Abstract
Recent studies suggest that surprisingly many mammalian genes have alternative promoters (APs); however, their biological roles, and the characteristics that distinguish them from single promoters (SPs), remain poorly understood. We constructed a large data set of evolutionarily conserved promoters, and used it to identify sequence features, functional associations, and expression patterns that differ by promoter type. The four promoter categories CpG-rich APs, CpG-poor APs, CpG-rich SPs, and CpG-poor SPs each show characteristic strengths and patterns of sequence conservation, frequencies of putative transcription-related motifs, and tissue and developmental stage expression preferences. APs display substantially higher sequence conservation than SPs and CpG-poor promoters than CpG-rich promoters. Among CpG-poor promoters, APs and SPs show sharply contrasting developmental stage preferences and TATA box frequencies. We developed a discriminator to computationally predict promoter type, verified its accuracy through experimental tests that incorporate a novel method for deconvolving mixed sequence traces, and used it to find several new APs. The discriminator predicts that almost half of all mammalian genes have evolutionarily conserved APs. This high frequency of APs, together with the strong purifying selection maintaining them, implies a crucial role in expanding the expression diversity of the mammalian genome.
Alternative promoters (APs) are important in regulating gene expression and generating protein diversity (Landry et al. 2003); however, the genome-wide prevalence of APs, their biological roles, and the regulatory mechanisms that govern their usage remain for the most part poorly understood. Recent large-scale studies that identify promoters via ChIP-chip analysis (Kim et al. 2005) or analysis of cDNA 5′-ends (Zavolan et al. 2002; Landry et al. 2003; Trinklein et al. 2003; Sharov et al. 2005; Carninci et al. 2006; Cooper et al. 2006; Kimura et al. 2006) suggest that 14%–58% of human genes may have APs. While such approaches are powerful, their rates of false positives (due to aberrant, likely nonfunctional mRNA transcripts [Sorek and Safer 2003; Dike et al. 2004]) and false negatives (due to incomplete sampling of tissues and developmental stages) remain uncertain. Consequently, it is useful to have additional approaches that incorporate more stringent criteria and to examine sequence characteristics that, in addition to illuminating molecular mechanisms, may permit computational prediction and directed experimental detection of additional promoters.
A powerful method to enrich for functional mRNA isoforms is to require their structures to be evolutionarily conserved (Sorek and Ast 2003; Sorek et al. 2004; Sugnet et al. 2004; Baek and Green 2005; Yeo et al. 2005). We used genomic alignments of full-length cDNAs and ESTs to construct a large data set of APs and single promoters (SPs) evolutionarily conserved in human and mouse, and used it to identify sequence features, functional associations, and expression patterns that differ by promoter type. We find that, relative to SPs, APs are in general accompanied by substantially higher sequence conservation upstream and downstream of the transcription start site; consistent with this, a number of short motifs are relatively overrepresented in APs. More broadly, subclassifying promoters by presence or absence of a CpG island, we find that each promoter category shows a characteristic strength and pattern of sequence conservation, frequencies of putative transcription-related motifs, and tissue and developmental-stage expression preferences. APs also display higher sequence conservation in the first intron; analysis of first exon splice donor sites suggests that this may reflect splicing regulatory signals. Expression analysis indicates that APs are more abundantly expressed in brain, heart, liver, and related tissues in embryonic and fetal stages, and gene ontology and TRANSFAC analyses confirm a broad linkage of APs to development. In contrast to CpG-poor APs, CpG-poor SPs show a strong bias toward post-embryonic expression and are more likely to have TATA boxes. CpG-rich SPs are more strongly associated with “housekeeping” genes than CpG-rich APs.
We used the characterized sequence differences between APs and SPs to construct a nonparametric approximate log-likelihood ratio discriminator that computationally predicts promoter type. Prediction accuracy was assessed by experiments that combined 5′ RACE with a novel sequencing trace analysis procedure that can identify a mixture of amplification products within a single trace without cloning. This permits a directed approach to identifying new instances of APs. Applying our discriminator to evolutionarily conserved promoters, we estimate that roughly 40%–50% of human and mouse genes have APs. This high frequency of APs, together with the strong purifying selection maintaining them, implies they play a crucial role in expanding the expression diversity of the mammalian genome.
Results and Discussion
Identification of evolutionarily conserved APs and SPs
Using genomic alignments of cDNAs and ESTs and the transcription start-site database DBTSS (Suzuki et al. 2002), we identified 12,025 promoter regions that are evolutionarily conserved between mouse and human. Of these, 1080 could be confidently assigned as alternative and 3109 as single. We focus on the “mutually exclusive first exon” category of APs (Supplemental Table S1), which constitute roughly 85% of all mammalian APs (Kimura et al. 2006), and exclude a small, previously identified subclass of APs that originate from duplication of first exons (Zhang et al. 2004), as these tend to have characteristics atypical of most APs (Zhang et al. 2004; Supplemental Methods). We additionally classified each promoter as CpG-rich if the flanking genomic region significantly overlaps one or more CpG islands, or as CpG-poor otherwise.
Sequence conservation in APs and SPs
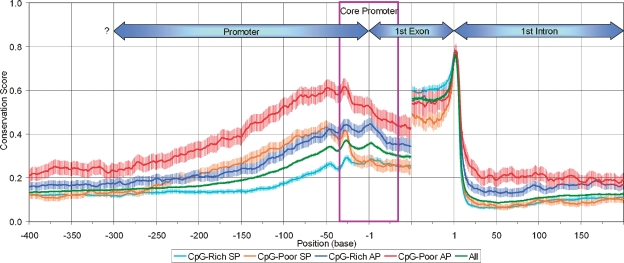
Studies of evolutionarily conserved alternative splicing have found higher sequence conservation in alternatively spliced exons and their flanking introns relative to constitutively spliced exons, likely reflecting strong purifying selection on splicing regulatory signals (Sorek and Ast 2003; Sugnet et al. 2004; Baek and Green 2005). We hypothesized that APs may similarly be enriched, relative to SPs, for transcriptional regulatory signals under purifying selection. Sequence conservation in APs is indeed substantially higher than in SPs (Fig. 1) over a region of several hundred bases that includes both the “core promoter” (Butler and Kadonaga 2002; Cooper et al. 2006) (where the pre-initiation complex binds, extending ∼35 bp upstream and ∼35 bp downstream of the transcription start site [TSS]) and the regulatory or control region upstream of the core promoter (where many activator and repressor proteins bind). Within each promoter type, CpG-poor promoters tend to be more highly conserved than CpG-rich ones, consistent with Carninci et al. (2006). Strong purifying selection implies in particular that most evolutionarily conserved APs are functionally important to the organism.
Figure 1.
Average sequence conservation score (using UCSC 17-vertebrate alignment [Siepel et al. 2005]) at each nucleotide position in promoter, first exon, and first intron regions for each promoter type. Exon scores were computed for 50 exonic bases (or half the exon size, for exons <100 bp) from the 5′ or 3′ exon end. APs show on average higher conservation than SPs, and CpG-poor promoters higher conservation than CpG-rich promoters, over several hundred bases in the promoter and first intron, and in the 5′ half of first exon. Conservation patterns in the 3′ half of first exon largely reflect protein-coding constraints. The difference in sequence conservation becomes negligible further upstream of the TSS (which effectively eliminates the possibility that sequence-conservation differences near the TSS reflect large-scale variation in mutational rate rather than purifying selection). Uncertain TSS placement due to variable start sites, common in CpG-rich promoters, may cause some smearing of the conservation pattern for such promoters, but cannot by itself cause the overall weaker pattern. The boxed core promoter is bases −35 to +35 relative to the TSS.
Strikingly, each class—CpG-rich AP, CpG-poor AP, CpG-rich SP, and CpG-poor SP—has a characteristic strength and pattern of conservation (Fig. 1). For CpG-rich promoters, the most highly conserved region is the core promoter, with conservation dropping rapidly upstream from it; in CpG-poor promoters, the most highly conserved region is located just 5′ of the core promoter, and the upstream decline in conservation is more gradual. CpG-poor SPs show a hybrid conservation pattern, similar to CpG-rich SPs in most of the core, but similar to CpG-rich APs at the 5′ end of the core (where the TATA box is located when present) and further upstream. CpG-poor APs show the highest conservation throughout the core and upstream control regions. Variation in conservation levels within the core promoter may be related to recent observations that components of the preinitiation complex (particularly TFIID) can vary by promoter and cell type (Hochheimer and Tjian 2003).
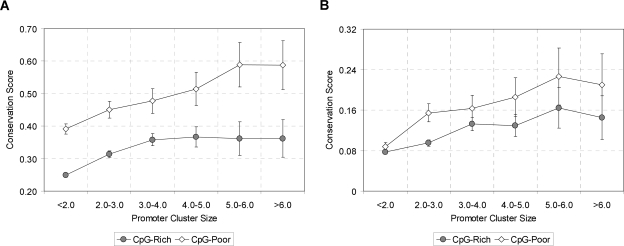
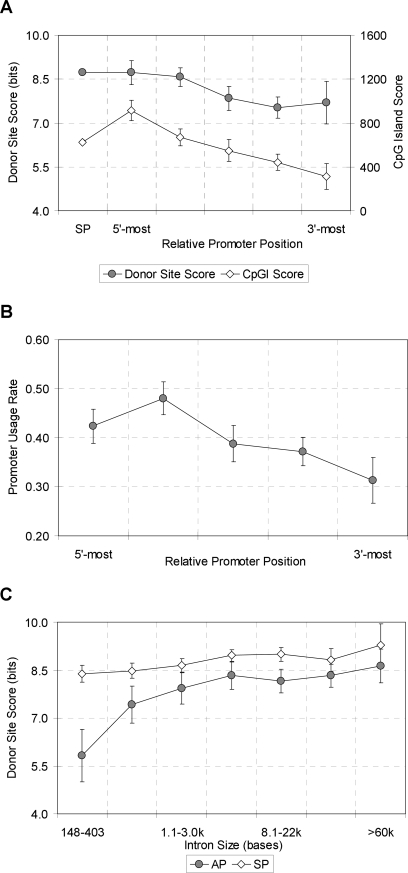
An interesting question is whether the higher conservation of APs reflects “intrinsic” signals (those needed to specify the particular tissues and developmental stages in which the promoter must be expressed or repressed) or “competitive” signals (those that regulate choice among different promoters in the same gene). To investigate this, we examined sequence conservation as a function of the number of promoters in a gene. Promoters in larger clusters tend to have higher sequence conservation (Fig. 2A), suggesting that some of the higher conservation reflects competitive signals. Upstream promoters are in general more highly expressed, and more likely to be CpG-rich, than downstream promoters (Fig. 3A,B).
Figure 2.
Sequence conservation in bases 16–100 upstream of TSS (A) and bases 16–100 in first intron (B) as a function of promoter cluster size (estimated number of promoters in gene, averaged between human and mouse) in 12,025 conserved promoters. Cluster size <2.0 includes SPs.
Figure 3.
Donor site score (A), CpG island score (A), and promoter usage rate (B) as a function of relative promoter position, and donor-site score as a function of intron size (C). In A and B, AP cases with the promoter cluster size of ≥2 in both human and mouse were analyzed.
Putative transcription-factor binding sites in APs and SPs
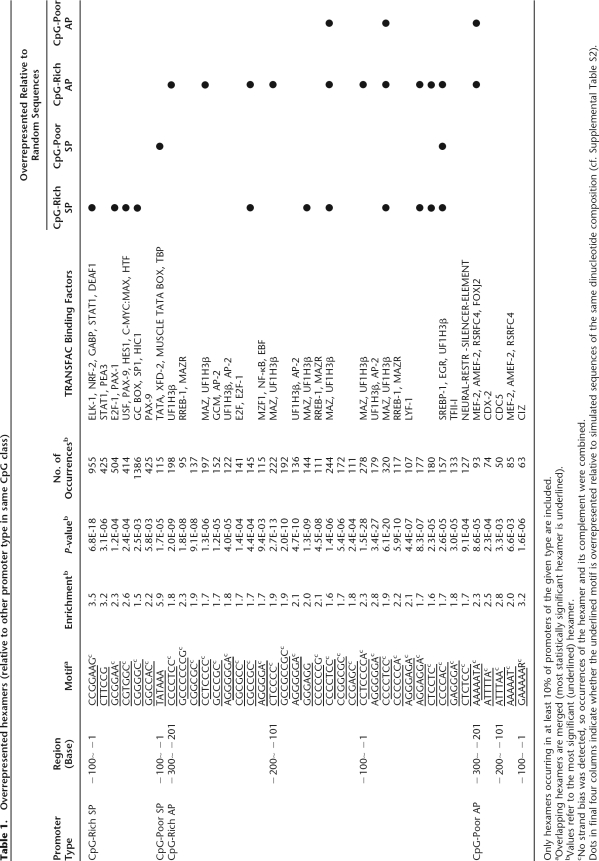
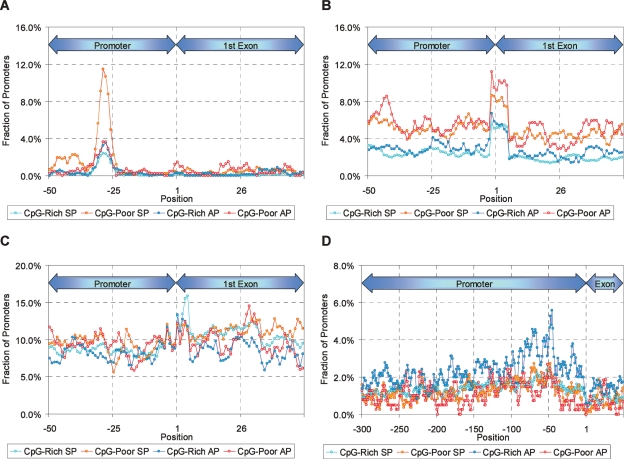
Since these conservation patterns likely reflect transcription-factor binding sites, we looked for hexamer motifs relatively overrepresented in APs or SPs within a CpG class, and searched them against known transcription-factor binding sites (Table 1). Several hexamers are overrepresented specifically in APs. Most of these appear related to binding sites for known tissue-specific transcriptional activators or context-dependent activator/repressors. An interesting example is the site for CTCF (Filippova et al. 1996; Kanduri et al. 2000) (overrepresented in CpG-rich APs), which is believed to play a role in delineating repressed domains via insulators and may help repress some APs. A positional analysis shows that this motif shows a strong bias toward the region just upstream of the core promoter (Fig. 4D). We also identified hexamers overrepresented relative to randomly generated sequences having the same dinucleotide composition (Supplemental Table S2). Many, but not all of the motifs listed in Table 1 are also overrepresented compared with random sequence.
Table 1.
Overrepresented hexamers (relative to other promoter type in same CpG class)
Only hexamers occurring in at least 10% of promoters of the given type are included.
aOverlapping hexamers are merged (most statistically significant hexamer is underlined).
bValues refer to the most significant (underlined) hexamer.
cNo strand bias was detected, so occurrences of the hexamer and its complement were combined.
Dots in final four columns indicate whether the underlined motif is overrepresented relative to simulated sequences of the same dinucleotide composition (cf. Supplemental Table S2).
Figure 4.
Positional distribution of TATA box motif, TATA (Butler and Kadonaga 2002) (A), initiator motif, YYANWYY (Butler and Kadonaga 2002) (B), downstream promoter element motif, RGWYV (Butler and Kadonaga 2002) (C), and CTCF binding site (combined count of CCCTCC [Filippova et al. 1996] and its complement) (D). For each nucleotide position, the number of promoters of a given type having a motif copy spanning that position is divided by the total number of promoters of that type. We used known promoters filtered as described in the Methods (Motif discovery and TRANSFAC search).
Several overrepresented motifs do not correspond to known binding sites, and a number of motifs are underrepresented in APs relative to SPs. Among CpG-poor promoters, the motif TATAAA is almost sixfold more common in SPs than in APs, raising the possibility that strong TATA boxes are incompatible with APs. Positional analysis of TATA and two other commonly found core promoter motifs (INR and DPE) indicates differing frequencies among the promoter types (Fig. 4A,B,C).
Possible splicing regulatory signals in first exon splice donor site in AP genes
Expression of alternate transcripts places unusual constraints not only on transcription initiation but also on transcript splicing, since transcripts from upstream promoters contain, but must not utilize, the donor splice sites of downstream promoter first exons. Such donor sites in fact are weaker than those of upstream promoter first exons (Fig. 3A). This pattern could reflect selection to avoid use of these sites in transcripts from the upstream promoter, or (since the first intron in the upstream transcript is in general larger) it could simply reflect selection for larger introns to have stronger splice sites. To distinguish these possibilities, we examined first donor site scores as a function of intron size (Fig. 3C). Although donor-site scores for both APs and SPs are positively correlated with intron size, AP donor sites are overall significantly weaker than SP donor sites for comparable intron sizes, suggesting that selection to avoid mis-splicing of the upstream promoter transcript does play a role. Note that APs also display higher conservation than SPs at the 5′ end of the first intron (Fig. 1), which may reflect the presence of splicing regulatory signals (enhancers and/or silencers) associated with the splicing constraints, although they could also reflect transcription-related signals in the first intron. A trend of higher conservation with more promoters is observed here as well (Fig. 2B).
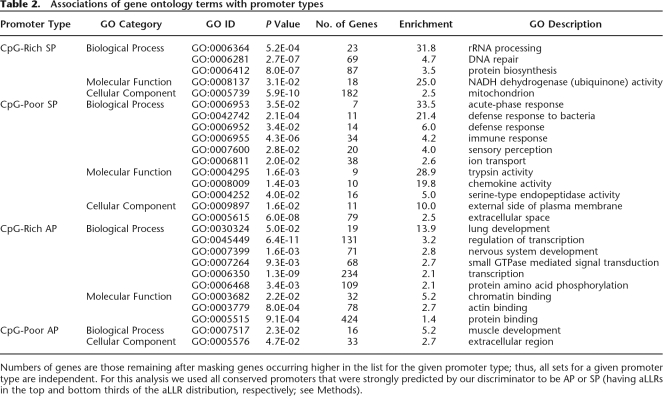
Functional and expression associations
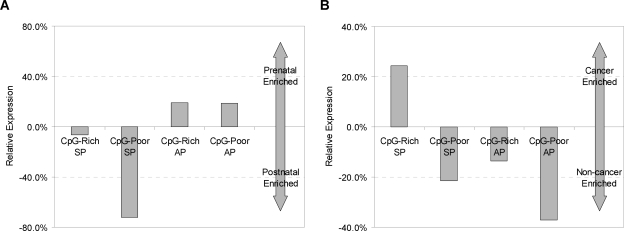
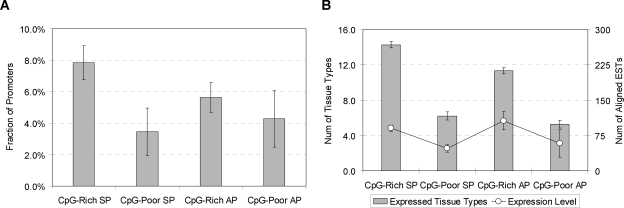
To illuminate the biological roles of the different promoter types, we tested for associations of promoter type with biological functions, and developmental and tissue expression specificity (Fig. 5; Table 2; Supplemental Table S3). CpG-rich SPs are linked to “housekeeping” functions required in most cell types, and show the strongest association with broadly expressed genes (Bird 1984; Ponger et al. 2001; Schug et al. 2005) (Fig. 6). Their overall lower sequence conservation (Fig. 1) is consistent with the idea that most such genes do not require elaborate expression regulation. CpG-rich SPs are also more frequently expressed in cancer cells (Fig. 5B), perhaps because a relative absence of repressive regulatory signals makes them more vulnerable to transcription initiation by the aberrantly activated transcription factors often found in cancer cells (Darnell Jr. 2002; Robertson 2005).
Figure 5.
Relative expression in early vs. late developmental stages (A) and in cancer vs. non-cancer cells (B) by promoter type. Expression level Ec in category c for each promoter type was measured by counting the number of aligned ESTs for that promoter type, and dividing by the sum over all four promoter types. Relative expression was computed by (EPrenatal-EPostnatal)/(EPostnatal) in A and (ECancer-ENon-cancer)/(ENon-cancer) in B. We used all conserved promoters that were strongly predicted by our discriminator to be AP or SP (having aLLRs in the top and bottom quartiles of the aLLR distribution, respectively).
Table 2.
Associations of gene ontology terms with promoter types
Numbers of genes are those remaining after masking genes occurring higher in the list for the given promoter type; thus, all sets for a given promoter type are independent. For this analysis we used all conserved promoters that were strongly predicted by our discriminator to be AP or SP (having aLLRs in the top and bottom thirds of the aLLR distribution, respectively; see Methods).
Figure 6.
Relative prevalence of putative housekeeping promoters (A), and the number of expressed tissue types and expression level (in noncancer cells) per promoter (B) by promoter type. CpG-rich SPs are enriched for putative housekeeping promoters (A) and tend to be expressed more broadly (B). We used all conserved promoters that were strongly predicted by our discriminator to be AP or SP (having aLLRs in the top and bottom quartiles of the aLLR distribution, respectively).
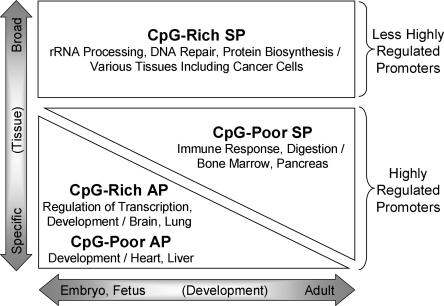
In contrast, APs are overrepresented among genes involved in transcription regulation and development, while CpG-poor SPs are overrepresented among immune response genes. The higher sequence conservation of these promoters likely reflects more narrowly defined spatial and temporal windows for expression that entail additional regulatory signals. Each promoter type shows significant tissue expression biases (Supplemental Table S3), which, in general, are consistent with the functional biases, and which tend to be stronger for the CpG-poor promoters (both SPs and APs) than for the CpG-rich ones. At a broad level, promoter types also show different developmental stage associations (Fig. 5A): relative to CpG-rich SP promoters, which tend to be expressed at all stages, CpG-poor SPs show a strong bias toward postnatal expression, whereas APs show a weak bias toward prenatal expression, in accord with the functional and tissue-association analyses. The functional and expression-association differences among the four promoter types are summarized in Figure 7.
Figure 7.
Summary of functions and expression specificity of different mammalian promoter types.
Predictive discovery of APs
To help in finding novel instances of APs, we developed a nonparametric, approximate log-likelihood ratio discriminator (aLLR), similar to one we previously used to distinguish alternatively and constitutively spliced exons (Baek and Green 2005), to predict, for a known conserved promoter, whether it is an AP or a SP. The discriminator uses sequence conservation, donor site score, expression level, exon size, and frequencies of short motifs as distinguishing criteria. (We only attempt to predict the mutually exclusive form of AP.) In experimental tests using oligo-capping RACE together with a novel method for deconvolving mixed sequence traces (see Methods), 34 of 46 (74%) predicted SPs showed evidence for a single promoter, while 28 of 44 (64%) predicted APs showed evidence for multiple promoters, for an overall accuracy of 69%. (In controls using known SPs and known APs, 21 of 24 known SPs and 20 of 24 known APs showed evidence for a single and multiple promoters, respectively, for an overall accuracy of 85%.) Our computational and experimental approach may be of value in identifying AP cases where high-throughput experimental methods have failed due to very low expression level or expression patterns highly specific to particular tissue types and/or developmental stages (Fig. 8).
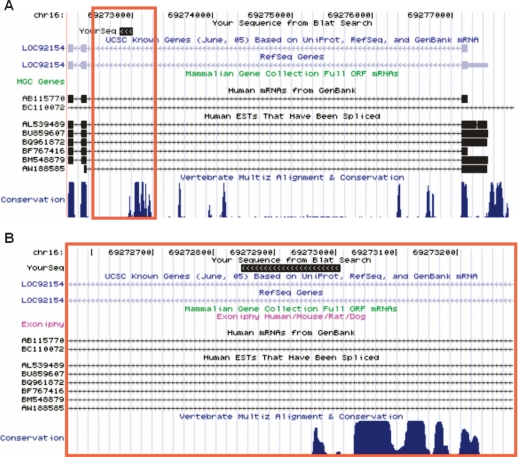
Figure 8.
A novel AP on human chromosome 16 discovered by our computational prediction and experimental verification. B is an expanded view of the red rectangle region in A (images captured from the UCSC Genome Browser). A conserved promoter with a representative isoform NM_138383 (shown as LOC92154 above) was predicted to be an AP with an approximate log-likelihood ratio of +21.2, although aligned cDNAs and ESTs only supported a single promoter (BC110072 lacks any exonic overlap with this gene, and thus, is unlikely to represent an upstream AP). The top black box denoted by “YourSeq” represents the genomic alignment of a first exonic sequence found in our oligo-capped RACE reads. In B, the bottom rows indicate highly conserved blocks in the region immediately upstream of the detected first exon. The direction of transcription is from right to left.
Applying our discriminator to the full set of 12,025 evolutionarily conserved promoters, we predict that roughly 40%–50% of genes have alternative promoters (Supplemental Fig. S1; Supplemental Table S4). This may be an underestimate because it does not include APs originating from duplicated first exons or APs not of the mutually exclusive type, but it is toward the high end of the range of other recent studies (Zavolan et al. 2002; Landry et al. 2003; Trinklein et al. 2003; Kim et al. 2005; Sharov et al. 2005; Carninci et al. 2006; Cooper et al. 2006; Kimura et al. 2006). Roughly 55% of mammalian genes are predicted to have APs or CpG-poor SPs, while the remaining 45% are predicted to have less highly regulated CpG-rich SPs. It should be emphasized that all of these estimates are approximate as a result of the uncertainties mentioned above and others inherent in our experimental tests (e.g., there is an unknown false negative rate in identifying APs associated with the fact that we have not sampled all tissues and developmental stages). Nonetheless, this apparent high frequency of alternative promoters, together with the strong purifying selection maintaining them (Fig. 1), suggests that they play a crucial role in expanding the expression diversity of the mammalian genome.
Methods
(See Supplemental Methods for details regarding the following: identification of conserved promoters, putative housekeeping promoters, CpG islands and recently duplicated first exons, splice site scoring, and GO and tissue/development association analyses.)
Promoter usage rate, promoter cluster size, and relative promoter position
Promoter usage rate (PUR, with values between 0 and 1) was determined as follows. For each representative isoform p, we computed the number Np of cDNAs and ESTs that have the same genomic orientation as the gene, which overlap the first exon of p, and whose 5′ ends are within 500 bases of the 5′ end of p. The promoter usage rate of p is then Np/N, where N is the sum of the Nq over all representative isoforms q in the gene plus the number of cDNAs and ESTs from the gene whose 5′ ends are located ≥500 bases upstream from the 5′ end of the upstream-most representative isoform (since these may represent potential upstream undetected promoters). Since representative isoforms were not required to be conserved in human and mouse, some of them may represent transcriptional “noise,” and consequently, the promoter usage rate is likely a rough estimate of promoter usage relative to others in the same gene. A similar procedure was carried out for mouse genes.
The promoter cluster size of a gene is defined to be the number of representative isoforms it has.
Relative promoter position of an alternative promoter P is defined to be (2RP -1)/2N, where N is the promoter cluster size of the gene and RP is the positional rank of P (1 for the 5′-most promoter in a gene, N for the 3′-most). The ith column (i = 2, . . . , 6) in Figure 3A and (i = 1, . . . , 5) in Figure 3B includes AP cases with relative promoter positions, averaged between human and mouse, of [0.2(i-2), 0.2(i-1)] and [0.2(i-1), 0.2i], respectively.
Nonparametric approximate log-likelihood ratio (aLLR) discriminator
We attempt to predict the type of promoter (AP or SP) based on the following characteristics: average sequence conservation scores in 12 segments, donor-site score, exon size, expression level, and frequencies of overrepresented short motifs (monomer to hexamer) in bases 1–300 upstream of the TSS, in the first exon, and in bases 1–300 of the first intron. We developed one discriminator for CpG-rich promoters and a separate one for CpG-poor promoters.
Overrepresented short motifs for APs or SPs in a given CpG class (CpG-rich or CpG-poor) were identified using a Yates corrected 2 × 2 χ2 contingency table test with columns representing AP vs. SP, and rows representing the searched motif vs. all other motifs (combined) of the same length. Table entries represent (nonoverlapping) motif counts from promoters in the given CpG class. Sequence conservation scores were based on conservation scores computed from alignments of 16 vertebrate genomes with human (Siepel et al. 2005) from the UCSC Genome Bioinformatics Site. For each human promoter, we defined 12 segments as follows: promoter segments consisting of bases 16–50, 51–100, 101–200, and 201–400 upstream of the transcription start site; the transcription start site (bases −15 to +5); 5′ and 3′ halves of the first exon; the first exon donor site (5 exonic + 15 intronic bases); and first intron bases 16–50, 51–100, 101–200, and 201–400. For each segment, conservation scores for each base were added and divided by the total number of bases in the segment. We computed sequence conservation for mouse promoters in a similar fashion and took the mouse–human average.
Our discriminator constructs a nonparametric approximate log-likelihood ratio for each promoter, and is motivated by the Neyman-Pearson lemma (Neyman and Pearson 1933), which implies that log-likelihood ratios provide optimal discrimination. A similar approach was successfully used to discriminate alternatively spliced from constitutively spliced exons, and methodological details can be found in our previous study (Baek and Green 2005).
To validate discriminatory power, we partitioned our set of known APs and SPs into a randomly chosen “training” subset containing 80% of the promoters, and a test set consisting of the other 20%. We used accuracies estimated from leave-one-out cross-validation on the training set, using the 15 parameters specified above together with the frequencies of overrepresented short motifs of various sizes, to find the most discriminatory motif size. We then measured prediction accuracy using the 20% test set (Supplemental Table S5).
As another test of the validity of the aLLR, we looked at promoter usage rate (PUR) as a function of aLLR for our entire data set of 12,025 conserved promoters. For this purpose, we used a version of the aLLR discriminator that does not use expression data. If the discriminator is accurate, we expect that the average PUR in predicted SPs should be close to 1.0, while the average PUR in predicted APs should be <1.0. Supplemental Figure S2 confirms that promoters with positive aLLRs (i.e., predicted APs) tend to have lower PURs, while promoters with negative aLLRs (predicted SPs) tend to have PURs close to 1.0.
For genome-wide prediction we randomly partitioned the entire conserved promoter data set into 20 subgroups, and computed aLLR(P) for the promoters P in each subgroup using the known APs and SPs in the remaining 19 subgroups, with optimal motif size identified using leave-one-out cross-validation on a randomly chosen 95% of known promoters (Supplemental Table S5).
Motif discovery and TRANSFAC search
Our data set of conserved APs and SPs likely contains some functionally single promoters that are misclassified as APs because of aberrant transcripts that have occurred in both mouse and human, or alternative promoters misclassified as SPs because there is insufficient data to reveal multiple promoters. For purposes of the motif search, we considered conserved APs with positive aLLR(P) and conserved SPs with negative aLLR(P) to be the most likely to be correctly classified, using a modified version of aLLR(P) that omits overrepresented motif information. A total of 43% of APs and 27% of SPs failed this criterion and were not used for the motif search.
Each putative promoter was partitioned into three segments consisting of bases 1–100, 101–200, and 201–300 upstream of the TSS. For each of the 46 = 4096 hexamers, we first tested for strand bias by comparing the frequency of the hexamer with that of its complement via a χ2 test, in each promoter class; when there was no evidence of strand bias, counts of the motif and its complement were combined. We then counted the hexamer frequency (ignoring overlaps) in APs and SPs in each CpG class, and a χ2 P-value was determined as described in “Nonparametric approximate log-likelihood ratio (aLLR) discriminator,” above. Hexamers were then sorted by P-value. If a hexamer had a perfect 5-base overlap with a hexamer having a lower P-value, it was clustered with that hexamer.
We did a similar analysis to identify hexamers overrepresented with respect to random sequence. For each putative promoter, we determined the frequencies of dinucleotides within the region consisting of bases 1–300 upstream of the TSS, and then used a Markov chain to generate a random sequence of the same length having similar overall dinucleotide composition. The above procedures then were repeated, comparing now the set of actual promoter sequences from a given class to the set of corresponding simulated sequences.
We obtained the consensus sequences of 569 vertebrate transcription-factor binding sites from the TRANSFAC database (Knuppel et al. 1994) Release 10.1 and found TRANSFAC consensus sequences matching representative hexamers with no gaps or mismatches, allowing up to two twofold-degenerate or one fourfold-degenerate TRANSFAC residues in the alignment. For a given hexamer, we report all TRANSFAC matches having maximal alignment score (defined by assigning a match to an unambiguous TRANSFAC residue a score of 1.0, a twofold-degenerate TRANSFAC residue a score of 0.5, and 0.0 otherwise).
Statistical analyses
Where relevant, a single value for each characteristic (e.g., conservation score, exon size) was obtained by averaging the human and mouse values. All P-values are Bonferroni-corrected for multiple testing. Vertical error bars in figures represent 95% confidence intervals for the population mean, computed assuming normality.
Choice of genes to experimentally test
We chose 94 predicted AP and SP candidates (47 of each type) for experimental testing as follows. We first identified conserved CpG-rich promoters whose aLLR values lay in the top or bottom quartiles of the aLLR distribution; to facilitate the experimental tests, we also required the representative isoform to have four or more exons, and that its four 5′-most exons each have size ≥50 bases and total size ≤600 bases. From this subset, we randomly chose 24 predicted APs and 24 predicted SPs. We chose 23 predicted CpG-poor APs and 23 predicted CpG-poor SPs in a similar manner. RACE sequencing (as described below) was successful for 46 predicted SPs and 44 predicted APs.
RNA samples
A master panel (Catalog #636643) of human total RNA was purchased from Clontech Laboratories, Inc. The panel consisted of RNA from 20 different tissues, which were pooled into groups of four as follows:
Group 1: brain, adrenal gland, bone marrow, and heart.
Group 2: liver, kidney, lung, and placenta.
Group 3: testis, prostate, salivary gland, and skeletal muscle.
Group 4: fetal brain, thymus, thyroid gland, and trachea.
Group 5: fetal liver, uterus, colon with mucosa, and spinal cord.
In addition, human total RNA from fetal embryos at 6-, 9- and 12-wk of development (Catalog #011-E-6W-TR, 011-E-9W-TR and 011-E-12W-TR) was purchased from Virogen and pooled together to form RNA Group 6.
RACE-ready cDNA
Full-length cDNA was generated using an oligo-capping technique (Maruyama and Sugano 1994): GeneRacer (Catalog # L1502–01) from Invitrogen essentially as outlined in the kit protocol. Briefly, the six RNA pools were treated with calf intestinal phosphatase to remove 5′ phosphates from truncated RNA but not from full-length capped RNA, and then treated with tobacco acid pyrophosphatase to remove the 5′ cap structure, leaving a 5′ phosphate group available for ligation. A GeneRacer RNA primer (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGA AGGAGUAGAAA-3′) was ligated to the full-length, decapped RNA using T4 RNA ligase. The ligated RNA pools were reverse transcribed using SuperScript III at 50°C for 45 min. The cDNA samples were treated with RNAse H at 37°C for 20 min prior to PCR amplification.
(See Supplemental Methods for details regarding primer design, PCR amplification, and sequencing).
Merged base reads
A technological innovation in this experimental approach is analysis of mixed sequence traces, which allows detection of multiple cDNA isoforms in a single sequencing reaction without cloning (B. Ewing, C. Davis, and P. Green, in prep.). Phred (Ewing and Green 1998; Ewing et al. 1998) identifies a called and (in most cases) an uncalled base at each predicted peak location, and writes these to a .poly output file when it is run with the -d or -dd option. In general, the called base corresponds to the largest peak near the predicted peak location, while the uncalled base represents the next largest peak. We form a “merged base read” using IUPAC ambiguity codes to simultaneously represent the called and uncalled bases at each position, and store this in a FASTA file.
Analysis of sequencing results
We aligned each merged base read to the complement of the nested 5′ Gene Racer primer sequence and to the complement of an extensive genomic sequence (including 200 kb upstream of the first exon, the first four exons, and the first three introns) surrounding the first exon of the target gene, using the Smith-Waterman algorithm (Smith and Waterman 1981) with a score matrix that assigns +2 to a pair of matching unambiguous nucleotides, +1 to a match of a nucleotide to a twofold degenerate ambiguity code, −4 to mismatches, −6 to gap initiation, and −5 to gap extension.
If the highest-scoring alignment to the primer sequence had a score of at least 12, the alignment was “subtracted” from the merged base read as described below, and the subtracted read searched against the primer sequence again; this process was iterated until the alignment score dropped below 12. A similar alignment and subtraction process was carried out for the genomic sequence, using a score cutoff of 40.
For sequence subtraction, a twofold degenerate residue was converted into the corresponding unambiguous residue and a once-converted residue into a masked residue, “X.” For instance, a twofold degenerate residue, “Y” was converted into “C” or “T” when the first aligned residue was “T” or “C,” respectively; if the first aligned residue was neither “T” nor “C,” “Y” was kept. After the second alignment, it was converted to “X” without regard to the aligned residue. Thus, each residue in a merged base read was allowed to match up to two residues of the searched sequence. This method is expected to effectively detect a mixture of up to two cDNA isoforms in a single sequencing reaction. In cases where the called residue was an unambiguous nucleotide, the unambiguous nucleotide was used both in the first and the second alignments.
For each target gene, the merged base reads from RACE of all six tissue groups (“RNA samples,” above) were compared with the primer sequence and the genomic sequence of the target gene. First exons were identified as genomic segments that matched the read, such that the matching read segment was adjacent to, and upstream of, a segment of the read that matched the RACE primer. If a single first exon was identified in this way, it was considered to be experimental evidence for a SP; if at least two nonoverlapping first exons whose 5′ ends differed by ≥500 bases were found, they were considered to be experimental evidence for an AP.
Data availability
The data set of conserved APs and SPs is shown in Supplemental Table S6. Sequence traces from our experimental tests are available from the NCBI Trace Archive, TI numbers 1540412808–1540413659, and the corresponding “merged base” reads are given in FASTA format as Supplemental Table S7.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5872707
References
- Baek D., Green P., Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.P. DNA methylation–how important in gene control? Nature. 1984;307:503–504. doi: 10.1038/307503a0. [DOI] [PubMed] [Google Scholar]
- Butler J.E., Kadonaga J.T., Kadonaga J.T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes & Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Taylor M.S., Engstrom P.G., Frith M.C., Engstrom P.G., Frith M.C., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cooper S.J., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Anton E.D., Nguyen L., Myers R.M., Nguyen L., Myers R.M., Myers R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E., Jr. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- Dike S., Balija V.S., Nascimento L.U., Xuan Z., Ou J., Zutavern T., Palmer L.E., Hannon G., Zhang M.Q., McCombie W.R., Balija V.S., Nascimento L.U., Xuan Z., Ou J., Zutavern T., Palmer L.E., Hannon G., Zhang M.Q., McCombie W.R., Nascimento L.U., Xuan Z., Ou J., Zutavern T., Palmer L.E., Hannon G., Zhang M.Q., McCombie W.R., Xuan Z., Ou J., Zutavern T., Palmer L.E., Hannon G., Zhang M.Q., McCombie W.R., Ou J., Zutavern T., Palmer L.E., Hannon G., Zhang M.Q., McCombie W.R., Zutavern T., Palmer L.E., Hannon G., Zhang M.Q., McCombie W.R., Palmer L.E., Hannon G., Zhang M.Q., McCombie W.R., Hannon G., Zhang M.Q., McCombie W.R., Zhang M.Q., McCombie W.R., McCombie W.R. The mouse genome: Experimental examination of gene predictions and transcriptional start sites. Genome Res. 2004;14:2424–2429. doi: 10.1101/gr.3158304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P., Hillier L., Wendl M.C., Green P., Wendl M.C., Green P., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V., Neiman P.E., Collins S.J., Lobanenkov V.V., Collins S.J., Lobanenkov V.V., Lobanenkov V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A., Tjian R., Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes & Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V., Wolffe A., Ohlsson R., Lobanenkov V.V., Ohlsson R., Lobanenkov V.V., Lobanenkov V.V. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., Sekine M., Tsuritani K., Wakaguri H., Tsuritani K., Wakaguri H., Wakaguri H., et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuppel R., Dietze P., Lehnberg W., Frech K., Wingender E., Dietze P., Lehnberg W., Frech K., Wingender E., Lehnberg W., Frech K., Wingender E., Frech K., Wingender E., Wingender E. TRANSFAC retrieval program: A network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol. 1994;1:191–198. doi: 10.1089/cmb.1994.1.191. [DOI] [PubMed] [Google Scholar]
- Landry J.R., Mager D.L., Wilhelm B.T., Mager D.L., Wilhelm B.T., Wilhelm B.T. Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Sugano S., Sugano S. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- Neyman J., Pearson E., Pearson E. On the problem of the most efficient tests of statistical hypotheses. Philos. Trans. R. Soc. Lond. Ser. A. 1933;231:289–337. [Google Scholar]
- Ponger L., Duret L., Mouchiroud D., Duret L., Mouchiroud D., Mouchiroud D. Determinants of CpG islands: Expression in early embryo and isochore structure. Genome Res. 2001;11:1854–1860. doi: 10.1101/gr.174501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Salbaum J.M., Bucan M., Stoeckert C.J., Jr., Bucan M., Stoeckert C.J., Jr., Stoeckert C.J., Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov A.A., Dudekula D.B., Ko M.S., Dudekula D.B., Ko M.S., Ko M.S. Genome-wide assembly and analysis of alternative transcripts in mouse. Genome Res. 2005;15:748–754. doi: 10.1101/gr.3269805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Waterman M.S., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Sorek R., Ast G., Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Safer H.M., Safer H.M. A novel algorithm for computational identification of contaminated EST libraries. Nucleic Acids Res. 2003;31:1067–1074. doi: 10.1093/nar/gkg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Shamir R., Ast G., Shamir R., Ast G., Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Sugnet C.W., Kent W.J., Ares M., Jr., Haussler D., Kent W.J., Ares M., Jr., Haussler D., Ares M., Jr., Haussler D., Haussler D. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yamashita R., Nakai K., Sugano S., Yamashita R., Nakai K., Sugano S., Nakai K., Sugano S., Sugano S. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–331. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein N.D., Aldred S.J., Saldanha A.J., Myers R.M., Aldred S.J., Saldanha A.J., Myers R.M., Saldanha A.J., Myers R.M., Myers R.M. Identification and functional analysis of human transcriptional promoters. Genome Res. 2003;13:308–312. doi: 10.1101/gr.794803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Van Nostrand E., Holste D., Poggio T., Burge C.B., Van Nostrand E., Holste D., Poggio T., Burge C.B., Holste D., Poggio T., Burge C.B., Poggio T., Burge C.B., Burge C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavolan M., van Nimwegen E., Gaasterland T., van Nimwegen E., Gaasterland T., Gaasterland T. Splice variation in mouse full-length cDNAs identified by mapping to the mouse genome. Genome Res. 2002;12:1377–1385. doi: 10.1101/gr.191702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Haws P., Wu Q., Haws P., Wu Q., Wu Q. Multiple variable first exons: A mechanism for cell- and tissue-specific gene regulation. Genome Res. 2004;14:79–89. doi: 10.1101/gr.1225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data set of conserved APs and SPs is shown in Supplemental Table S6. Sequence traces from our experimental tests are available from the NCBI Trace Archive, TI numbers 1540412808–1540413659, and the corresponding “merged base” reads are given in FASTA format as Supplemental Table S7.