et al. 2004)—and tissue-specific cofactors—GATA4, GATA6, NKX2-5, and myocardin and its related transcription factors (MKL1 and MKL2) (for review, see Cen et al. 2004). Although some factors are tissue specific, most do not bind DNA in the absence of SRF, which suggests binding may also be affected by epigenetics (Shaw et al. 1989; Verger and Duterque-Coquillaud 2002). Finally, SRF itself can be modified by phosphorylation to alter its affinity for DNA (Iyer et al. 2006). Each of these variables contributes to expression of the downstream gene.
The study we describe significantly improves our knowledge of SRF binding sites and demonstrates the variability of binding in diverse cell types. By using chromatin immunoprecipitation (ChIP) in combination with human promoter microarrays, we identified more than 200 binding sites of SRF in three human cell lines. We confirmed binding at known sites but also identified many new sites. Our study is also unique in its extensive validation by quantitative PCR (qPCR), resulting in positive validation for 86 of 116 tested sites. These validation data allow us to assert that an additional 60 binding sites are real at a 90% confidence level. In an effort to understand the mechanism of tissue-specific SRF binding observed in our data, we characterized binding of the ELK4 cofactor as well as epigenetic modifications at validated SRF binding sites. Together, these data provide the most comprehensive view of gene regulation by SRF to date.
Results
Identification of over 200 genome-wide SRF binding sites
By using ChIP and human promoter microarrays, we characterized genome-wide binding sites of SRF in three cell lines: Jurkat cells (a transformed lymphoblast cell line), T/G HA-VSMC (primary aortic smooth muscle cells), and Be(2)-C (a neuroblastoma cell line). In Jurkat cells, we assayed binding before and after induction with serum, although we did not observe many differences between the two treatments. To detect SRF binding sites, we applied each ChIP sample to arrays tiling 1.3 kb each (1 kb upstream of the transcription start site [TSS] and 0.3 kb downstream) of ∼19,000 human promoters. Based on the average DNA shearing size, we expected to detect binding sites ranging from ∼2 kb upstream of the TSS to 1 kb downstream. Because of random shearing of genomic DNA, we observed the greatest DNA enrichment at the SRF binding sites with reduced but detectable enrichment by using probes up to a few kilobases away from the binding site. Our ability to detect SRF occupancy is proportional to the distance between the probes and the binding site (Trinklein et al. 2004). On the basis of the ratio of ChIP-enriched sample to a control mock immunoprecipitation, we determined a Z-score resulting in a score and confidence for SRF binding at each promoter (see Methods).
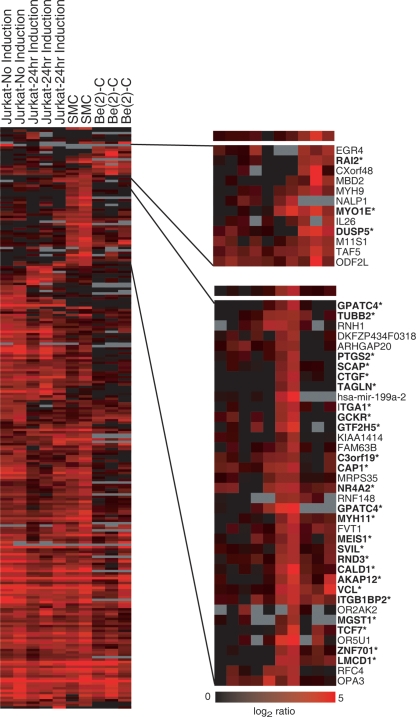
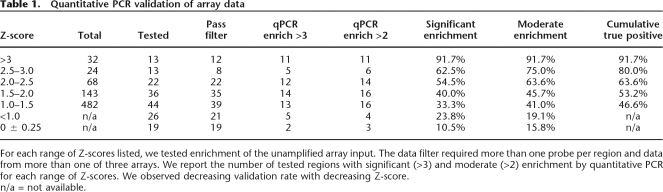
We identified a total of 216 putative targets of SRF in the cell lines and conditions described (Fig. 1). These sites all have Z-scores >2 in multiple cell lines or a Z-score >2.5, to improve confidence, if they appear in only one cell line. These thresholds were chosen to maximize true positives and minimize false negatives based on the validation study described (Methods). (An additional 144 putative sites were identified in only one cell type with a Z-score between 2 and 2.5.) In two phases, we independently measured the ChIP enrichment at SRF binding sites as determined by the array. In the first phase, we tested 163 amplicons identified in Jurkat cells with Z-scores ranging from 0 to >3, allowing a careful evaluation of the accuracy of the array. Classifying putative sites by Z-score, we observed 92% validation of binding sites with a Z-score >3 and a cumulative validation of 64% for sites with a Z-score >2 (Table 1). In the second phase of validation, we tested an additional 64 sites by qPCR, emphasizing validation of binding sites identified in only one cell type or condition. Between the two rounds of validation, 86 binding sites were successfully validated. Of the untested putative binding sites, 47 have a Z-score >3. We expect 92% of these to validate based on the validation rate of other sites with similar Z-scores (see Methods and Table 1). An additional 13 untested sites are inferred to be true with high confidence based on conservation of human and mouse SRF consensus sites in the region (see below). Only 29 of the binding sites we identified were common to all cell types and conditions of cell growth. Common binding sites include those regulating genes involved in cell proliferation such as early growth response 1 (EGR1), early growth response 2 (EGR2), FOS, FOSB, and SRF itself, where SRF is known to bind constitutively (Herrera et al. 1989). By using the EASE software package, we annotated the function of genes whose promoters were bound in all three cell types and found that about half of these genes are transcription factors and DNA binding proteins, which is significant compared with the genome-wide distribution (P < 0.05).
Figure 1.
SRF binding in three cell types. For each cell type and each putative target, the intensity of red indicates size of the Z-score obtained from the arrays. The right panel displays a subset of genes bound specifically by SRF in neurons (top) and smooth muscle cells (bottom). Gene names followed by an asterisk have been validated by quantitative PCR or were previously known.
Table 1.
Quantitative PCR validation of array data
For each range of Z-scores listed, we tested enrichment of the unamplified array input. The data filter required more than one probe per region and data from more than one of three arrays. We report the number of tested regions with significant (>3) and moderate (>2) enrichment by quantitative PCR for each range of Z-scores. We observed decreasing validation rate with decreasing Z-score.
n/a = not available.
SRF occupancy varies by cell type
Previous work characterizing binding sites of SRF focused on a single cell type, such as rat or mouse cardiomyocytes (Manabe and Owens 2001; Nelson et al. 2005; Zhang et al. 2005) or a single human tissue culture cell line (Rene et al. 2005). We found that many SRF binding sites depend on tissue type. Over half of the targets we identified were unique to a particular cell type or condition (Fig. 1). Because of the implication for disease and development, binding sites specific to smooth muscle cells and neurons were especially interesting. We used qPCR to test binding at 65 tissue-dependent SRF binding sites with a Z-score >2.5 on the array. Of the 65 tested, 43 promoters validated (enrichment greater than threefold by qPCR) as tissue-dependent SRF-binding sites. Because of the array false-negative rate, we independently confirmed by qPCR that these sites were not bound by SRF in the remaining cell types for 12 cell-type–dependent targets (Supplemental Fig. 1).
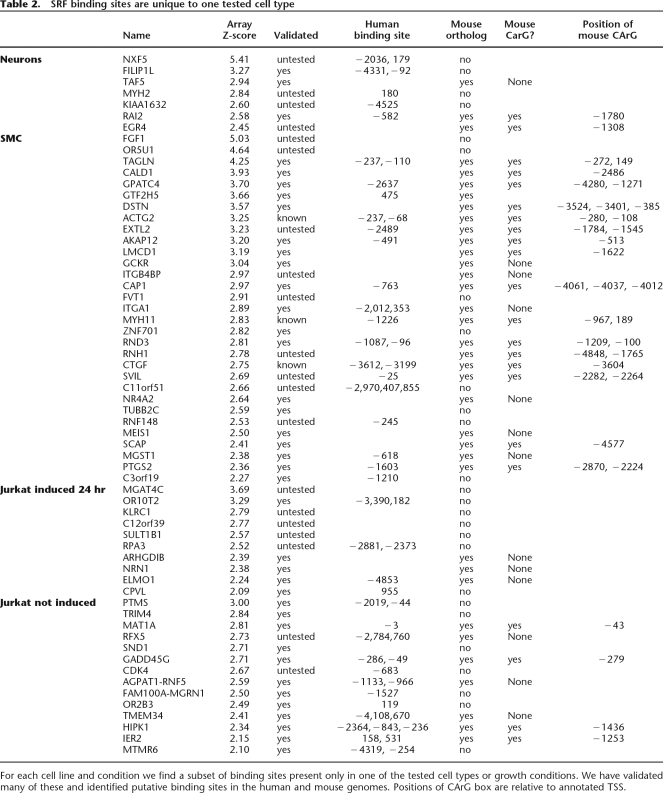
We identified 27 targets specific to smooth muscle cells (tested and validated 19 of 19 successful amplicons) (Table 2). These data confirmed known binding sites in promoters of genes involved in muscle development and cytoskeleton including transgelin (TAGLN), caldesmon (CALD1), vinculin (VCL), smooth muscle myosin heavy chain 11 (MYH11), and melusin (ITGB1BP2). We observed SRF binding at promoters of genes encoding additional cytoskeletal proteins such as myosin 1e (MYO1E) and beta tubulin (TUBB). Most interestingly, we identified binding sites in genes not previously known to be regulated by SRF. For example, adenylyl cyclase-associated protein 1 (CAP1) is involved in the cyclic AMP cycle, is expressed in smooth muscle and cardiomyocytes, and binds actin (Diehn et al. 2003; Hsu et al. 2006). Another SRF-regulated gene, heart alpha-kinase 2 (ALPK2), is expressed in heart, skeletal muscle, and cardiomyocytes (Jongeneel et al. 2005). ALPK2 is one member of a protein kinase-like family whose other members phosphorylate myosin heavy chains. We also discovered a binding site in the muscle-specific supervillin (SVIL) isoform 2 promoter (Gangopadhyay et al. 2004). SVIL binds actin and vinculin in skeletal muscle and is thought to be important to cell adhesion and motility (Ting et al. 2002). Finally, two sites we identified are in promoters of genes involved in prostaglandin synthesis: prostaglandin-endoperoxide synthase 2 (PTGS2) and microsomal glutathione S-transferase 1 (MGST1), which have been previously described for their role in inflammation; however, both genes are highly expressed in smooth muscle (Su et al. 2004). The expression of these genes in muscle and their interactions with cytoskeletal proteins make them good candidates for genes contributing to the phenotypes of SRF developmental knockouts.
Table 2.
SRF binding sites are unique to one tested cell type
For each cell line and condition we find a subset of binding sites present only in one of the tested cell types or growth conditions. We have validated many of these and identified putative binding sites in the human and mouse genomes. Positions of CArG box are relative to annotated TSS.
Another class of promoters is bound by SRF only in the neuronal cell line. These sites include the promoter of another cytoskeletal protein, non-muscle myosin (MYH9), and the EGR4 promoter. The Egr family is partially redundant (Tourtellotte et al. 2000) and previous work demonstrates a role for Egr1 in synaptic plasticity (Li et al. 2005). Egr4 may compensate for or complement the role of Egr1 in the brain. We also show SRF binding in the nuclear export factor (NPAS4) promoter. This gene directly regulates expression of drebrin, a protein involved in dendritic-cytoskeleton modulation at synapses (Ooe et al. 2004). The FILIP1L promoter was also identified as a brain-specific binding site. Although this protein is normally associated with cell cycle transition, recent data implicate the complex containing FILIP1L in axonal growth and synaptic function (for review, see Teng and Tang 2005). Identification of neuron-specific binding sites of SRF is particularly important since the role of SRF in the brain is poorly understood.
SRF binding sites contain CArG boxes
Since SRF binding changes with cell type, we searched for differences in sequence content between promoters bound uniquely in each cell type. We observed two interesting differences between promoters bound by SRF in all cell types and promoters bound in only one cell type. Promoters bound by SRF in only a single cell type are nearly twice as likely as common SRF binding sites to contain a TATA box within 500 bp of the predicted TSS (χ2 = 7.68, P = 0.053). On the basis of this observation, it is not surprising that promoters bound by SRF under only a single condition are less likely to overlap a CpG island (χ2 = 11.08, P = 0.01) (Gardiner 1995).
Next, we hoped to determine whether the binding sites for SRF differed between the two classes of promoters. To do this, we first characterized the presence of the known consensus site near SRF binding sites. The consensus binding site for SRF, often called the CArG box, is well established from a large number of biochemical and mutagenesis experiments: CC(A/T)TATA(A/T)GG (Vlieghe et al. 2006). We searched promoters with SRF binding sites up to 4 kb upstream and 1 kb downstream of the putative TSS for the known consensus site allowing one base pair of mismatch. We found that 61% of the 216 total binding sites contain a CArG box. Of these, 32% (43 sites) contain multiple CArG boxes. Despite a larger search window, 82.5% of these sites are within 2 kb upstream and 1 kb downstream of the TSS. Of all promoters represented on the array, 30% contain this sequence, and 20% of those contain multiple targets. While the degeneracy of the consensus results in a high incidence of this sequence in all promoters, the enrichment we observe in our sites is significant at P < 10−23. Within the 86 targets validated by qPCR, a similar number, 67%, contain a CArG box. This shows that some bona fide binding sites do not have a canonical CArG box in the sequence we searched. If we search for the best available match in these sequences, we find that four promoters contain a sequence resembling a CArG box that does not match the consensus within 1-bp mismatch but may be able to bind SRF. Other sequences contain an AT-rich sequence that may be able to bind SRF under the right conditions. Subdividing the sites based on cell type did not significantly change the occurrence or sequence of the motif. By using MEME, CisModule, and BioProspector, our searches for alternate motifs specific to individual cell types did not reveal any novel motifs (Liu et al. 2001; Zhou and Wong 2004; Bailey et al. 2006).
While almost all previously published SRF binding sites are within 1 kb of the TSS, Sun and colleagues (2006) estimate that 60% of functional CArG boxes are within 2 kb of the start site and nearly all SRF binding sites are within 4 kb. In the SRF-bound promoters we identified, the median distance from the consensus site to TSS is 400 bp, with over 80% of consensus sites within 2 kb of the TSS; therefore, the sites we have identified are skewed toward the TSS. This is expected since the arrays contain probes only up to 1 kb upstream of the TSS. While we can capture more distant binding sites, it is more difficult to distinguish these signals from noise; therefore, we preferentially identify sites close to the TSS. Overall, the presence of CArG boxes near SRF binding sites provides further evidence for our successful search. The absence of a consensus in some cases is also interesting because it suggests that the accepted consensus site may not describe all SRF binding sites. By using the motif finders described above, we were unable to characterize a motif in these regions that differed significantly from the known consensus; however, even the known motif could not be identified by these algorithms if we provided the full sequence of the putative binding site. The CG-rich nature of these sequences and the amount of input sequence confounds all three motif-finders.
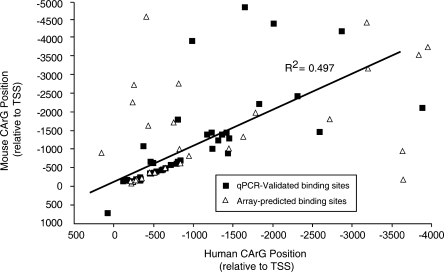
Because functionally important CArG boxes should be evolutionarily conserved, we searched the orthologous mouse promoters for the consensus binding site. In many cases, the mouse promoters also contain a CArG box. We identified orthologous promoters for 96 of the 133 human promoters containing a CArG box. Of these, 56 (58%) contain a sequence matching the CArG consensus site in the mouse genome. We have plotted the position of each predicted CArG box relative to the predicted start site and see a significant correlation between the positions in human and mouse (Fig. 2). This suggests that the orientation relative to the TSS may be important. Interestingly, the presence of a CArG box in both human and mouse promoters is a strong indication of SRF binding. Seventy-three percent of validated SRF-bound promoters with a CArG box also contained an orthologous consensus site in mouse. While the presence of the CArG box in mouse is not required, in the cases in which an orthologous CArG box existed, validation rate was very high. For 32 of 33 promoters with orthologous human and mouse CArG boxes, we successfully validated SRF binding indicated by arrays. On the basis of these analyses, we predict that the 23 binding sites with a conserved CArG box that were not validated by qPCR are also true SRF binding sites with high confidence (>95%). In addition, 10 of these sites also have a Z-score >3. Indeed, the untested promoters include genes previously identified to be regulated by SRF, such as smooth muscle α- and γ-actins (ACTA2 and ACTG2), connectin tissue growth factor (CTGF), and smooth muscle myosin heavy chain (MYH11) (for review, see Miano 2003; Sun et al. 2006).
Figure 2.
CArG position is conserved between human and mouse. We compared positions of the SRF consensus binding site in mouse and human promoters and observed a strong correlation (R2 = 0.497). This correlation is driven primarily by consensus sites within 1–2 kb of the TSS. We display sites validated by quantitative PCR as squares and untested sites as triangles.
Correlating cell-type–dependent SRF binding with epigenetic modifications
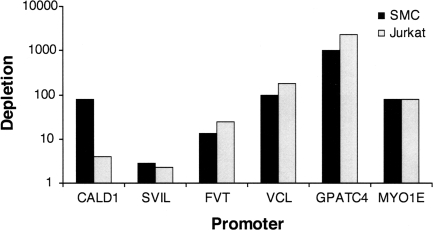
Because SRF binding is often dependent on cell type, we wished to explore the mechanism determining SRF binding sites in vivo. We proposed that differences in chromatin structure between cell types could control SRF binding. We assayed three modifications affecting chromatin structure and asked whether differences in these modifications correlated with differences in SRF binding. First, we assayed DNA methylation of the six promoters bound by SRF in smooth muscle cells but not Jurkat cells that overlapped with CpG islands (Fig. 3). By using methyl-sensitive enzymes, we digested unmethylated DNA and removed the regions cut into small pieces by the six enzymes using size selection. We assayed depletion of these regions, which represent unmethylated DNA by qPCR, normalizing to an uncut sample prepared in parallel (see Methods). In 5 of 6 cases, we found that promoters were unmethylated in both cell types indicating that differential methylation of these promoters does not account for the difference we observed in SRF binding. In the case of the caldesmon gene (CALD1), there is a difference in depletion between the two cell types, suggesting that while this is not the typical mechanism for regulation, DNA methylation may play a role in some cases.
Figure 3.
DNA methylation SRF binding sites found only in smooth muscle cells (SMC). In SMC (black) and Jurkat cells (gray) we observed a depletion of unmethylated DNA in the promoter of six genes. Except for the caldesmon gene (CALD1), depletion is similar between the two cell types. Depletion is a measure of fold difference between uncut and cut sample (see Methods).
Histone modifications provide another indication of chromatin structure and are generally considered to be more dynamic than DNA methylation. We assayed acetylation of histone H3 and trimethylation of histone H3 at lysine 4 (H3K4), both modifications present in active chromatin (Santos-Rosa et al. 2002), by using ChIP and qPCR in smooth muscle and Jurkat cells. As expected, we detected both of these modifications at active genes such as GAPDH and RPL30; however, we only observed significant enrichment due to these modifications at three of the nine promoters bound by SRF in smooth muscle cells but not Jurkat cells (data not shown). In cases where we observed acetylation and trimethyl modification of histone H3, it was present in both cell types. It remains a possibility that other histone modifications, specifically those resulting in silencing, would correlate with the lack of SRF binding in Jurkat cells compared with smooth muscle cells, but we have not identified a correlation with the tests described here.
Tissue-independent SRF binding sites are bound by ELK4
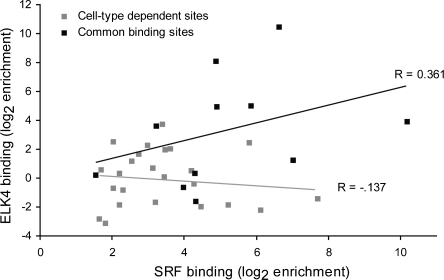
At the binding sites we tested, epigenetic modifications did not correlate well with cell-specific binding sites for SRF. An alternative hypothesis to explain the differential binding we observed involves the activity of SRF cofactors, which can be expressed broadly or tissue-specifically to determine the binding sites of SRF in each cell type. ELK4, a previously characterized SRF cofactor, is ubiquitously expressed and associates with SRF to activate transcription of downstream genes. On the basis of its expression pattern, we expected ELK4 to control SRF binding sites common to multiple cell types and conditions. We assayed binding of ELK4 at all 37 validated SRF binding sites in Jurkat cells. We demonstrated binding of ELK4 to 13 of 37 SRF binding sites including EGR1, EGR2, and FOS, all of which are bound by SRF in all tested cell lines (see Supplemental materials for all data). Eleven of these 13 promoters bound by both ELK4 and SRF are bound by SRF in at least two cell lines tested. In fact, we see a correlation between SRF binding and ELK4 binding only at sites that are cell-type independent (Fig. 4). In addition, we tested ELK4 binding in SMC and neurons at four targets unique to each cell type and found ELK4 binding at only one of the eight SRF binding sites. These data support the hypothesis that SRF cofactors play a role in determining SRF binding sites, particularly for ubiquitously bound targets.
Figure 4.
Cell-type dependent binding and the ELK4 cofactor. For SRF binding sites independent of cell type (black squares), binding of SRF and ELK4 at the site is correlated. For cell-type dependent SRF binding sites (grey squares) no correlation is observed. Binding of each factor was determined by quantitative PCR as described (Methods) and is displayed as a log of the enrichment observed at each site compared to a panel of negative controls.
Discussion
These data and analyses provide a significant improvement in knowledge of previously determined roles for SRF. We identified a total of 216 SRF binding sites and completed one of the most comprehensive ChIP–chip validation studies for any transcription factor to date. The result was the identification of 86 validated binding sites and 60 additional high-confidence binding sites for a total of 146 sites at a 90% or higher confidence level (see Supplemental Table 4). (High-confidence binding sites were not tested by qPCR but have an array Z-score >3 or a Z-score >2 with CArG box conserved in human and mouse.) The total number of sites identified in the genome is relatively small compared with other cell proliferation transcription factors such as MYC; however, SRF may regulate this process by controlling a smaller number of genes, whereas MYC is known to act generally to promote transcription. Compared to previous work, our study included an extensive validation study absent in many other data sets, leaving the possibility that other groups may have high false-positive rates. Previous studies have used ChIP sequencing and ChIP with semi-quantitative PCR to identify over 100 in vivo SRF binding sites; however, these binding sites were identified almost exclusively in cardiomyocytes (Philippar et al. 2004; Zhang et al. 2005; Balza Jr. and Misra 2006; Sun et al. 2006). While over 90 of these genes were present on the arrays, our experiments identified only 30 previously known SRF binding sites. Nearly half of the overlap is from sites we found in smooth muscle that were previously found in cardiomyocytes, a similar cell type (Zhang et al. 2005). Twenty sites we did not identify are more than 10 kb upstream of the TSS in mouse and a few are located in the first intron. Many of these would not be identified by our method. Additional binding sites may have been missed because of false-negative array results; however, the evidence we have presented supporting the degree of cell-type dependent binding suggests these binding sites may be specific to the cell types tested in previous experiments. Indeed, we tested cell types not previously assayed and were able to verify binding at these sites. These new targets were identified in cell lines where other groups have not searched. Future studies including additional tissues and cell lines will provide interesting insight into SRF function throughout the body.
We identified 84 (60 qPCR validated, 24 untested) SRF binding sites that changed with cell type. These genes are key to understanding the role that SRF plays in determining cell fate during development and maintaining a chosen cell fate. A more complete list of downstream molecules that control muscle and neuronal development will be valuable for a more detailed understanding of these processes. In neurons, for example, SRF is necessary for migration and circuitry formation. SRF was previously thought to exert its control on this pathway primarily through regulation of cytoskeletal proteins; however, we have uncovered a connection between SRF and FILIP1L, a key component of the APC complex and a molecule not previously known to be under the control of SRF but vital to normal neuronal function. Likewise, in smooth muscle cells, where the target genes of SRF have been more fully explored, we have identified a number of previously unknown binding sites, including one in the promoter of CAP1. The CAP1 protein is involved in regulating actin dynamics and binds cofilin, another target gene of SRF (Bertling et al. 2004). Knowledge of these genes and others regulated by SRF in smooth muscle provide a key resource for understanding the function of SRF in these cells. To understand all the roles of SRF in development and cell maintenance, a complete study of human tissues will be necessary.
Here we explored two potential mechanisms by which SRF binding sites are influenced: chromatin structure and cofactor presence. In the cases we explored, the main factor correlated to SRF binding was the presence of SRF cofactors. Our data provide one example in which CpG island methylation seems to affect SRF binding. In each of the other five cases we tested, the promoters of genes differentially bound were unmethylated in both cell types. We also assayed trimethylation at histone H3K4 and acetylation of histone H3, and we did not observe a difference between Jurkat cells and smooth muscle cells (SMC) at promoters bound by SRF in SMC but not Jurkat cells. Our favored hypothesis explaining the ability of SRF to bind differentially is the presence of SRF cofactors. GATA4, for example, is expressed only in muscle and is known to bind with SRF at a few smooth muscle-specific targets (Small and Krieg 2003). Alternatively, ELK4 is a ubiquitously expressed SRF cofactor. We have shown that ELK4 binds with SRF at promoters bound by SRF in multiple cell types, such as EGR1, EGR2, and FOS. Although we have not collected binding data for other SRF cofactors because of lack of ChIP-quality antibodies for these proteins, our data are consistent with hypotheses supporting the role of cofactors in determining SRF binding sites.
The goal of our study was to gain a more complete understanding of elements that regulate genes in this network and ultimately predict their effects, individually and cumulatively, on gene expression. The work presented here provides, first and foremost, a more comprehensive and accurate catalog of genes that are regulated by SRF: 216 total binding sites, the vast majority of which were previously unknown. Our data include 86 validated and 60 high-confidence sites. These data, in combination with studies characterizing chromatin structure and cofactor occupancy at SRF binding sites, provide the beginnings of a network that will ultimately lead to a more complete understanding of the roles of SRF.
As technology improves and large-scale studies become increasingly feasible, data sets identifying binding sites in different tissues, binding sites of SRF cofactors, and expression levels of downstream genes will allow us to complete the regulatory network involving SRF. Understanding the regulation of these genes will provide insight into processes as diverse as embryonic development, heart disease, cancer, and learning.
Methods
Cell culture
We obtained each of the cell lines from American Type Culture Collection (www.atcc.org). Jurkat (TIB-152), BE(2)-C (CRL-2268), and T/G HA-VSMC (CRL-1999) were grown as directed. To induce the serum response, we grew Jurkat cells in media with 0.1% FBS for 48 h and then induced the serum response by replacing the low-serum media with media containing 10% FBS for 24 h.
Chromatin immunoprecipitation
We performed ChIP reactions as previously described with modifications (Trinklein et al. 2004). Briefly, we collected 10–20 million cells for each chromatin preparation of each cell type. We prepared biological duplicates for the primary smooth muscle cells and the uninduced Jurkat cells and triplicates for the serum-induced Jurkat cells and neuroblastoma cell line. We added formaldehyde to 1% final concentration to cross-link proteins and DNA. We scraped adherent cells from plates and spun down suspension cells, then lysed and collected nuclei. We sonicated chromatin to a final size of ∼500–1000 base pairs. We coupled 5 μg of each antibody (anti-SRF [Santa Cruz Biotech sc-335], anti-SAP1a [sc-13030], anti-H3K4me3 [Abcam ab8580], and H3ac [upstate 06–599]) to sheep anti-mouse magnetic beads (Invitrogen 112–02D) by overnight incubation at 4°C in 5 mg/mL BSA. We added the coupled beads to chromatin and precipitated DNA–protein complexes by overnight incubation at 4°C. After immunoprecipitation, we reversed cross-linking by incubation at 65°C overnight and purified DNA with phenol–chloroform extraction followed by a PCR clean-up column (Qiagen cat. no. 28104). Each of the biological replicates was amplified and hybridized to arrays individually.
Human promoter microarrays
Biological replicates of DNA samples recovered from the ChIP were amplified using ligation-mediated PCR as previously described (Kim et al. 2005). After amplification, we labeled 1 μg of DNA for each slide by using the Bioprime Plus Array CGH labeling Module (Invitrogen Cat. no. 18095–014). We labeled each ChIP sample with the red channel dye (Alexa Fluor-647) and Mock IP DNA (ChIP reaction without antibody) with the green channel dye (Alexa Fluor-555). We hybridized these samples competitively to proximal human promoter arrays. The two-array set, each with about 44,000 features, tiled ∼19,000 human promoters (Agilent cat no. G4481A). The TSS predictions for each promoter are based almost exclusively on RefSeq genes, with a few additions from the microRNA registry, the Mammalian Gene Collection (MGC), and Ensembl (Gerhard et al. 2004; Griffiths-Jones 2004; Birney et al. 2006). For each promoter, there is an average of 4 probes of 45–60 nucleotides each tiling ∼1000 bp upstream and 300 bp downstream. We hybridized samples to the array by using the Agilent Array CGH protocol, with one modification (http://chem.agilent.com). Briefly, we combined labeled DNA samples with Cot-1 DNA, blocking reagent, and hybridization buffer, applied the sample to the arrays, and sealed them with gasket slides. We hybridized arrays in a rotating incubator at 60°C, rather than the recommended 65°C. We also conducted washes as recommended by the Agilent CGH protocol. We scanned arrays by using the Agilent dual laser scanner (part no G2565BA) and used Agilent Feature Extraction software (version 8) to analyze the images obtained from scanning. This software invokes a number of algorithms to normalize the data including dye normalization, identification of statistical outliers, and background subtraction. (For more information, see Feature Extraction Software Manual available for download at www.chem.agilent.com.) Raw data are available online at Gene Expression Omnibus (GEO accession number GSE5998).
Microarray analysis
We analyzed output from the feature extraction software, eliminating probes that did not significantly exceed background intensities for both channels. (Supplemental Table 1 contains the raw ratio of experimental to control sample for each probe on each array.) For each probe, we calculated a log2 ratio of experimental to control signals and averaged replicates. This number was converted to a Z-score by assuming a normal distribution of ratios of all probes; thus, high Z-scores have the most significant enrichment ratios. (Z = (x − xbar)/σ, where x is the experimental ratio, xbar is the mean ratio for all probes, and σ is the standard deviation.) By using the annotations provided by Agilent for each probe, we grouped probes by accession number and chromosomal position, allowing for multiple promoters per gene (see Supplemental Table 2 for list of all promoters and Z-scores calculated). To determine SRF binding sites, we calculated a weighted average Z-score for each cluster by down-weighting the lowest and highest Z-score in each cluster to half the weight of the remaining probes.
Microarray validation
With data from the serum-treated Jurkat cells, we used quantitative PCR to determine the enrichment of each of 162 amplicons with a range of Z-scores from near 0 (not significant) to >3 (highly significant) (Table 1). For each putative site, we tested enrichment by using the same replicate DNA samples that were hybridized to the arrays (preamplification). We required putative binding sites to have data for more than one probe within the promoter and at least two of the three array replicates have Z-scores >2. We defined significant enrichment by qPCR as average enrichment across three replicates of greater than threefold. Enrichment of greater than twofold was considered moderate but not significant. By testing a wide range of Z-scores, we estimated the false-negative rate for our array. At a Z-score threshold of 2, we estimated 12% false negative and with a threshold of Z >3, ∼25% false negative. By using these data, we can appropriately assign a Z-score threshold for the remaining experiments. Unless otherwise noted, the Z-score threshold is 2 for sites identified in more than one cell line or 2.5 for sites in only a single cell line.
GO term analysis and gene annotation
We used the EASE software package to identify overrepresented GO terms in our gene lists. The software is available for download at http://david.niaid.nih.gov/david/ease.htm (Hosack et al. 2003). We used the EASE score to determine significance.
Quantitative PCR
We used the BioRad iCycler for all quantitative PCR (qPCR) validation as previously described (Trinklein et al. 2004). Briefly, we compared threshold cycles of each putative binding site to a standard curve and calculated fold-enrichment compared with negative control regions designed to amplify the middle exons of genes. All PCR reactions were 40 cycles and amplicons were 60–100 base pairs. Primer sequences are available in the Supplemental materials (Supplemental Table 3).
Preparation of genomic DNA
We prepared DNA from Jurkat and T/G HA-VSMC cell pellets containing ∼4 × 106 cells. After lysis we homogenized the sample by using Qiashredder columns (Qiagen cat. no. 79654). We used the Qiagen Allprep RNA/DNA Mini Kit (cat. no. 80204) as instructed to purify total RNA and genomic DNA. Genomic DNA was used as input for the methylation assay.
Methylation assay
To determine the methylation status at the described targets, we digested 2 μg of genomic DNA from each cell line with six methyl sensitive enzymes in sequential digests (HpaII, HgaI, HpyCH4IV, AciI, HhaI, and BstuI). Digestion with this cocktail of enzymes depletes over 99% of unmethylated CpG islands (A.L. Brown, N.D. Trinklein, and R.M. Myers, unpubl.). We detected depletion of unmethylated regions compared with an uncut sample by using qPCR as described above.
Acknowledgments
We thank members of the Myers Laboratory for helpful discussion and encouragement. We thank Chris Hopkins for his advice and discussions as we established the Agilent array technology in our laboratory. S.J.C. is supported by the Stanford Genome Training Program (Training Grant NIH 5 T32 HG00044). This work was supported by NIH Grant 1 U01 HG 03162-01 (to R.M.M) from the National Human Genome Research Institute.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5875007
References
- Alberti S., Krause S.M., Kretz O., Philippar U., Lemberger T., Casanova E., Wiebel F.F., Schwarz H., Frotscher M., Schutz G., Krause S.M., Kretz O., Philippar U., Lemberger T., Casanova E., Wiebel F.F., Schwarz H., Frotscher M., Schutz G., Kretz O., Philippar U., Lemberger T., Casanova E., Wiebel F.F., Schwarz H., Frotscher M., Schutz G., Philippar U., Lemberger T., Casanova E., Wiebel F.F., Schwarz H., Frotscher M., Schutz G., Lemberger T., Casanova E., Wiebel F.F., Schwarz H., Frotscher M., Schutz G., Casanova E., Wiebel F.F., Schwarz H., Frotscher M., Schutz G., Wiebel F.F., Schwarz H., Frotscher M., Schutz G., Schwarz H., Frotscher M., Schutz G., Frotscher M., Schutz G., Schutz G., et al. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc. Natl. Acad. Sci. 2005;102:6148–6153. doi: 10.1073/pnas.0501191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenian S., Weinhold B., Oelgeschlager M., Ruther U., Nordheim A., Weinhold B., Oelgeschlager M., Ruther U., Nordheim A., Oelgeschlager M., Ruther U., Nordheim A., Ruther U., Nordheim A., Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W., Williams N., Misleh C., Li W.W., Misleh C., Li W.W., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balza R.O., Jr., Misra R.P., Misra R.P. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J. Biol. Chem. 2006;281:6498–6510. doi: 10.1074/jbc.M509487200. [DOI] [PubMed] [Google Scholar]
- Belaguli N.S., Schildmeyer L.A., Schwartz R.J., Schildmeyer L.A., Schwartz R.J., Schwartz R.J. Organization and myogenic restricted expression of the murine serum response factor gene. A role for autoregulation. J. Biol. Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- Bertling E., Hotulainen P., Mattila P.K., Matilainen T., Salminen M., Lappalainen P., Hotulainen P., Mattila P.K., Matilainen T., Salminen M., Lappalainen P., Mattila P.K., Matilainen T., Salminen M., Lappalainen P., Matilainen T., Salminen M., Lappalainen P., Salminen M., Lappalainen P., Lappalainen P. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell. 2004;15:2324–2334. doi: 10.1091/mbc.E04-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Cox T., Cunningham F., Curwen V., Cutts T., Cunningham F., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter G., Gross C., Wasylyk B., Gross C., Wasylyk B., Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Cen B., Selvaraj A., Prywes R., Selvaraj A., Prywes R., Prywes R. Myocardin/MKL family of SRF coactivators: Key regulators of immediate early and muscle specific gene expression. J. Cell. Biochem. 2004;93:74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- Chang J., Wei L., Otani T., Youker K.A., Entman M.L., Schwartz R.J., Wei L., Otani T., Youker K.A., Entman M.L., Schwartz R.J., Otani T., Youker K.A., Entman M.L., Schwartz R.J., Youker K.A., Entman M.L., Schwartz R.J., Entman M.L., Schwartz R.J., Schwartz R.J. Inhibitory cardiac transcription factor, SRF-N, is generated by caspase 3 cleavage in human heart failure and attenuated by ventricular unloading. Circulation. 2003;108:407–413. doi: 10.1161/01.CIR.0000084502.02147.83. [DOI] [PubMed] [Google Scholar]
- Chang H.Y., Sneddon J.B., Alizadeh A.A., Sood R., West R.B., Montgomery K., Chi J.T., de van Rijn M., Botstein D., Brown P.O., Sneddon J.B., Alizadeh A.A., Sood R., West R.B., Montgomery K., Chi J.T., de van Rijn M., Botstein D., Brown P.O., Alizadeh A.A., Sood R., West R.B., Montgomery K., Chi J.T., de van Rijn M., Botstein D., Brown P.O., Sood R., West R.B., Montgomery K., Chi J.T., de van Rijn M., Botstein D., Brown P.O., West R.B., Montgomery K., Chi J.T., de van Rijn M., Botstein D., Brown P.O., Montgomery K., Chi J.T., de van Rijn M., Botstein D., Brown P.O., Chi J.T., de van Rijn M., Botstein D., Brown P.O., de van Rijn M., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.Y., Nuyten D.S., Sneddon J.B., Hastie T., Tibshirani R., Sorlie T., Dai H., He Y.D., van’t Veer L.J., Bartelink H., Nuyten D.S., Sneddon J.B., Hastie T., Tibshirani R., Sorlie T., Dai H., He Y.D., van’t Veer L.J., Bartelink H., Sneddon J.B., Hastie T., Tibshirani R., Sorlie T., Dai H., He Y.D., van’t Veer L.J., Bartelink H., Hastie T., Tibshirani R., Sorlie T., Dai H., He Y.D., van’t Veer L.J., Bartelink H., Tibshirani R., Sorlie T., Dai H., He Y.D., van’t Veer L.J., Bartelink H., Sorlie T., Dai H., He Y.D., van’t Veer L.J., Bartelink H., Dai H., He Y.D., van’t Veer L.J., Bartelink H., He Y.D., van’t Veer L.J., Bartelink H., van’t Veer L.J., Bartelink H., Bartelink H., et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl. Acad. Sci. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Sherlock G., Binkley G., Jin H., Matese J.C., Hernandez-Boussard T., Rees C.A., Cherry J.M., Botstein D., Brown P.O., Sherlock G., Binkley G., Jin H., Matese J.C., Hernandez-Boussard T., Rees C.A., Cherry J.M., Botstein D., Brown P.O., Binkley G., Jin H., Matese J.C., Hernandez-Boussard T., Rees C.A., Cherry J.M., Botstein D., Brown P.O., Jin H., Matese J.C., Hernandez-Boussard T., Rees C.A., Cherry J.M., Botstein D., Brown P.O., Matese J.C., Hernandez-Boussard T., Rees C.A., Cherry J.M., Botstein D., Brown P.O., Hernandez-Boussard T., Rees C.A., Cherry J.M., Botstein D., Brown P.O., Rees C.A., Cherry J.M., Botstein D., Brown P.O., Cherry J.M., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O., et al. SOURCE: A unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin A., Alarcon J.M., Weisberg S.P., Touzani K., Huang Y.Y., Nordheim A., Kandel E.R., Alarcon J.M., Weisberg S.P., Touzani K., Huang Y.Y., Nordheim A., Kandel E.R., Weisberg S.P., Touzani K., Huang Y.Y., Nordheim A., Kandel E.R., Touzani K., Huang Y.Y., Nordheim A., Kandel E.R., Huang Y.Y., Nordheim A., Kandel E.R., Nordheim A., Kandel E.R., Kandel E.R. A role in learning for SRF: Deletion in the adult forebrain disrupts LTD and the formation of an immediate memory of a novel context. Neuron. 2006;50:127–143. doi: 10.1016/j.neuron.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Gangopadhyay S.S., Takizawa N., Gallant C., Barber A.L., Je H.D., Smith T.C., Luna E.J., Morgan K.G., Takizawa N., Gallant C., Barber A.L., Je H.D., Smith T.C., Luna E.J., Morgan K.G., Gallant C., Barber A.L., Je H.D., Smith T.C., Luna E.J., Morgan K.G., Barber A.L., Je H.D., Smith T.C., Luna E.J., Morgan K.G., Je H.D., Smith T.C., Luna E.J., Morgan K.G., Smith T.C., Luna E.J., Morgan K.G., Luna E.J., Morgan K.G., Morgan K.G. Smooth muscle archvillin: A novel regulator of signaling and contractility in vascular smooth muscle. J. Cell Sci. 2004;117:5043–5057. doi: 10.1242/jcs.01378. [DOI] [PubMed] [Google Scholar]
- Gardiner K. Human genome organization. Curr. Opin. Genet. Dev. 1995;5:315–322. doi: 10.1016/0959-437x(95)80045-x. [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere C., Cavadore J.C., Blanchard J.M., Lamb N.J., Fernandez A., Cavadore J.C., Blanchard J.M., Lamb N.J., Fernandez A., Blanchard J.M., Lamb N.J., Fernandez A., Lamb N.J., Fernandez A., Fernandez A. p67SRF is a constitutive nuclear protein implicated in the modulation of genes required throughout the G1 period. Cell Regul. 1991;2:575–588. doi: 10.1091/mbc.2.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard D.S., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Klein S.L., Old S., Rasooly R., Good P., Old S., Rasooly R., Good P., Rasooly R., Good P., Good P., et al. The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera R.E., Shaw P.E., Nordheim A., Shaw P.E., Nordheim A., Nordheim A. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature. 1989;340:68–70. doi: 10.1038/340068a0. [DOI] [PubMed] [Google Scholar]
- Hosack D.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Dennis G., Jr., Sherman B.T., Lane H.C., Lempicki R.A., Sherman B.T., Lane H.C., Lempicki R.A., Lane H.C., Lempicki R.A., Lempicki R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kuhn R.M., Diekhans M., Haussler D., Diekhans M., Haussler D., Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Iyer V.R., Eisen M.B., Ross D.T., Schuler G., Moore T., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Eisen M.B., Ross D.T., Schuler G., Moore T., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Ross D.T., Schuler G., Moore T., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Schuler G., Moore T., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Moore T., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Staudt L.M., Hudson J., Jr., Boguski M.S., Hudson J., Jr., Boguski M.S., Boguski M.S., et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Iyer D., Chang D., Marx J., Wei L., Olson E.N., Parmacek M.S., Balasubramanyam A., Schwartz R.J., Chang D., Marx J., Wei L., Olson E.N., Parmacek M.S., Balasubramanyam A., Schwartz R.J., Marx J., Wei L., Olson E.N., Parmacek M.S., Balasubramanyam A., Schwartz R.J., Wei L., Olson E.N., Parmacek M.S., Balasubramanyam A., Schwartz R.J., Olson E.N., Parmacek M.S., Balasubramanyam A., Schwartz R.J., Parmacek M.S., Balasubramanyam A., Schwartz R.J., Balasubramanyam A., Schwartz R.J., Schwartz R.J. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc. Natl. Acad. Sci. 2006;103:4516–4521. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel C.V., Delorenzi M., Iseli C., Zhou D., Haudenschild C.D., Khrebtukova I., Kuznetsov D., Stevenson B.J., Strausberg R.L., Simpson A.J., Delorenzi M., Iseli C., Zhou D., Haudenschild C.D., Khrebtukova I., Kuznetsov D., Stevenson B.J., Strausberg R.L., Simpson A.J., Iseli C., Zhou D., Haudenschild C.D., Khrebtukova I., Kuznetsov D., Stevenson B.J., Strausberg R.L., Simpson A.J., Zhou D., Haudenschild C.D., Khrebtukova I., Kuznetsov D., Stevenson B.J., Strausberg R.L., Simpson A.J., Haudenschild C.D., Khrebtukova I., Kuznetsov D., Stevenson B.J., Strausberg R.L., Simpson A.J., Khrebtukova I., Kuznetsov D., Stevenson B.J., Strausberg R.L., Simpson A.J., Kuznetsov D., Stevenson B.J., Strausberg R.L., Simpson A.J., Stevenson B.J., Strausberg R.L., Simpson A.J., Strausberg R.L., Simpson A.J., Simpson A.J., et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15:1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Qu C., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Barrera L.O., Qu C., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Qu C., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Van Calcar S., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Trinklein N.D., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Cooper S.J., Luna R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Luna R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Glass C.K., Rosenfeld M.G., Myers R.M., Rosenfeld M.G., Myers R.M., Myers R.M., et al. Direct isolation and identification of promoters in the human genome. Genome Res. 2005;15:830–839. doi: 10.1101/gr.3430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll B., Kretz O., Fiedler C., Alberti S., Schutz G., Frotscher M., Nordheim A., Kretz O., Fiedler C., Alberti S., Schutz G., Frotscher M., Nordheim A., Fiedler C., Alberti S., Schutz G., Frotscher M., Nordheim A., Alberti S., Schutz G., Frotscher M., Nordheim A., Schutz G., Frotscher M., Nordheim A., Frotscher M., Nordheim A., Nordheim A. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat. Neurosci. 2006;9:195–204. doi: 10.1038/nn1627. [DOI] [PubMed] [Google Scholar]
- Lazo P.S., Dorfman K., Noguchi T., Mattei M.G., Bravo R., Dorfman K., Noguchi T., Mattei M.G., Bravo R., Noguchi T., Mattei M.G., Bravo R., Mattei M.G., Bravo R., Bravo R. Structure and mapping of the fosB gene. FosB downregulates the activity of the fosB promoter. Nucleic Acids Res. 1992;20:343–350. doi: 10.1093/nar/20.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Carter J., Gao X., Whitehead J., Tourtellotte W.G., Carter J., Gao X., Whitehead J., Tourtellotte W.G., Gao X., Whitehead J., Tourtellotte W.G., Whitehead J., Tourtellotte W.G., Tourtellotte W.G. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol. Cell. Biol. 2005;25:10286–10300. doi: 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Brutlag D.L., Liu J.S., Brutlag D.L., Liu J.S., Liu J.S. BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001;6:127–138. [PubMed] [Google Scholar]
- Manabe I., Owens G.K., Owens G.K. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ. Res. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- Miano J.M. Serum response factor: Toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- Miano J.M., Ramanan N., Georger M.A., de Mesy Bentley K.L., Emerson R.L., Balza R.O., Jr., Xiao Q., Weiler H., Ginty D.D., Misra R.P., Ramanan N., Georger M.A., de Mesy Bentley K.L., Emerson R.L., Balza R.O., Jr., Xiao Q., Weiler H., Ginty D.D., Misra R.P., Georger M.A., de Mesy Bentley K.L., Emerson R.L., Balza R.O., Jr., Xiao Q., Weiler H., Ginty D.D., Misra R.P., de Mesy Bentley K.L., Emerson R.L., Balza R.O., Jr., Xiao Q., Weiler H., Ginty D.D., Misra R.P., Emerson R.L., Balza R.O., Jr., Xiao Q., Weiler H., Ginty D.D., Misra R.P., Balza R.O., Jr., Xiao Q., Weiler H., Ginty D.D., Misra R.P., Xiao Q., Weiler H., Ginty D.D., Misra R.P., Weiler H., Ginty D.D., Misra R.P., Ginty D.D., Misra R.P., Misra R.P. Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. 2004;101:17132–17137. doi: 10.1073/pnas.0406041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T.J., Balza R., Jr., Xiao Q., Misra R.P., Balza R., Jr., Xiao Q., Misra R.P., Xiao Q., Misra R.P., Misra R.P. SRF-dependent gene expression in isolated cardiomyocytes: Regulation of genes involved in cardiac hypertrophy. J. Mol. Cell. Cardiol. 2005;39:479–489. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Ooe N., Saito K., Mikami N., Nakatuka I., Kaneko H., Saito K., Mikami N., Nakatuka I., Kaneko H., Mikami N., Nakatuka I., Kaneko H., Nakatuka I., Kaneko H., Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol. Cell. Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlakian A., Charvet C., Escoubet B., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Charvet C., Escoubet B., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Escoubet B., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Ludosky M.A., Paulin D., Daegelen D., Paulin D., Daegelen D., Daegelen D., et al. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 2005;112:2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778. [DOI] [PubMed] [Google Scholar]
- Perez-Albuerne E.D., Schatteman G., Sanders L.K., Nathans D., Schatteman G., Sanders L.K., Nathans D., Sanders L.K., Nathans D., Nathans D. Transcriptional regulatory elements downstream of the JunB gene. Proc. Natl. Acad. Sci. 1993;90:11960–11964. doi: 10.1073/pnas.90.24.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U., Schratt G., Dieterich C., Muller J.M., Galgoczy P., Engel F.B., Keating M.T., Gertler F., Schule R., Vingron M., Schratt G., Dieterich C., Muller J.M., Galgoczy P., Engel F.B., Keating M.T., Gertler F., Schule R., Vingron M., Dieterich C., Muller J.M., Galgoczy P., Engel F.B., Keating M.T., Gertler F., Schule R., Vingron M., Muller J.M., Galgoczy P., Engel F.B., Keating M.T., Gertler F., Schule R., Vingron M., Galgoczy P., Engel F.B., Keating M.T., Gertler F., Schule R., Vingron M., Engel F.B., Keating M.T., Gertler F., Schule R., Vingron M., Keating M.T., Gertler F., Schule R., Vingron M., Gertler F., Schule R., Vingron M., Schule R., Vingron M., Vingron M., et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell. 2004;16:867–880. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- Qureshi S.A., Cao X.M., Sukhatme V.P., Foster D.A., Cao X.M., Sukhatme V.P., Foster D.A., Sukhatme V.P., Foster D.A., Foster D.A. v-Src activates mitogen-responsive transcription factor Egr-1 via serum response elements. J. Biol. Chem. 1991;266:10802–10806. [PubMed] [Google Scholar]
- Rene C., Taulan M., Iral F., Doudement J., L’Honore A., Gerbon C., Demaille J., Claustres M., Romey M.C., Taulan M., Iral F., Doudement J., L’Honore A., Gerbon C., Demaille J., Claustres M., Romey M.C., Iral F., Doudement J., L’Honore A., Gerbon C., Demaille J., Claustres M., Romey M.C., Doudement J., L’Honore A., Gerbon C., Demaille J., Claustres M., Romey M.C., L’Honore A., Gerbon C., Demaille J., Claustres M., Romey M.C., Gerbon C., Demaille J., Claustres M., Romey M.C., Demaille J., Claustres M., Romey M.C., Claustres M., Romey M.C., Romey M.C. Binding of serum response factor to cystic fibrosis transmembrane conductance regulator CArG-like elements, as a new potential CFTR transcriptional regulation pathway. Nucleic Acids Res. 2005;33:5271–5290. doi: 10.1093/nar/gki837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T., Schreiber S.L., Mellor J., Kouzarides T., Mellor J., Kouzarides T., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Shaw P.E., Schroter H., Nordheim A., Schroter H., Nordheim A., Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989;56:563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- Small E.M., Krieg P.A., Krieg P.A. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev. Biol. 2003;261:116–131. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Chen G., Streb J.W., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Chen G., Streb J.W., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Streb J.W., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Yang Y., Stoeckert C.J., Jr., Miano J.M., Stoeckert C.J., Jr., Miano J.M., Miano J.M. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F.Y., Tang B.L., Tang B.L. APC/C regulation of axonal growth and synaptic functions in postmitotic neurons: The Liprin-αconnection. Cell. Mol. Life Sci. 2005;62:1571–1578. doi: 10.1007/s00018-005-5043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting H.J., Yeh S., Nishimura K., Chang C., Yeh S., Nishimura K., Chang C., Nishimura K., Chang C., Chang C. Supervillin associates with androgen receptor and modulates its transcriptional activity. Proc. Natl. Acad. Sci. 2002;99:661–666. doi: 10.1073/pnas.022469899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourtellotte W.G., Nagarajan R., Bartke A., Milbrandt J., Nagarajan R., Bartke A., Milbrandt J., Bartke A., Milbrandt J., Milbrandt J. Functional compensation by Egr4 in Egr1-dependent luteinizing hormone regulation and Leydig cell steroidogenesis. Mol. Cell. Biol. 2000;20:5261–5268. doi: 10.1128/mcb.20.14.5261-5268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Treisman R., Marais R., Wynne J., Marais R., Wynne J., Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N.D., Chen W.C., Kingston R.E., Myers R.M., Chen W.C., Kingston R.E., Myers R.M., Kingston R.E., Myers R.M., Myers R.M. Transcriptional regulation and binding of heat shock factor 1 and heat shock factor 2 to 32 human heat shock genes during thermal stress and differentiation. Cell Stress Chaperones. 2004;9:21–28. doi: 10.1379/481.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A., Duterque-Coquillaud M., Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Vlieghe D., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Wasserman W.W., van Roy F., Lenhard B., van Roy F., Lenhard B., Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.X., Garcia-Gras E., Wycuff D.R., Marriot S.J., Kadeer N., Yu W., Olson E.N., Garry D.J., Parmacek M.S., Schwartz R.J., Garcia-Gras E., Wycuff D.R., Marriot S.J., Kadeer N., Yu W., Olson E.N., Garry D.J., Parmacek M.S., Schwartz R.J., Wycuff D.R., Marriot S.J., Kadeer N., Yu W., Olson E.N., Garry D.J., Parmacek M.S., Schwartz R.J., Marriot S.J., Kadeer N., Yu W., Olson E.N., Garry D.J., Parmacek M.S., Schwartz R.J., Kadeer N., Yu W., Olson E.N., Garry D.J., Parmacek M.S., Schwartz R.J., Yu W., Olson E.N., Garry D.J., Parmacek M.S., Schwartz R.J., Olson E.N., Garry D.J., Parmacek M.S., Schwartz R.J., Garry D.J., Parmacek M.S., Schwartz R.J., Parmacek M.S., Schwartz R.J., Schwartz R.J. Identification of direct serum-response factor gene targets during Me2SO-induced P19 cardiac cell differentiation. J. Biol. Chem. 2005;280:19115–19126. doi: 10.1074/jbc.M413793200. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Wong W.H., Wong W.H. CisModule: De novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc. Natl. Acad. Sci. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]