Abstract
Two evolutionarily unrelated superfamilies of G-protein coupled receptors, V1Rs and V2Rs, bind pheromones and “ordinary” odorants to initiate vomeronasal chemical senses in vertebrates, which play important roles in many aspects of an organism’s daily life such as mating, territoriality, and foraging. To study the macroevolution of vomeronasal sensitivity, we identified all V1R and V2R genes from the genome sequences of 11 vertebrates. Our analysis suggests the presence of multiple V1R and V2R genes in the common ancestor of teleost fish and tetrapods and reveals an exceptionally large among-species variation in the sizes of these gene repertoires. Interestingly, the ratio of the number of intact V1R genes to that of V2R genes increased by ∼50-fold as land vertebrates evolved from aquatic vertebrates. A similar increase was found for the ratio of the number of class II odorant receptor (OR) genes to that of class I genes, but not in other vertebrate gene families. Because V1Rs and class II ORs have been suggested to bind to small airborne chemicals, whereas V2Rs and class I ORs recognize water-soluble molecules, these increases reflect a rare case of adaptation to terrestrial life at the gene family level. Several gene families known to function in concert with V2Rs in the mouse are absent outside rodents, indicating rapid changes of interactions between vomeronasal receptors and their molecular partners. Taken together, our results demonstrate the exceptional evolutionary fluidity of vomeronasal receptors, making them excellent targets for studying the molecular basis of physiological and behavioral diversity and adaptation.
Olfaction, or nasal chemoreception, plays a critical role in the daily life of vertebrates. The nasal cavity of most air-breathing vertebrates contains two distinct olfactory tissues/organs: the main olfactory epithelium (MOE) and the vomeronasal organ (VNO) (Dulac and Torello 2003). MOE-mediated olfaction and VNO-mediated olfaction use completely different receptors and signal transduction pathways, and excite different regions of the brain (Dulac and Torello 2003). It was initially thought that MOE and VNO have distinct functions, as MOE detects “ordinary” odorants whereas VNO is specialized for detecting pheromones (Dulac 1997; Buck 2000). This view is changing, as several studies suggested that the MOE can also detect pheromones, whereas the VNO can also detect ordinary odorants (Sam et al. 2001; Boehm et al. 2005; Mandiyan et al. 2005; Yoon et al. 2005; Baxi et al. 2006). Here, pheromones refer to a loosely defined class of chemicals that are emitted and sensed by individuals of the same species to elicit sexual/social behaviors and physiological changes. Examples of pheromone-related behaviors and physiological changes include individual recognition, induction of early puberty, block of pregnancy, and male–male aggression (Keverne 1999).
The molecular biology of vertebrate olfaction is best understood in the laboratory mouse Mus musculus. Mouse has about 1000 odorant receptors (ORs) responsible for the MOE-mediated olfaction (Buck and Axel 1991; Mombaerts 2004) and has over 200 vomeronasal receptors (V1Rs and V2Rs) for VNO-mediated olfaction (Dulac and Axel 1995; Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997; Mombaerts 2004). The evolution of ORs has been extensively studied (Hughes and Hughes 1993; Issel-Tarver and Rine 1997; Rouquier et al. 1998; Sharon et al. 1999; Gilad et al. 2000; Newman and Trask 2003; Niimura and Nei 2005), while that of V1Rs and V2Rs is not well researched. We therefore conduct a comparative genomic analysis of vertebrate V1Rs and V2Rs. V1Rs and V2Rs form two evolutionarily unrelated superfamilies of seven-transmembrane G-protein coupled receptors (Dulac and Axel 1995; Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997). Previous analyses identified 187 intact V1R genes (Shi et al. 2005) and 61 intact V2R genes (Yang et al. 2005) in the mouse genome. Most, if not all V1R and V2R genes, are expressed in the VNO of terrestrial vertebrates (Dulac and Torello 2003) or the MOE of teleost fishes (Cao et al. 1998; Pfister and Rodriguez 2005), as fishes do not have the VNO (Grus and Zhang 2006). It is believed that each vomeronasal sensory neuron expresses only one allele of either a V1R or a V2R gene (Dulac and Torello 2003). V1Rs and V2Rs differ in a number of properties that suggest their different functions. First, V1Rs are coexpressed with the G-protein subunit Gαi2 in sensory neurons whose cell bodies are located in the apical part of the vomeronasal epithelium (Dulac and Torello 2003; Mombaerts 2004). In contrast, V2Rs are expressed in GαO-positive neurons whose cell bodies are located basally in the vomeronasal epithelium (Dulac and Torello 2003; Mombaerts 2004). Neurons expressing V1Rs and V2Rs project to the anterior and posterior accessory olfactory bulb, respectively, where they form multiple glomeruli in spatially conserved domains (Dulac and Torello 2003). Second, most V2Rs, but not V1Rs, are coexpressed with M10 and M1 families of Major-Histocompatibility-Complex (MHC) class Ib molecules (Ishii et al. 2003; Loconto et al. 2003; Silvotti et al. 2005). It appears that M10 proteins function as escort molecules in the transport of V2Rs (with the exception of V2R2) to the cell membrane of vomeronasal sensory neurons (Loconto et al. 2003; Silvotti et al. 2005). Third, V1Rs have a short N-terminal extracellular region, and the entire protein is encoded by one exon. In contrast, V2Rs have a long N-terminal extracellular region with high sequence variability, and the entire protein is encoded in six exons. The difference in protein structure suggests that V1Rs and V2Rs bind to different groups of ligands. Indeed, it has been suggested (Emes et al. 2004) that V1Rs bind to small volatile chemicals such as 2-heptanone, a known pheromone in mice (Boschat et al. 2002), whereas V2Rs bind to water-soluble molecules such as the peptide ligands of MHC class I molecules (Leinders-Zufall et al. 2004) and exocrine gland peptides (ESPs) (Kimoto et al. 2005).
The suggested functional differences between V1Rs and V2Rs predict that their evolutionary patterns should be different. In particular, because terrestrial vertebrates tend to encounter volatile chemicals while aquatic vertebrates encounter water-soluble chemicals, we hypothesize that the relative size of the V1R repertoire compared to the V2R repertoire should have increased in the vertebrate transition from water to land. This hypothesis can be tested by examining the complete V1R and V2R gene repertoires in sequenced vertebrate genomes. To date, the V1R gene repertoire has been described in the human, mouse, rat, cow, dog, opossum, chicken, and several fishes (Rodriguez and Mombaerts 2002; Rodriguez et al. 2002; Grus and Zhang 2004; Hillier et al. 2004; Zhang et al. 2004; Grus et al. 2005; Pfister and Rodriguez 2005; Shi et al. 2005; Young et al. 2005). However, the V2R repertoire has been reported only in the mouse, rat, zebrafish, and fugu (Bjarnadottir et al. 2005; Hashiguchi and Nishida 2005; Yang et al. 2005), although a small number of V2R genes are also known for the human (Kouros-Mehr et al. 2001), African clawed frog (Hagino-Yamagishi et al. 2004), goldfish (Cao et al. 1998), and salmon (Dukes et al. 2006). This scarcity in V2R information is largely due to the complexity of the V2R gene structure. We recently developed a computational pipeline for fast and accurate identification of V2R genes from genome sequences (Yang et al. 2005). Using this and other methods, we here identify the nearly complete V1R and V2R gene repertoires of 11 vertebrates and analyze the evolutionary patterns of these gene repertoires. We then provide evidence supporting the hypothesis of an evolutionary shift from V2R to V1R gene repertoires in the emergence of land vertebrates. Finally, we show that several gene families that function in concert with V2Rs in mice are absent outside rodents, suggesting a high fluidity in the evolution of interactions between vomeronasal receptors and their functional partners.
Results
Evolution of the vertebrate V1R gene repertoire
At the time of this study, 12 vertebrate genomes have been sequenced, including seven mammals (human, chimpanzee, mouse, rat, dog, cow, and opossum), one bird (chicken), one amphibian (western clawed frog), and three teleost fishes (fugu, green spotted pufferfish, and zebrafish). The V1R gene repertoire has been described for some of these species. However, due to a rapid improvement in the quality of many of these genome sequences, it is important to update previous descriptions. Here we describe or update the V1R gene repertoires of the mouse, rat, dog, cow, opossum, chicken, frog, fugu, green spotted pufferfish, and zebrafish. We did not study the chimpanzee because of its close evolutionary relationship to the human. We did not update the human V1R gene repertoire, because the genome sequence has not been improved since the last report of the repertoire. For each genome, we classified the identified V1R sequences into two categories, intact and disrupted genes. Intact genes refer to those that contain full-length open reading frames (ORFs) with seven complete transmembrane domains. Disrupted genes are those with incomplete ORFs due to incomplete genome sequencing or with disrupted ORFs due to pseudogenization. In addition, those sequences that lack seven intact transmembrane domains are also considered disrupted. Our stringent criteria make the estimate of intact gene number conservative. The amino acid sequences of newly identified intact V1R genes are provided in Supplemental data set 1.
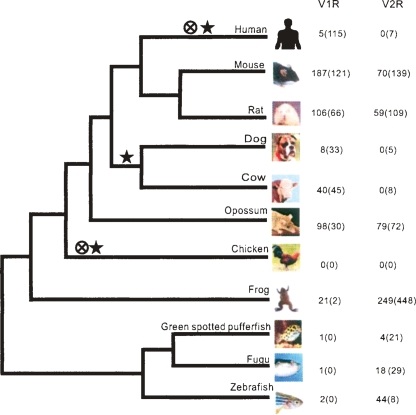
The numbers of intact and disrupted V1R genes vary tremendously among the 11 vertebrates (Fig. 1). The largest V1R repertoire is found in the mouse, rat, and opossum, with 187, 106, and 98 intact genes, respectively. Cow has a considerably smaller number (Grus et al. 2005) of intact V1R genes. Dog was previously reported to have only eight intact V1R genes (Grus et al. 2005; Young et al. 2005) and this number remains unchanged even in the updated genome sequence. Amphibian V1R genes were unknown before, and here we identified 21 intact and two disrupted genes from the western clawed frog. An earlier study reported one V1R gene in several teleost fishes (Pfister and Rodriguez 2005). Here we identified an additional V1R gene in the zebrafish and also confirmed its presence in the medaka fish Oryzias latipes (http://dolphin.lab.nig.ac.jp/medaka/). No V1R genes were found in the chicken, which is not surprising, because birds are not known to have VNO or VNO-mediated olfaction (Keverne 1999). It is likely that the pseudogenization of various genes involved in the chicken vomeronasal transduction pathway took place so long ago that even V1R pseudogenes are unidentifiable from the chicken genome. Similarly, the gene encoding TRPC2, an ion channel indispensable for vomeronasal transduction, is also absent in the chicken genome (Grus and Zhang 2006). In parallel to the ancient loss of the VNO in birds, an independent loss in catarrhine primates (humans, apes, and Old World monkeys) occurred about 23 million years ago (Zhang and Webb 2003). Because of the relatively short time since this loss, the TRPC2 pseudogene is still present in the human and other catarrhine genomes (Liman and Innan 2003; Zhang and Webb 2003). Similarly, 120 human V1R sequences have been identified (Young et al. 2005), although only five of them retain intact ORFs (Rodriguez and Mombaerts 2002). These five V1R genes are probably relics of an ongoing pseudogenization process (Zhang and Webb 2003).
Figure 1.
Evolution of vertebrate V1R and V2R gene repertoires. The tree shows the well established phylogeny of the 11 vertebrates considered here. The numbers of intact V1R and V2R genes are given with the numbers of disrupted genes in parentheses. A cross with a circle indicates the loss of the entire V1R repertoire, whereas a star indicates the loss of the entire V2R repertoire. Although humans have five intact V1R genes, they are likely to be relics of an ongoing pseudogenization process (Zhang and Webb 2003). Thus, the entire V1R repertoire is considered to be lost in humans. Based on the parsimony principle, we infer that the loss of the V2R repertoire in the cow and dog occurred in their common ancestor.
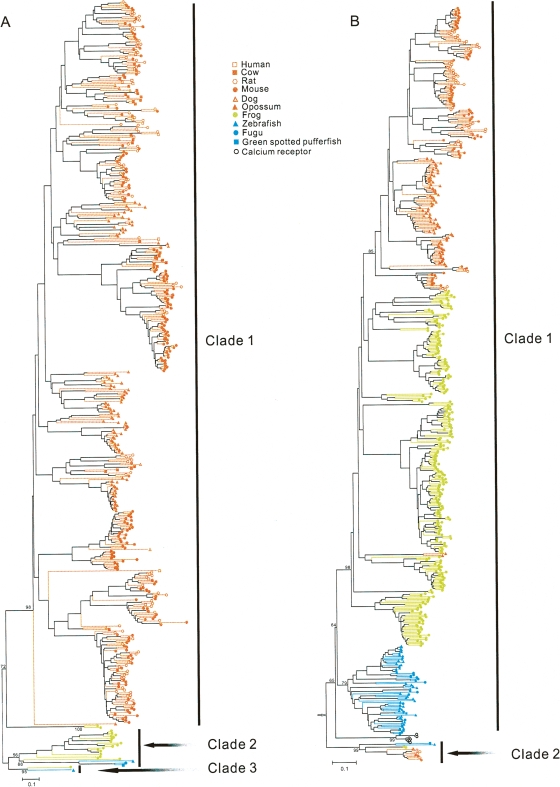
We reconstructed a neighbor-joining (NJ) tree of all intact V1Rs from the 11 vertebrates (Fig. 2A). The root of the tree is difficult to determine because even the closest outgroup (T2R bitter taste receptors) is too divergent to provide accurate root information. We tentatively root the tree in such a way that the numbers of gene losses and gains required to explain the evolution of V1R genes are minimized. The tree shows that vertebrate V1Rs can be divided into three major clades that diverged from each other prior to the separation of tetrapods and teleosts (Fig. 2A). Clade 1 now includes genes from the frog and mammals, whereas clade 2 and clade 3 contain frog and fish genes. It is likely that there was at least one V1R gene in each of the three clades in the common ancestor of tetrapods and teleosts. Clade 1 was lost in fishes, whereas clades 2 and 3 were lost in mammals. In addition, clade 3 was lost in fugu and green spotted pufferfish, although it is retained in the zebrafish and medaka fish. Major expansions of the V1R gene repertoire occurred in some mammals (clade 1), whereas minor expansions occurred in the frog (clades 1 and 2) (Fig. 2A). It should be pointed out that, because the level of sequence divergence is high and the number of amino acid sites useful for the phylogenetic analysis is small, the bootstrap percentages at many nodes of the tree are not high, suggesting that many details of the phylogeny are unresolved.
Figure 2.
Neighbor-joining trees of intact vomeronasal receptors from vertebrates. (A) The V1R tree. (B) The V2R tree. The trees were reconstructed with protein Poisson distances and the scale bars show 0.1 amino acid substitutions per site. The open arrow in the V2R tree indicates the root of the tree, which is determined using vertebrate T1R taste receptors as an outgroup. Bootstrap percentages for some major groups are presented. Vomeronasal receptors from the mammals, frog, and fishes are shown by orange, green, and blue symbols, respectively. The numbers of amino acid sites used for the two trees are 138 and 246, respectively, after the removal of alignment gaps.
Evolution of the vertebrate V2R gene repertoire
The V2R gene repertoire has been described in the mouse, rat, zebrafish, and fugu. Here we update the results for these four species and describe for the first time the complete V2R gene repertoires of the human, dog, cow, opossum, chicken, western clawed frog, and green spotted pufferfish. Note that, although a few human V2R pseudogenes were previously known (Kouros-Mehr et al. 2001), no systematic genomic surveys have been undertaken to identify the human V2R gene repertoire and to verify the prevailing hypothesis that humans lack any intact V2R genes. Again, we classified V2R sequences into two categories, intact and disrupted genes, using the same definitions as used for V1Rs. Surprisingly, the across-vertebrate variation in V2R gene number is even greater than that for V1R genes. The largest V2R repertoire is found in the frog, with 249 intact genes and 408 disrupted genes. In contrast, no intact V2R genes are detected in the chicken, cow, dog, and human. Four, 18, 79, 70, and 59 intact V2R genes are identified from the green spotted pufferfish, fugu, opossum, mouse, and rat, respectively. A recent study described 70 putatively functional genes and 18 pseudogenes in an old version (Zv4) of the zebrafish genome sequence (Hashiguchi and Nishida 2005). We searched a new version (Zv5) and found 44 intact genes and eight disrupted genes. The difference is likely due to the improved quality of the new genome assembly and the increased stringency of our search criteria. Considering the incompleteness of the zebrafish genome assembly, we counted those sequences in chromosome assemblies, but not those merely in contig assemblies. The amino acid sequences of newly described intact V2R genes are provided in Supplemental data set 2.
To understand the evolutionary history of the V2R genes, we reconstructed a protein NJ tree of all vertebrate V2Rs (Fig. 2B). Vertebrate Ca2+-sensing receptors (CasRs) are also included in the tree because they are closely related to some V2Rs (Yang et al. 2005). Sweet/Umami taste receptors (T1Rs) are used as an outgroup to root the tree. Consistent with our previous results (Yang et al. 2005), V2Rs appear to have two origins, as some V2R genes are more closely related to CasRs than to other V2Rs (previously named family C), although the bootstrap support to this dichotomy is not high (65%). The phylogeny suggests the presence of at least two V2R genes, represented by clade 1 and clade 2, in the common ancestor of teleosts and tetrapods (Fig. 2B). The tree also shows multiple lineage-specific expansions of the V2R repertoire in the fishes, frog, opossum, and rodents. We did not find any intact V2R genes, but identified seven, five, and eight disrupted genes, respectively, from the human, dog, and cow genomes. Because the human genome sequence has a high quality with 99% coverage and 99.9% accuracy (International Human Genome Sequencing Consortium 2004) and the dog and cow genome sequences also have high coverages (7.6× for dog and 6× for cow), the absence of intact V2R genes in these genomes is probably real and the identified disrupted genes are likely to be pseudogenes. In the NJ tree of these disrupted genes and all intact V2R genes from the mouse, rat, and opossum (Supplemental Fig. 1), it can be seen that the lineages leading to the disrupted genes tend to have long branches, as expected from the neutral theory (Kimura 1983). More interestingly, the disrupted genes of each species do not cluster in the phylogenetic tree, suggesting a relative diverse V2R repertoire in the human, dog, and cow, respectively, before the start of pseudogenization. Based on the mammalian phylogeny, primates are closely related to rodents and they both belong to the superorder Euarchontoglires, which is the sister group of Laurasiatheria, to which carnivores and artiodactyls belong (Murphy et al. 2004). Thus, the entire V2R repertoire must have been lost at least three times (in chicken, human, and cow/dog) among the 11 vertebrates considered here (Fig. 1).
A shift from V2Rs to V1Rs in the vertebrate transition to land
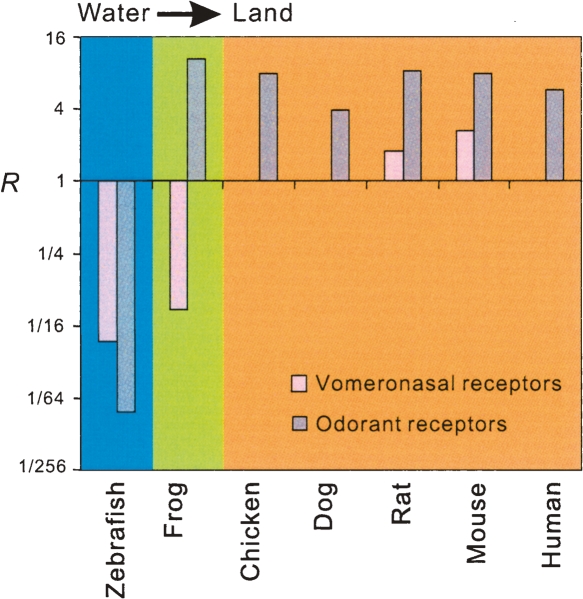
As aforementioned, some empirical evidence suggests that V1Rs bind to small airborne chemicals whereas V2Rs recognize water-soluble molecules. This functional dichotomy, if true, may predict an evolutionary enhancement of the ratio (RV) of the number of intact V1R genes to that of V2R genes in the vertebrate transition from water to land. We computed RV in seven vertebrates whose odorant receptor repertoire sizes are known for comparison (Fig. 3). The zebrafish has RV < 1, as it has fewer V1R genes than V2R genes. In contrast, all mammals have RV > 1, due to an excess of V1R genes over V2R genes. Some mammals such as the dog and cow have RV values of infinity, as they have no intact V2Rs. Amphibians represent an intermediate stage in the vertebrate transition from water to land, because the larvae live in water and breathe with gills, whereas adults live in or near water and breathe with lungs. We thus predict that the RV of the frog is between those of the zebrafish and mammals. Our result is consistent with this prediction (Fig. 3).
Figure 3.
Increase of the ratio (R) of the number of vomeronasal (or odorant) receptors for airborne chemicals to the number of receptors for water-soluble molecules in the vertebrate transition from water to land. For vomeronasal receptors, R is the number of intact V1R genes divided by the number of intact V2R genes in a species. For odorant receptors, R is the number of intact class II OR genes divided by the number of intact class I OR genes in a species. The numbers of odorant receptor genes in various vertebrates are from Olender et al. (2004), Niimura and Nei (2005), and Quignon et al. (2005). R is not shown if it is infinity.
If our observation of an increase in RV during the vertebrate transition from water to land is indeed caused by this transition, we may predict that other families of receptors that bind water-soluble and airborne molecules have also experienced a similar change. In fact, two classes of odorant receptors (ORs) are known in vertebrates (Freitag et al. 1995). Heterologous cells expressing frog class II ORs and class I ORs have been shown to respond to volatile and water-solvable molecules, respectively (Mezler et al. 2001). Thus, similar to vomeronasal receptors, we hypothesize that the ratio (RO) of the number of intact class II OR genes to that of class I OR genes increased in the vertebrate transition to land. Indeed, such an evolutionary pattern is observed (Fig. 3), based on the available data of OR gene repertoires of various vertebrates (Olender et al. 2004; Niimura and Nei 2005; Quignon et al. 2005). Note that the frog appears more similar to fish in RV, but more similar to mammals in RO. The underlying cause of this discrepancy is unclear, but it may be related to the relative numbers of water-soluble and airborne chemicals that the frog VNO and MOE detect. The observed pattern would suggest that the majority of water-soluble chemicals are detected by the VNO and the majority of airborne chemicals are detected by the MOE in the frog.
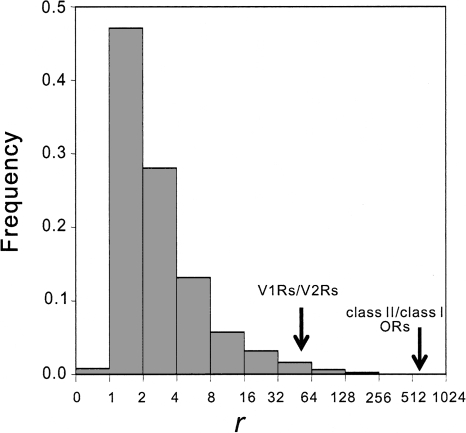
To examine whether the observed increases in RV and RO can be simply explained by typical random turnovers of gene lineages in the evolution of large gene families (Zhang et al. 2000), we conducted a genomewide analysis between all gene families of the zebrafish and mouse, based on the gene family annotations in Ensembl (http://www.ensembl.org). There are 6001 gene families shared between the two species (i.e., same Ensembl family ID). To reduce the influence of transposons, evolutionarily highly conserved ribosomal proteins, and ambiguous gene families, we excluded 1 LINE1 family, 121 ribosomal protein families, and 1431 gene families with ambiguous or unknown functions. Considering that we are interested in the evolution of large gene families, we used only those families that have at least 10 genes in either zebrafish or mouse. A total of 462 such gene families were identified for subsequent analysis (Supplemental Table 1). For any two gene families i and j, we computed r = [nm(i)/nm(j)]/[nz(i)/nz(j)] when r ≥ 1, or r = [nz(i)/nz(j)]/[nm(i)/nm(j)] when r < 1. Here, nm(i) and nz(i) are the numbers of (putatively functional) genes in gene family i of the mouse and zebrafish, respectively. By enumerating all possible pairs of families among the 462 gene families, we obtained the frequency distribution of r (Fig. 4). The mean of this distribution is 5.06. For V1R and V2R families, r equals 56.8. Thus, it is unlikely that the observed amount of size changes in V1R and V2R families during the vertebrate transition to land can be explained by typical random turnovers of gene lineages in large families (P = 0.009; Fig. 4). Similarly, for class II and class I ORs, r equals 665, significantly greater than the random expectation (P = 0.0001; Fig. 4). To generate the above distribution of r we limited our analysis to gene families of sizes >10 genes in either zebrafish or mouse. Our results for vomeronasal and olfactory receptors become more significant when smaller gene families are also used in generating the distribution of r. Qualitatively similar results are obtained when we use the cutoff size of 20 genes, but the number of gene families left (125) for analysis becomes too small to give a reliable distribution of r.
Figure 4.
Frequency distribution of the ratio (r) between mouse and zebrafish for the relative size of two randomly picked gene families from the genomes. More specifically, for gene families i and j, r = [nm(i)/nm(j)]/[nz(i)/nz(j)] when r ≥ 1, and r = [nz(i)/nz(j)]/[nm(i)/nm(j)] when r < 1. Here, nm(i) and nz(i) are the numbers of putatively functional genes in gene family i of the mouse and zebrafish, respectively. Arrows show the r value for V1Rs/V2Rs and that for class II/class I ORs.
Evolution of protein families that interact with V2Rs
In mice, there are two gene families that are known to function in concert with V2Rs. One of them is the M10 family of MHC class Ib molecules, which serve as escort molecules in the transport of some V2Rs to the cell membrane of vomeronasal sensory neurons (Loconto et al. 2003). The second is the exocrine gland peptide (ESP) family, which can activate the V2R-expressing vomeronasal sensory neurons and have been suggested to be ligands of some V2Rs (Kimoto et al. 2005). To study whether M10 and ESP families coevolve with V2Rs, we examined these families in each of the 11 vertebrates studied here. In addition to the six previously described mouse M10 genes (Ishii et al. 2003; Loconto et al. 2003), we identified four M10 genes in the rat. Furthermore, nine intact and 11 disrupted mouse ESP genes and five intact and three disrupted rat ESP genes were identified. The amino acid sequences of the newly identified intact M10 and ESP genes are provided in Supplemental data sets 3 and 4, respectively. Consistent with the absence of V2R genes in the dog, cow, and human, neither M10 nor ESP genes are present in these species. Unexpectedly, however, M10 and ESP genes are not found in the opossum genome, despite the presence of numerous intact V2R genes. Nevertheless, genes flanking the M10 and ESP genes in the mouse genome are present in the opossum genome, suggesting that the absence of opossum M10 and ESP genes is unlikely due to gaps in the draft genome sequence. Rather, these genes most likely do not exist in the opossum. An interesting question is whether the opossum lost the M10 and ESP genes or rodents acquired them in evolution. The latter scenario is more likely because M10 and ESP genes are found in rodents only. Hence, the requirement of M10 molecules for the transport of some V2Rs to cell membranes is probably a rodent-specific phenomenon and the use of ESPs as potentially V2R-recognizing pheromones are also rodent-specific.
Previous phylogenetic analyses of V1R and V2R genes showed that only a few one-to-one orthologous gene pairs exist even between closely related species such as the mouse and rat, due to rapid birth and death of vomeronasal receptor genes (Lane et al. 2002, 2004; Grus and Zhang 2004; Yang et al. 2005). Interestingly, similar patterns are found in the M10 and ESP genes. The six mouse M10 genes form a cluster that is separated from the four rat M10 genes in the gene tree (Supplemental Fig. 2). In the ESP tree, only two one-to-one orthologous pairs are found among the nine mouse intact genes and five rat intact genes (Supplemental Fig. 3). These results further suggest that M10 and ESP families are subject to a rapid gene birth and death process in rodents (Nei and Rooney 2005).
Discussion
In this study, we performed evolutionary analyses of V1R and V2R vomeronasal receptor gene repertoires of 11 vertebrates including three fishes, one frog, one bird, and six mammals, based on the available genome sequences. We found that the ratio of the number of intact V1R genes to that of intact V2R genes increased by ∼50-fold in the evolutionary transition from water to land (Fig. 3). Although the ligands of most V1Rs and V2Rs are yet to be identified, the available functional data suggest that V1Rs bind to small volatile chemicals, whereas V2Rs bind to water-soluble molecules (Boschat et al. 2002; Emes et al. 2004; Leinders-Zufall et al. 2004; Kimoto et al. 2005). For example, 2-heptanone, a small volatile pheromone, activates the V1Rb2-expressing vomeronasal sensory neurons (Boschat et al. 2002). In addition, V2R-expressing vomeronasal sensory neurons respond to the peptide ligands of MHC class I molecules (Leinders-Zufall et al. 2004) and ESPs (Kimoto et al. 2005). If the majority of V1Rs and V2Rs bind to volatile and water-soluble molecules, respectively, as suggested by the limited functional data, our observation of an evolutionary shift from V2Rs to V1Rs in the vertebrate transition from water to land is easy to understand, because aquatic vertebrates encounter and need to detect more water-soluble molecules, whereas land vertebrates need to sense more volatile chemicals. Interestingly, there is a similar evolutionary trend in the ratio of the number of class II ORs to that of class I ORs (Fig. 3), which have been suggested to bind to volatile and water-solvable molecules, respectively, at least in frogs (Mezler et al. 2001). Recent evolutionary studies showed that the phylogenetic diversity of class I ORs exceeds that of class II ORs and that class II ORs form a monophyletic group within class I ORs (Alioto and Ngai 2005; Niimura and Nei 2005). These findings suggest that class II ORs may have originated from class I ORs, consistent with their proposed functional distinction (Freitag et al. 1995; Mezler et al. 2001). Thus, the two nasal chemosensory systems appear to show a consistent pattern of a shift from receptors for water-soluble molecules to those for volatiles in the vertebrate transition from water to land, reflecting a rare case of adaptation to terrestrial life at the gene family level. Our genome-wide comparison of mouse and zebrafish gene families further demonstrated that such dramatic family-size changes are unlikely due to random gene-lineage turnovers during gene family evolution. We emphasize that the functional distinction between V1Rs and V2Rs and that between class I ORs and class II ORs are yet to be firmly established and thus our interpretation of the observed evolutionary patterns of these receptors requires further scrutiny in the future. In this regard, it would be interesting to examine if receptors for presumably water-soluble molecules (V2Rs and class I ORs) are expressed during the larval stage of amphibian development, whereas receptors for presumably airborne molecules (V1Rs and class II ORs) are expressed in the adult stage. Such developmental shift of vomeronasal and odorant receptor repertoires would be another evidence for adaptation to terrestrial life. Indeed, temporal patterns of OR gene expression in frogs support this view (Mezler et al. 1999). One caveat of our analysis is that only one species (western clawed frog) representing the water-to-land transition in vertebrate evolution is examined. Additional species of amphibians would help verify our results. Furthermore, there are some terrestrial vertebrates that returned to water during their evolution (e.g., sea snakes, the platypus, and whales) and it would be interesting to examine their vomeronasal and odorant receptors. However, it should be noted that all these animals, although living in water, are air-breathers. Thus, their vomeronasal and odorant receptors may still be similar to those of terrestrial vertebrates.
As is clear from our phylogenetic analyses (Fig. 2), the differences in intact gene number between V1Rs and V2Rs among vertebrates likely arose from the asymmetric evolutionary dynamics of the two gene superfamilies. First, terrestrial vertebrates possess more V1R genes than fishes do. In terms of the number of intact V1R genes, only one or two genes are found in teleosts, whereas over 100 genes exist in the mouse and rat. Even when the dog V1R repertoire, the smallest among those mammals with functional VNOs, is considered, there is still a four- to eightfold increase in repertoire size compared to the fishes. This increase is apparently due to the extensive duplications of clade 1 V1R genes in mammals (Fig. 2A). Second, in comparison to the V1R gene repertoire, the V2R repertoire is frequently lost in terrestrial vertebrates (Fig. 1). For example, the V2R gene repertoire is lost in the cow and dog, where V1R genes are present. Furthermore, an experimental study failed to find any intact V2R genes from the goat (Wakabayashi et al. 2002). A recent study identified two types of vomeronasal systems in mammals (Takigami et al. 2004). The segregated system is found in rodents and opossums, which have both Gαi2- and GαO-expressing vomeronasal sensory neurons that separately project their axons to segregated (anterior and posterior) regions of the accessory olfactory bulb (Takigami et al. 2004). The uniform system is found in all other examined mammals that have VNOs (goat, dog, horse, musk shrew, and marmoset). These organisms contain only Gαi2-expressing vomeronasal sensory neurons (Takigami et al. 2004). Because V1Rs are expressed in Gαi2-positive neurons and V2Rs are expressed in GαO-positive neurons (Mombaerts 2004), we hypothesize that only a small number of mammalian lineages such as rodents and opossums possess functional V2Rs. Consistent with this hypothesis, to date no functional V2Rs are known to exist outside rodents and opossums. However, it is unclear why V2R repertoires expanded in rodents and opossums if V2Rs are primarily used for detecting water-soluble molecules. It is possible that these species use peptide pheromones (e.g., MHC ligands) more often than other terrestrial vertebrate do. Third, in contrast to the multiple losses of the entire V2R repertoire in mammals, the V2R gene repertoire expanded in teleosts with prominent patterns of lineage-specific gene duplication (Fig. 2B). Among all gene families, the V1R family was previously found to have the highest variation in gene number across mammals (Grus et al. 2005). Our present analysis shows a similar pattern for the V2R family. Even when all vertebrate gene families are considered, V1R and V2R families are certainly among the most extreme ones in terms of gene number variation among species (Fig. 1).
It is interesting to note that both V1R and V2R repertoires expanded in the frog. The frog V1R repertoire expansion mainly occurred in clade 2, which contains no mammalian members and only one teleost member. The frog V2R repertoire expansion was even more substantive than that of V1R, making the frog V2R repertoire largest among all vertebrates. The frog expansions of both vomeronasal receptor gene families may be related to the origin of the VNO. Although teleosts have V1R and V2R genes, they do not have VNOs (Grus and Zhang 2006). Instead, teleost V1R and V2R genes are expressed in the microvillar sensory neurons in the olfactory epithelium, which has been suggested to be homologous to tetrapod vomeronasal sensory neurons (Grus and Zhang 2006). The morphologically identifiable VNO first appeared in amphibians, which may have resulted from a substantial improvement of vomeronasal functions, as suggested by the great expansions of the V1R and V2R repertoires. It was recently reported that the frog species examined here has no T1R sweet/umami taste receptor genes although it has numerous bitter taste receptor genes (Shi and Zhang 2006). Because fish V2Rs can detect amino acids (Speca et al. 1999), it is possible that some frog V2Rs are responsible for detecting amino acids, which provide sweet/umami tastes. In fact, some snakes are known to use VNOs in mediating responses to foraging cues (Baxi et al. 2006).
Surprisingly, the M10 family of MHC proteins, necessary for escorting V2Rs to the membrane of vomeronasal sensory neurons in mice, is absent outside rodents. Similarly, ESPs, which can activate mouse V2R-expressing vomeronasal sensory neurons, are also absent outside rodents. These results show that not only V1R and V2R repertoires vary substantially across vertebrates, the molecular partners of vomeronasal receptors also differ immensely in different species. Unfortunately, almost the entire molecular neurobiology of the vomeronasal system is based on the study of mice and rats, which are not typical among vertebrates, or even within mammals, in features such as the morphological complexity of the VNO, size of the vomeronasal receptor gene repertoires, and molecular partners of vomeronasal receptors. On one hand, our findings of the exceptional evolutionary fluidity of vomeronasal receptors and their molecular partners demonstrate the need to explore a diverse group of vertebrates for a full understanding of the organization and function of the vomeronasal system. On the other hand, they suggest that genes functioning in the vomeronasal system may be excellent targets for studying the molecular basis of vertebrate behavioral and physiological diversity and adaptation.
Methods
Identification of V1R, V2R, M10, and ESP genes
V1R genes were searched in the genome sequences of the mouse (Mus musculus), rat (Rattus norvegicus), dog (Canis familiaris), cow (Bos taurus), opossum (Monodelphis domestica), chicken (Gallus gallus), frog (Xenopus tropicalis), fugu (Takifugu rubripes), green spotted pufferfish (Tetraodon nigroviridis), and zebrafish (Danio rerio), by the method described before (Grus et al. 2005; Shi et al. 2005). V2R genes were searched in these species and in the human (Homo sapiens) by the method of Yang et al. (2005). M10 and ESP genes were also searched using Yang et al.’s method. The genome sequences were obtained from Ensembl (www. ensembl.org). The assemblies used were human NCBI 35, mouse NCBI m35, rat RGSC 3.4, dog Canfam 2.0, cow Btau 2.0, opossum Mondom 2.0, chicken WASHUC 1.0, frog JGI 4.0, fugu Fugu 4.0, green spotted pufferfish Tetraodon 7.0, and zebrafish ZV5.
Briefly, the method of Yang et al. (2005) for identifying V2R genes involves three steps. First, we used all known intact V2R genes as queries to BLAST against a genome sequence to identify the genomic locations for candidate V2R genes. Second, the genomic DNA sequences of the homologous genes and the known V2R protein sequences were used to conduct protein-to-genomic sequence alignment on GeneWise (http://www.ebi.ac.uk/wise2/), which has been used widely in gene prediction and genome annotation (Birney et al. 2004). Third, the identified putative V2R genes were BLASTed against the nr database of GenBank to ensure that the best hit was a V2R. This step was necessary because some other receptors (e.g., Ca2+-sensing receptors and T1R taste receptors) are known to be homologous to V2Rs. A V2R sequence was regarded as a disrupted gene if its disrupted open reading frame was longer than 200 nucleotides, which could usually code for two transmembrane domains and a connecting loop. Sequences shorter than 200 nucleotides were discarded. To reduce the influence of assembly errors, we required that the intergenic region between tandem disrupted genes should be longer than that of an average intact V2R gene. Otherwise, the consecutive homologous segments were regarded as one disrupted gene. This stringent criterion led to a low rate of false detection of disrupted V2R genes, although some true cases might have been missed. The method for identifying V1Rs is similar to the above description, except that the second step is omitted, because V1R genes contain no introns in the coding region. Although the majority of the genome sequences used here are draft sequences, which inevitably sets a limit on the completeness and accuracy of the receptor repertoires we can identify, we believe that our conclusions are unaffected, as it is the ratio of the number of V1R genes to that of V2R genes, not the absolute numbers of genes, that form the basis of our conclusion.
Evolutionary analyses
Deduced amino acid sequences of V1Rs and V2Rs were respectively aligned by CLUSTAL_X (Thompson et al. 1997) with manual adjustments. Phylogenetic trees of V1Rs and V2Rs were reconstructed using the neighbor-joining method (Saitou and Nei 1987) with protein Poisson distances (Nei and Kumar 2000). Gap sites in the alignment were not used in the phylogenetic reconstruction (complete-deletion option). The reliability of the estimated tree was evaluated by the bootstrap method (Felsenstein 1985) with 1000 pseudo-replications. MEGA3 (Kumar et al. 2004) was used for the phylogenetic analysis.
Acknowledgments
We thank Soochin Cho and Wendy Grus for valuable comments. We thank the Human Genome Sequencing Center at Baylor College of Medicine, Broad Institute, DOE Joint Genome Institute, and National Institute of Genetics and University of Tokyo for making the cow, opossum, frog, and medaka fish draft genome sequences available. This work was supported by research grants from National Institutes of Health and University of Michigan to J.Z.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6040007
References
- Alioto T.S., Ngai J., Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxi K.N., Dorries K.M., Eisthen H.L., Dorries K.M., Eisthen H.L., Eisthen H.L. Is the vomeronasal system really specialized for detecting pheromones? Trends Neurosci. 2006;29:1–7. doi: 10.1016/j.tins.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R., Clamp M., Durbin R., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnadottir T.K., Fredriksson R., Schioth H.B., Fredriksson R., Schioth H.B., Schioth H.B. The gene repertoire and the common evolutionary history of glutamate, pheromone (V2R), taste(1) and other related G protein-coupled receptors. Gene. 2005;362:70–84. doi: 10.1016/j.gene.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Boehm U., Zou Z., Buck L.B., Zou Z., Buck L.B., Buck L.B. Feedback loops link odor and pheromone signaling with reproduction. Cell. 2005;123:683–695. doi: 10.1016/j.cell.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Boschat C., Pelofi C., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Pelofi C., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Randin O., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Roppolo D., Luscher C., Broillet M.C., Rodriguez I., Luscher C., Broillet M.C., Rodriguez I., Broillet M.C., Rodriguez I., Rodriguez I. Pheromone detection mediated by a V1r vomeronasal receptor. Nat. Neurosci. 2002;5:1261–1262. doi: 10.1038/nn978. [DOI] [PubMed] [Google Scholar]
- Buck L.B. The molecular architecture of odor and pheromone sensing in mammals. Cell. 2000;100:611–618. doi: 10.1016/s0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R., Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Cao Y., Oh B.C., Stryer L., Oh B.C., Stryer L., Stryer L. Cloning and localization of two multigene receptor families in goldfish olfactory epithelium. Proc. Natl. Acad. Sci. 1998;95:11987–11992. doi: 10.1073/pnas.95.20.11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukes J.P., Deaville R., Gottelli D., Neigel J.E., Bruford M.W., Jordan W.C., Deaville R., Gottelli D., Neigel J.E., Bruford M.W., Jordan W.C., Gottelli D., Neigel J.E., Bruford M.W., Jordan W.C., Neigel J.E., Bruford M.W., Jordan W.C., Bruford M.W., Jordan W.C., Jordan W.C. Isolation and characterisation of main olfactory and vomeronasal receptor gene families from the Atlantic salmon (Salmo salar) Gene. 2006;371:257–267. doi: 10.1016/j.gene.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Dulac C. Molecular biology of pheromone perception in mammals. Semin. Cell Dev. Biol. 1997;8:197–205. doi: 10.1006/scdb.1997.0140. [DOI] [PubMed] [Google Scholar]
- Dulac C., Axel R., Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Dulac C., Torello A.T., Torello A.T. Molecular detection of pheromone signals in mammals: From genes to behaviour. Nat. Rev. Neurosci. 2003;4:551–562. doi: 10.1038/nrn1140. [DOI] [PubMed] [Google Scholar]
- Emes R.D., Beatson S.A., Ponting C.P., Goodstadt L., Beatson S.A., Ponting C.P., Goodstadt L., Ponting C.P., Goodstadt L., Goodstadt L. Evolution and comparative genomics of odorant- and pheromone-associated genes in rodents. Genome Res. 2004;14:591–602. doi: 10.1101/gr.1940604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Freitag J., Krieger J., Strotmann J., Breer H., Krieger J., Strotmann J., Breer H., Strotmann J., Breer H., Breer H. Two classes of olfactory receptors in Xenopus laevis. Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Gilad Y., Segre D., Skorecki K., Nachman M.W., Lancet D., Sharon D., Segre D., Skorecki K., Nachman M.W., Lancet D., Sharon D., Skorecki K., Nachman M.W., Lancet D., Sharon D., Nachman M.W., Lancet D., Sharon D., Lancet D., Sharon D., Sharon D. Dichotomy of single-nucleotide polymorphism haplotypes in olfactory receptor genes and pseudogenes. Nat. Genet. 2000;26:221–224. doi: 10.1038/79957. [DOI] [PubMed] [Google Scholar]
- Grus W.E., Zhang J., Zhang J. Rapid turnover and species-specificity of vomeronasal pheromone receptor genes in mice and rats. Gene. 2004;340:303–312. doi: 10.1016/j.gene.2004.07.037. [DOI] [PubMed] [Google Scholar]
- Grus W.E., Zhang J., Zhang J. Origin and evolution of the vertebrate vomeronasal system viewed through system-specific genes. Bioessays. 2006;28:709–718. doi: 10.1002/bies.20432. [DOI] [PubMed] [Google Scholar]
- Grus W.E., Shi P., Zhang Y.P., Zhang J., Shi P., Zhang Y.P., Zhang J., Zhang Y.P., Zhang J., Zhang J. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc. Natl. Acad. Sci. 2005;102:5767–5772. doi: 10.1073/pnas.0501589102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagino-Yamagishi K., Moriya K., Kubo H., Wakabayashi Y., Isobe N., Saito S., Ichikawa M., Yazaki K., Moriya K., Kubo H., Wakabayashi Y., Isobe N., Saito S., Ichikawa M., Yazaki K., Kubo H., Wakabayashi Y., Isobe N., Saito S., Ichikawa M., Yazaki K., Wakabayashi Y., Isobe N., Saito S., Ichikawa M., Yazaki K., Isobe N., Saito S., Ichikawa M., Yazaki K., Saito S., Ichikawa M., Yazaki K., Ichikawa M., Yazaki K., Yazaki K. Expression of vomeronasal receptor genes in Xenopus laevis. J. Comp. Neurol. 2004;472:246–256. doi: 10.1002/cne.20073. [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y., Nishida M., Nishida M. Evolution of vomeronasal-type odorant receptor genes in the zebrafish genome. Gene. 2005;362:19–28. doi: 10.1016/j.gene.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Herrada G., Dulac C., Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Bork P., Burt D.W., Groenen M.A., Delany M.E., Burt D.W., Groenen M.A., Delany M.E., Groenen M.A., Delany M.E., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hughes A.L., Hughes M.K., Hughes M.K. Adaptive evolution in the rat olfactory receptor gene family. J. Mol. Evol. 1993;36:249–254. doi: 10.1007/BF00160480. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Ishii T., Hirota J., Mombaerts P., Hirota J., Mombaerts P., Mombaerts P. Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr. Biol. 2003;13:394–400. doi: 10.1016/s0960-9822(03)00092-7. [DOI] [PubMed] [Google Scholar]
- Issel-Tarver L., Rine J., Rine J. The evolution of mammalian olfactory receptor genes. Genetics. 1997;145:185–195. doi: 10.1093/genetics/145.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keverne E.B. The vomeronasal organ. Science. 1999;286:716–720. doi: 10.1126/science.286.5440.716. [DOI] [PubMed] [Google Scholar]
- Kimoto H., Haga S., Sato K., Touhara K., Haga S., Sato K., Touhara K., Sato K., Touhara K., Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Kouros-Mehr H., Pintchovski S., Melnyk J., Chen Y.J., Friedman C., Trask B., Shizuya H., Pintchovski S., Melnyk J., Chen Y.J., Friedman C., Trask B., Shizuya H., Melnyk J., Chen Y.J., Friedman C., Trask B., Shizuya H., Chen Y.J., Friedman C., Trask B., Shizuya H., Friedman C., Trask B., Shizuya H., Trask B., Shizuya H., Shizuya H. Identification of non-functional human VNO receptor genes provides evidence for vestigiality of the human VNO. Chem. Senses. 2001;26:1167–1174. doi: 10.1093/chemse/26.9.1167. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Nei M., Tamura K., Nei M., Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lane R.P., Cutforth T., Axel R., Hood L., Trask B.J., Cutforth T., Axel R., Hood L., Trask B.J., Axel R., Hood L., Trask B.J., Hood L., Trask B.J., Trask B.J. Sequence analysis of mouse vomeronasal receptor gene clusters reveals common promoter motifs and a history of recent expansion. Proc. Natl. Acad. Sci. 2002;99:291–296. doi: 10.1073/pnas.012608399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane R.P., Young J., Newman T., Trask B.J., Young J., Newman T., Trask B.J., Newman T., Trask B.J., Trask B.J. Species specificity in rodent pheromone receptor repertoires. Genome Res. 2004;14:603–608. doi: 10.1101/gr.2117004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinders-Zufall T., Brennan P., Widmayer P., S P.C., Maul-Pavicic A., Jager M., Li X.H., Breer H., Zufall F., Boehm T., Brennan P., Widmayer P., S P.C., Maul-Pavicic A., Jager M., Li X.H., Breer H., Zufall F., Boehm T., Widmayer P., S P.C., Maul-Pavicic A., Jager M., Li X.H., Breer H., Zufall F., Boehm T., S P.C., Maul-Pavicic A., Jager M., Li X.H., Breer H., Zufall F., Boehm T., Maul-Pavicic A., Jager M., Li X.H., Breer H., Zufall F., Boehm T., Jager M., Li X.H., Breer H., Zufall F., Boehm T., Li X.H., Breer H., Zufall F., Boehm T., Breer H., Zufall F., Boehm T., Zufall F., Boehm T., Boehm T. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science. 2004;306:1033–1037. doi: 10.1126/science.1102818. [DOI] [PubMed] [Google Scholar]
- Liman E.R., Innan H., Innan H. Relaxed selective pressure on an essential component of pheromone transduction in primate evolution. Proc. Natl. Acad. Sci. 2003;100:3328–3332. doi: 10.1073/pnas.0636123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loconto J., Papes F., Chang E., Stowers L., Jones E.P., Takada T., Kumanovics A., Fischer Lindahl K., Dulac C., Papes F., Chang E., Stowers L., Jones E.P., Takada T., Kumanovics A., Fischer Lindahl K., Dulac C., Chang E., Stowers L., Jones E.P., Takada T., Kumanovics A., Fischer Lindahl K., Dulac C., Stowers L., Jones E.P., Takada T., Kumanovics A., Fischer Lindahl K., Dulac C., Jones E.P., Takada T., Kumanovics A., Fischer Lindahl K., Dulac C., Takada T., Kumanovics A., Fischer Lindahl K., Dulac C., Kumanovics A., Fischer Lindahl K., Dulac C., Fischer Lindahl K., Dulac C., Dulac C. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 2003;112:607–618. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Mandiyan V.S., Coats J.K., Shah N.M., Coats J.K., Shah N.M., Shah N.M. Deficits in sexual and aggressive behaviors in Cnga2 mutant mice. Nat. Neurosci. 2005;8:1660–1662. doi: 10.1038/nn1589. [DOI] [PubMed] [Google Scholar]
- Matsunami H., Buck L.B., Buck L.B. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- Mezler M., Konzelmann S., Freitag J., Rossler P., Breer H., Konzelmann S., Freitag J., Rossler P., Breer H., Freitag J., Rossler P., Breer H., Rossler P., Breer H., Breer H. Expression of olfactory receptors during development in Xenopus laevis. J. Exp. Biol. 1999;202:365–376. doi: 10.1242/jeb.202.4.365. [DOI] [PubMed] [Google Scholar]
- Mezler M., Fleischer J., Breer H., Fleischer J., Breer H., Breer H. Characteristic features and ligand specificity of the two olfactory receptor classes from Xenopus laevis. J. Exp. Biol. 2001;204:2987–2997. doi: 10.1242/jeb.204.17.2987. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pevzner P.A., O’Brien S.J., Pevzner P.A., O’Brien S.J., O’Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S., Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Nei M., Rooney A.P., Rooney A.P. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T., Trask B.J., Trask B.J. Complex evolution of 7E olfactory receptor genes in segmental duplications. Genome Res. 2003;13:781–793. doi: 10.1101/gr.769003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y., Nei M., Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olender T., Fuchs T., Linhart C., Shamir R., Adams M., Kalush F., Khen M., Lancet D., Fuchs T., Linhart C., Shamir R., Adams M., Kalush F., Khen M., Lancet D., Linhart C., Shamir R., Adams M., Kalush F., Khen M., Lancet D., Shamir R., Adams M., Kalush F., Khen M., Lancet D., Adams M., Kalush F., Khen M., Lancet D., Kalush F., Khen M., Lancet D., Khen M., Lancet D., Lancet D. The canine olfactory subgenome. Genomics. 2004;83:361–372. doi: 10.1016/j.ygeno.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Pfister P., Rodriguez I., Rodriguez I. Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in fish species. Proc. Natl. Acad. Sci. 2005;102:5489–5494. doi: 10.1073/pnas.0402581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon P., Giraud M., Rimbault M., Lavigne P., Tacher S., Morin E., Retout E., Valin A.S., Lindblad-Toh K., Nicolas J., Giraud M., Rimbault M., Lavigne P., Tacher S., Morin E., Retout E., Valin A.S., Lindblad-Toh K., Nicolas J., Rimbault M., Lavigne P., Tacher S., Morin E., Retout E., Valin A.S., Lindblad-Toh K., Nicolas J., Lavigne P., Tacher S., Morin E., Retout E., Valin A.S., Lindblad-Toh K., Nicolas J., Tacher S., Morin E., Retout E., Valin A.S., Lindblad-Toh K., Nicolas J., Morin E., Retout E., Valin A.S., Lindblad-Toh K., Nicolas J., Retout E., Valin A.S., Lindblad-Toh K., Nicolas J., Valin A.S., Lindblad-Toh K., Nicolas J., Lindblad-Toh K., Nicolas J., Nicolas J., et al. The dog and rat olfactory receptor repertoires. Genome Biol. 2005;6:R83. doi: 10.1186/gb-2005-6-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I., Mombaerts P., Mombaerts P. Novel human vomeronasal receptor-like genes reveal species-specific families. Curr. Biol. 2002;12:R409–R411. doi: 10.1016/s0960-9822(02)00909-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez I., Del Punta K., Rothman A., Ishii T., Mombaerts P., Del Punta K., Rothman A., Ishii T., Mombaerts P., Rothman A., Ishii T., Mombaerts P., Ishii T., Mombaerts P., Mombaerts P. Multiple new and isolated families within the mouse superfamily of V1r vomeronasal receptors. Nat. Neurosci. 2002;5:134–140. doi: 10.1038/nn795. [DOI] [PubMed] [Google Scholar]
- Rouquier S., Taviaux S., Trask B.J., Brand-Arpon V., van den Engh G., Demaille J., Giorgi D., Taviaux S., Trask B.J., Brand-Arpon V., van den Engh G., Demaille J., Giorgi D., Trask B.J., Brand-Arpon V., van den Engh G., Demaille J., Giorgi D., Brand-Arpon V., van den Engh G., Demaille J., Giorgi D., van den Engh G., Demaille J., Giorgi D., Demaille J., Giorgi D., Giorgi D. Distribution of olfactory receptor genes in the human genome. Nat. Genet. 1998;18:243–250. doi: 10.1038/ng0398-243. [DOI] [PubMed] [Google Scholar]
- Ryba N.J., Tirindelli R., Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sam M., Vora S., Malnic B., Ma W., Novotny M.V., Buck L.B., Vora S., Malnic B., Ma W., Novotny M.V., Buck L.B., Malnic B., Ma W., Novotny M.V., Buck L.B., Ma W., Novotny M.V., Buck L.B., Novotny M.V., Buck L.B., Buck L.B. Neuropharmacology. Odorants may arouse instinctive behaviours. Nature. 2001;412:142. doi: 10.1038/35084137. [DOI] [PubMed] [Google Scholar]
- Sharon D., Glusman G., Pilpel Y., Khen M., Gruetzner F., Haaf T., Lancet D., Glusman G., Pilpel Y., Khen M., Gruetzner F., Haaf T., Lancet D., Pilpel Y., Khen M., Gruetzner F., Haaf T., Lancet D., Khen M., Gruetzner F., Haaf T., Lancet D., Gruetzner F., Haaf T., Lancet D., Haaf T., Lancet D., Lancet D. Primate evolution of an olfactory receptor cluster: Diversification by gene conversion and recent emergence of pseudogenes. Genomics. 1999;61:24–36. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- Shi P., Zhang J., Zhang J. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol. Biol. Evol. 2006;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
- Shi P., Bielawski J.P., Yang H., Zhang Y.P., Bielawski J.P., Yang H., Zhang Y.P., Yang H., Zhang Y.P., Zhang Y.P. Adaptive diversification of vomeronasal receptor 1 genes in rodents. J. Mol. Evol. 2005;60:566–576. doi: 10.1007/s00239-004-0172-y. [DOI] [PubMed] [Google Scholar]
- Silvotti L., Giannini G., Tirindelli R., Giannini G., Tirindelli R., Tirindelli R. The vomeronasal receptor V2R2 does not require escort molecules for expression in heterologous systems. Chem. Senses. 2005;30:1–8. doi: 10.1093/chemse/bjh250. [DOI] [PubMed] [Google Scholar]
- Speca D.J., Lin D.M., Sorensen P.W., Isacoff E.Y., Ngai J., Dittman A.H., Lin D.M., Sorensen P.W., Isacoff E.Y., Ngai J., Dittman A.H., Sorensen P.W., Isacoff E.Y., Ngai J., Dittman A.H., Isacoff E.Y., Ngai J., Dittman A.H., Ngai J., Dittman A.H., Dittman A.H. Functional identification of a goldfish odorant receptor. Neuron. 1999;23:487–498. doi: 10.1016/s0896-6273(00)80802-8. [DOI] [PubMed] [Google Scholar]
- Takigami S., Mori Y., Tanioka Y., Ichikawa M., Mori Y., Tanioka Y., Ichikawa M., Tanioka Y., Ichikawa M., Ichikawa M. Morphological evidence for two types of Mammalian vomeronasal system. Chem. Senses. 2004;29:301–310. doi: 10.1093/chemse/bjh032. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Plewniak F., Jeanmougin F., Higgins D.G., Jeanmougin F., Higgins D.G., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y., Mori Y., Ichikawa M., Yazaki K., Hagino-Yamagishi K., Mori Y., Ichikawa M., Yazaki K., Hagino-Yamagishi K., Ichikawa M., Yazaki K., Hagino-Yamagishi K., Yazaki K., Hagino-Yamagishi K., Hagino-Yamagishi K. A putative pheromone receptor gene is expressed in two distinct olfactory organs in goats. Chem. Senses. 2002;27:207–213. doi: 10.1093/chemse/27.3.207. [DOI] [PubMed] [Google Scholar]
- Yang H., Shi P., Zhang Y.P., Zhang J., Shi P., Zhang Y.P., Zhang J., Zhang Y.P., Zhang J., Zhang J. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics. 2005;86:306–315. doi: 10.1016/j.ygeno.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Yoon H., Enquist L.W., Dulac C., Enquist L.W., Dulac C., Dulac C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell. 2005;123:669–682. doi: 10.1016/j.cell.2005.08.039. [DOI] [PubMed] [Google Scholar]
- Young J.M., Kambere M., Trask B.J., Lane R.P., Kambere M., Trask B.J., Lane R.P., Trask B.J., Lane R.P., Lane R.P. Divergent V1R repertoires in five species: Amplification in rodents, decimation in primates, and a surprisingly small repertoire in dogs. Genome Res. 2005;15:231–240. doi: 10.1101/gr.3339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Webb D.M., Webb D.M. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc. Natl. Acad. Sci. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Dyer K.D., Rosenberg H.F., Dyer K.D., Rosenberg H.F., Rosenberg H.F. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl. Acad. Sci. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Rodriguez I., Mombaerts P., Firestein S., Rodriguez I., Mombaerts P., Firestein S., Mombaerts P., Firestein S., Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004;83:802–811. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]