Abstract
We present here the annotation of the complete genome of rice Oryza sativa L. ssp. japonica cultivar Nipponbare. All functional annotations for proteins and non-protein-coding RNA (npRNA) candidates were manually curated. Functions were identified or inferred in 19,969 (70%) of the proteins, and 131 possible npRNAs (including 58 antisense transcripts) were found. Almost 5000 annotated protein-coding genes were found to be disrupted in insertional mutant lines, which will accelerate future experimental validation of the annotations. The rice loci were determined by using cDNA sequences obtained from rice and other representative cereals. Our conservative estimate based on these loci and an extrapolation suggested that the gene number of rice is ∼32,000, which is smaller than previous estimates. We conducted comparative analyses between rice and Arabidopsis thaliana and found that both genomes possessed several lineage-specific genes, which might account for the observed differences between these species, while they had similar sets of predicted functional domains among the protein sequences. A system to control translational efficiency seems to be conserved across large evolutionary distances. Moreover, the evolutionary process of protein-coding genes was examined. Our results suggest that natural selection may have played a role for duplicated genes in both species, so that duplication was suppressed or favored in a manner that depended on the function of a gene.
The majority of the world’s population depends on cereal crops as their primary source of carbohydrate. Among the cultivated cereal crops, rice makes up ∼20% of the total calorific intake for the human population as a whole (http://www.irri.org/science/ricestat/index.asp). In order to cope with increasing global demand for food and because of its importance as a staple, many agrobiological studies have been performed with the aim of developing more efficient rice cultivars.
With the completion of the rice genome (Oryza sativa L. ssp. japonica cultivar Nipponbare) by the international consortium on rice genome sequencing (International Rice Genome Sequencing Project 2005), it has become possible to elucidate the layers of information encoded by the sequence. Analyses of rice full-length cDNAs and the rice proteome are in progress (Kikuchi et al. 2003; Komatsu et al. 2004; Komatsu and Tanaka 2005). Additionally, construction of integrative annotations for the rice genome, transcriptome, and proteome is being undertaken. In order to standardize the annotation of the genome data for Nipponbare, we organized an international consortium for rice genome annotation, the Rice Annotation Project (RAP), with the aim of allowing more efficient analysis of genomic information and accelerating post-sequencing research activities. It is also hoped that the annotation will provide a comparative data resource for cereal genomics researchers working on other species and contribute to their endeavors.
To cope with the enormous amount of information produced by large-scale sequencing, several automated annotation methods have been developed for the purpose of efficient data processing. However, it is acknowledged that automated annotation alone tends to result in a high proportion of erroneous annotations, and therefore annotation data results should be carefully curated by experts before any public release in order to cut down on the amount of these erroneous annotations. Currently, manual curation remains a necessary process for developing an accurate biological database (Misra et al. 2002; Camon et al. 2003). With this in mind, we brought together a large group of specialists to curate the results of our automated rice gene functional assignment. By bringing individuals from complementary disciplines together, the amount of time required to perform the manual curation was significantly reduced.
There are a large number of full-length cDNAs and expressed sequence tags (ESTs) available for rice and other cereals (Fernandes et al. 2002; Wu et al. 2002; Kikuchi et al. 2003; Gardiner et al. 2004; Lai et al. 2004; Zhang et al. 2004; Jantasuriyarat et al. 2005). This wealth of information is a boon for genome annotation because it provides excellent support for transcribed regions, which, in turn, allows more precise predictions than current ab initio methods can provide. By using the annotation data set based on our curation and mapping of cDNAs to the genome, we were able to approximate the number of genes in the rice genome, classify transcribed sequences by probable function, and identify other features pertinent to the rice genome.
Arabidopsis thaliana is one of the most well-studied model organisms. Comparison of rice with the dicotyledon may assist in developing a greater understanding of intrinsic mechanisms among cereals at the molecular level. Use of knowledge accumulated about A. thaliana genes to quantify their counterparts in rice is one example of such a comparative study (Izawa et al. 2003; Yamaguchi et al. 2006). Additionally, clues as to the evolution of these two flowering plants could be obtained. Here we describe a comparative analysis of the genomes and protein sequences of O. sativa and A. thaliana on the basis of manually curated data. This analysis focuses on the number of genes, composition of functional domains, and patterns of gene duplication.
Results
Number of loci in the rice genome
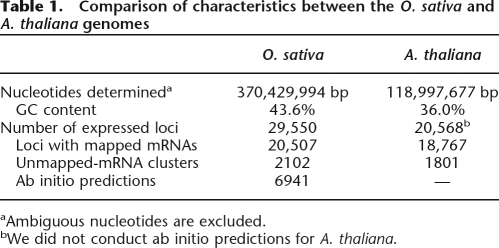
Early estimates of the total number of rice genes by various teams indicated that the rice genome probably contained between 40,000 and 60,000 protein-coding genes, many of which did not have any counterparts in the A. thaliana genome (Goff et al. 2002; Sasaki et al. 2002; Yu et al. 2002; Kikuchi et al. 2003; Bennetzen et al. 2004). However, it was later suggested that this figure had probably been overestimated because of inaccuracies inherent in the ab initio gene prediction methods used (Bennetzen et al. 2004; Jabbari et al. 2004). Thus, when making our own estimate, we chose to focus on loci for which gene identification was supported by evidence of transcription. In this report, we refer to cDNA sequences registered in the plant division of the DNA databanks as mRNAs and to those in the EST division as ESTs, while mRNAs and ESTs are collectively called cDNAs. Of the 34,887 O. sativa mRNAs available, taken from the DDBJ (DNA Data Bank of Japan) release 59, a subset of 32,127 (92%) was produced by the Rice Full-Length cDNA Consortium (Kikuchi et al. 2003), which we designate as FLcDNAs. Our mapping of the 34,887 mRNAs resulted in 20,507 predicted gene loci being successfully mapped to the current rice genome assembly (build 3). However, there were 2257 mRNAs that could not be mapped to the genome, and these were grouped into 2102 clusters (Table 1). Therefore, ∼93% of the mRNAs could be mapped onto the genome. For details about the unmapped mRNAs, see Supplemental Methods.
Table 1.
Comparison of characteristics between the O. sativa and A. thaliana genomes
aAmbiguous nucleotides are excluded.
bWe did not conduct ab initio predictions for A. thaliana.
Despite advances in the field of ab initio gene-finding methods, it still remains a challenge to accurately predict the location of genes and exons among the genomes of higher eukaryotes including flowering plants (Schoof and Karlowski 2003; Yao et al. 2005). A further complication comes from pseudogenes, which are in many cases indistinguishable from functional or transcribed genes, and as a result they may be predicted to be functional by ab initio methods. When we compared results obtained from ab initio prediction and cDNA mapping, our analysis indicated that 40,523 genes had been predicted by the ab initio methods but had not been covered by any rice mRNAs, whereas only 6941 (17.1%) of those predicted genes overlapped EST(s) from rice or cDNA(s) from other monocots. Therefore, these results suggest that caution is advised when estimating the total number of genes for an organism based merely on ab initio gene prediction.
Thus, we decided to use the 6941 predicted loci to which rice mRNAs were not mapped but other cDNAs could be (Table 1). We did not use the loci that were supported only by ESTs and were detected by neither the ab initio prediction programs nor the mRNA-mapping, because there seem to be a multitude of aberrant transcripts that were possibly experimental artifacts. As a result, the candidate loci of our data set could be classified into two types: identified transcripts with mRNA (FLcDNA) clones and predicted transcripts with cDNA support. The number of loci predicted for the rice genome in this study was 29,550 including the unmapped mRNA clusters (Table 1).
However, loci may exist that ab initio predictions failed to detect or for which no cDNAs have been sequenced. In fact, 1728 (8.4%) of the 20,507 mapped-mRNA loci were not predicted in our analysis, suggesting that, in addition to the 6941 predicted loci (Table 1), there may be a further 637 loci that were not predicted. Furthermore, 3298 (16.1%) of the mapped-mRNA loci were not supported by any other cDNAs, so that 1332 predicted loci might be absent from our data set. Finally, 122 loci that were neither predicted nor supported by cDNAs should be added. If we consider all of these predicted loci, the estimated number of transcribed loci in the rice genome becomes 31,641. Recent total gene estimates have suggested that there are between 38,000 and 40,000 genes in rice (Yu et al. 2005), and 37,544 protein-coding genes in the same genome assembly as we used (International Rice Genome Sequencing Project 2005). Our conservative estimation based on support by physical clones indicates that the O. sativa genome possesses a smaller number of genes, ∼32,000. If we were to consider only the protein-coding genes, this number may be reduced to 30,000 or less.
Comparison of transcript diversity between O. sativa and A. thaliana
The transcribed regions in the O. sativa genome were found to span ∼72 Mb for the predicted loci with mRNA support and ∼23 Mb for the predicted loci without mRNA support (Supplemental Table 2). When combined, these regions would encompass ∼100 Mb because the ab initio predictions of the protein-coding regions did not consider the 5′- and 3′-untranslated regions (UTRs). Hence, more than a quarter of the rice genome appears to contain transcribed regions, while about half of the A. thaliana genome appears to be transcribed (Supplemental Table 2). This difference can perhaps be explained by the presence of transposable elements inserted among the intergenic regions of rice, which are greater in number than in A. thaliana. Our results suggested that ∼30% of the rice genome was composed of repetitive elements (Supplemental Table 3), which concurs with the results of another study that suggested a figure of ∼35% (International Rice Genome Sequencing Project 2005). In contrast, merely ∼10% of the A. thaliana genome is estimated to be made up of transposons (The Arabidopsis Genome Initiative 2000). Moreover, it appears that introns and UTRs of rice may have accepted more transposon inserts than those of A. thaliana (Supplemental Fig. 1).
Primary gene structures were found to be quite similar between the two species examined. There were on average five exons per transcript. The proportion of single-exon genes was ∼20% (Supplemental Table 2), which is in contrast to the “minced” exon–intron structures that have been observed in mammals (Lander et al. 2001). The similar structures observed in both A. thaliana and rice imply that the gene structures in these two species have remained relatively stable since their estimated date of divergence more than 100 million years ago (Chaw et al. 2004).
Curation of ORF functions
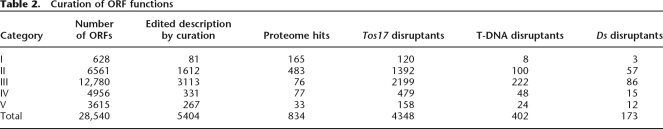
Of the 29,550 predicted loci with cDNA support, 28,540 were candidates for protein-coding genes. We could detect 834 open reading frames (ORFs) for which proteins were identified by comparison with the rice proteome data (Table 2; Komatsu et al. 2004; Komatsu and Tanaka 2005). The functions of these 28,540 proteins, which were inferred by BLASTX similarity searches against databases of proteins, were checked by manual curation. In order to avoid re-quotation of electronic annotation, a predicted function was only assigned to a sequence if the BLASTX search showed that the sequence had ≥50% identity to a protein in the database and that the protein in question had had its function confirmed experimentally (see Methods). Use of these guidelines led to 5404 (18.9%) of the automated functional predictions being altered during curation (Table 2). Curation therefore improves the accuracy levels of annotation for further experimental or computational studies.
Table 2.
Curation of ORF functions
The ORFs were classified into five categories according to their level of sequence similarity (see Methods). The probable protein products of 7189 loci had functions identified or inferred by BLASTX searches (Categories I and II of Table 2). Functional domains were detected in 12,780 ORFs (Category III) by InterProScan (Zdobnov and Apweiler 2001; Quevillon et al. 2005). In total, 70.0% of the protein functions could be inferred to a sufficient level of certainty by our curation methods.
For the remaining sequences, the functions could not be inferred, but similarity to proteins of unknown function in the databases was detected for Category IV proteins. Since the proteins of Category V did not show any homology with proteins contained in the databases, many of the sequences classified in this category may be novel. It is also suspected that this category may contain a high percentage of spurious ORFs, produced by false predictions (Das et al. 1997). In fact, the distribution of predicted sequence lengths indicates that both Categories IV and V, in general, have much shorter ORFs than those of the other categories (Supplemental Fig. 2). With this in mind, our estimate for the number of protein-coding genes may be slightly inflated by these possible false predictions.
Identification of non-protein-coding RNAs
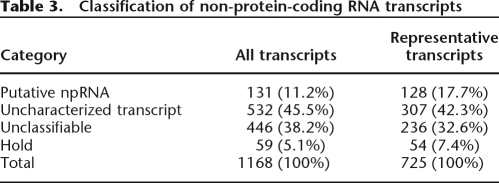
Over recent years, there has been a widely reported increase in the number of identified RNA transcripts with no apparent protein-coding potential (Huttenhofer et al. 2005; Sunkar et al. 2005). RNA genes play important roles in chromosomal silencing, transcriptional regulation, developmental control, and responses to stress (MacIntosh et al. 2001). The rice genome was reported to encode 763 transfer RNAs, 158 microRNAs, 215 small nucleolar RNAs, and 93 spliceosomal RNA genes (International Rice Genome Sequencing Project 2005). At the present time, no detailed study of RNA polymerase II-driven non-protein-coding RNAs (npRNAs) has been reported for rice. Analysis of the RAP data set identified 1168 transcripts that could be clustered into 725 predicted loci but either lacked an ORF or were predicted to encode a putative short peptide (≤80 amino acids). These transcripts were evaluated for various features such as exon structure, genomic context, canonical polyadenylation signal, polyadenosine tail, support by ESTs, and antisense transcripts (see Supplemental Methods). On the basis of these features, transcripts were classified into four categories: “npRNA” (putative non-protein-coding RNA), “uncharacterized transcript” (possible alternative 3′-UTR or isoform), “unclassifiable” (possible genomic fragment or incomplete transcript), and “hold” (transcripts that could not be mapped stringently to the rice genome) (Table 3).
Table 3.
Classification of non-protein-coding RNA transcripts
We identified 131 transcripts (11.2%) as putative npRNAs, and 108 of these were multi-exon transcripts with an average exon number of 2.8 (Supplemental Table 4). The remaining 23 npRNAs were single-exon transcripts with canonical 3′-end features and/or EST support. Interestingly, 55 putative npRNAs were found to overlap the exons and/or introns of sense genes (Supplemental Table 5) and may function as antisense npRNAs (as-npRNAs). For instance, the Os08g0103700 npRNA appears to overlap two predicted sense genes on the antisense strand. It overlaps the first exon of a BTP/POZ domain-containing protein (Os08g0103600) gene, and the last intron and exon of a NAM-like protein (Os08g0103900) gene (Supplemental Fig. 3). Previously, the NAC1 transcription factor gene, a member of the NAM family, was reported to be down-regulated by the small RNA gene miR164b (Guo et al. 2005). Control of both Os08g0103600 and Os08g0103900 via the RNAi (RNA interference) pathway seems very possible, and this potential relationship definitely warrants further study.
Most of the sense genes overlapped by as-npRNAs came under our classification of hypothetical proteins. However, using our annotation criteria, 27 predicted loci could be assigned a probable function. This set of candidates may constitute a good starting point for further analysis of plant as-npRNA mechanics.
Correlation between tRNA gene numbers and codon usage
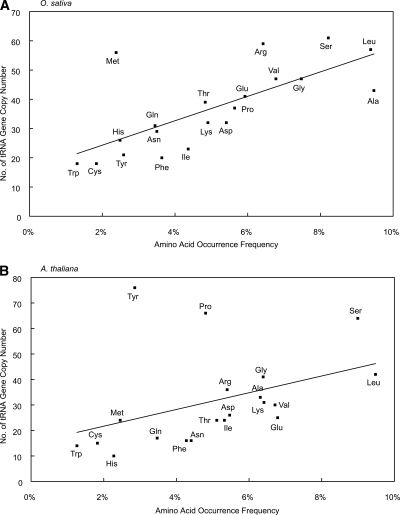
Isoacceptor tRNAs that correspond to frequently used codons are enriched for efficient translation in microbes, while such a tendency was not observed in higher eukaryotes such as chickens (Grantham et al. 1980; Ikemura 1981, 1985). It was later reported that the number of copies of isoacceptor tRNAs in the Caenorhabditis elegans genome correlated with the amount of relative synonymous codon usage (RSCU) (Duret 2000). This observation suggests that the abundance of a particular tRNA variety within the cell is proportional to the number of copies of that tRNA gene in the genome. Although the isoacceptor tRNA abundance has not been experimentally determined in rice, the relationship between the isoacceptor tRNAs and codon usage can be assessed by analyzing the complete genome sequence.
The number of isoacceptors in the rice genome was estimated on the basis of tRNAscan-SE predictions (Supplemental Table 6). First, we plotted the frequency of each amino acid obtained from the entire rice protein set against the number of corresponding tRNAs (Fig. 1A). We found a positive linear correlation between amino acid usage and the number of corresponding tRNA genes in the rice genome, which suggests that rice controls the expression of tRNAs vital for efficient protein synthesis via corresponding tRNA gene copy number, that is, tRNA gene copy numbers are proportional to individual amino acid biases. This is in contrast to current thinking that complex eukaryotes such as rice might have a complex gene regulation system. Moreover, the A. thaliana tRNA genes showed a similar pattern (Fig. 1B). Hence, it is strongly suggested that the tRNA abundance in both O. sativa and A. thaliana is determined simply by the number of gene copies rather than by complicated tRNA transcriptional regulation. Since the same tendency was found in C. elegans (Duret 2000), this type of tRNA control system may have been developed during the early stages of eukaryote evolution.
Figure 1.
Correlation between the number of tRNA gene copies and occurrence frequency of amino acids in (A) O. sativa and (B) A. thaliana. The horizontal axis indicates the occurrence frequency (%), and the vertical axis indicates the copy number. R2 values were 0.41 and 0.16 in O. sativa and A. thaliana, respectively.
Second, the numbers of isoacceptors and the RSCU were examined in rice, but a clear relationship between the two was not observed (Supplemental Table 6). It is currently thought that most tRNAs are modified after transcription, which allows two or more codons to be recognized by a single tRNA (Tranquilla et al. 1982). Additionally, synonymous codon usage variation in rice appears to be primarily due to mutational bias rather than natural selection (Liu et al. 2004). These factors may have led to masking of a correlation between the isoacceptors and RSCU. Nevertheless, in eight out of nine twofold degenerate codons, the major isoacceptor was the most abundant codon (Supplemental Table 6). Therefore, it is possible that an abundance of codon-specific tRNAs in the genome has influenced the evolution of the codon bias in rice, or vice versa.
Evolutionary process of the genes in O. sativa and A. thaliana
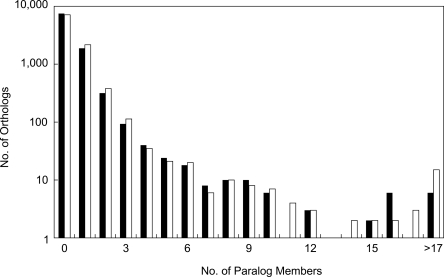
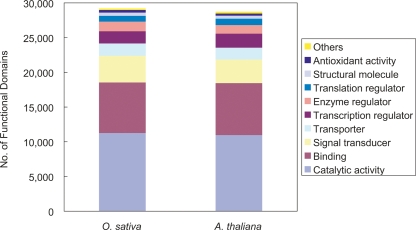
We compared the protein sets of O. sativa and A. thaliana by BLASTP and found that 9914 were possible ortholog pairs with an average evolutionary distance of 0.42 by p distance (Nei and Kumar 2000; Supplemental Fig. 4). The mean distance estimated by the Poisson-γ correction using a shape parameter of 2.25 was 0.70. In order to look for paralogous sequences that may have evolved after divergence from a common ancestor, when the paralogs were examined at the 5% significance level (see Methods), the estimated number of paralogs created after divergence from a common ancestor was 3828 in O. sativa and 4581 in A. thaliana, which corresponded to 0.39 and 0.46 duplication events on average, respectively. When detecting paralogs, we used a relatively conservative criterion. Therefore, if we included all the nonsignificant paralogs, our data indicated that O. sativa had acquired 5320 duplicate genes and A. thaliana had 5929. Although these observations indicate that the A. thaliana genome seems to have undergone more duplications than that of O. sativa, the duplications seem to be mainly attributable to a small number of large gene families having more than 30 members. The distribution of paralog clusters is similar between the two species (Fig. 2), which was unexpected because the two species seem to have experienced independent genome-wide duplication events (Lynch and Conery 2000; Yu et al. 2005). In fact, the process of genome evolution by duplication in each species appears to have been quite different (Supplemental Fig. 5). Functional classifications of the proteins based on Gene Ontology (GO) annotations are shown in Figure 3; the numbers of known functional domains were not significantly different between the two species. A similar tendency was observed in the frequencies of the top 40 InterPro IDs detected by the InterProScan search (Supplemental Table 7). Thus, as previously pointed out (Yu et al. 2002), it would appear that these distantly related flowering plants seem to share similar sets of known functional domains, although there may be several functionally important domains unique to each lineage and as yet uninvestigated.
Figure 2.
Number of orthologous clusters created by duplication after the speciation of O. sativa and A. thaliana. The vertical axis indicates the log-scaled number of orthologs (clusters), and the horizontal axis indicates the number of paralog members in a cluster. Black bars represent O. sativa, and white bars represent A. thaliana.
Figure 3.
Number of functional domains based on InterProScan and Gene Ontology. Note that the numbers do not correspond to the numbers of the proteins because a single protein can contain multiple domains.
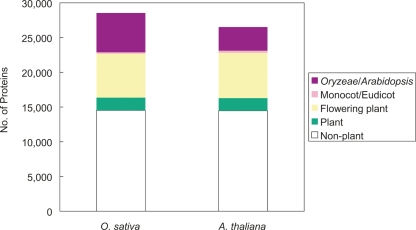
The protein sets still contained those lacking counterparts in the other species. In order to extensively examine the lineage-specific gene candidates for these proteins, all the proteins were compared with the UniProt Knowledgebase (UniProtKB). In both species, >14,000 proteins showed significant similarity to those obtained from nonplant species (Fig. 4), which implies that these have evolved so conservatively that the sequences did not alter drastically under strong purifying selection. In addition, the number of plant-specific homologs found in each species was similar, while there were several transcripts that were found to be specific to Oryzeae (5663 proteins) and Arabidopsis (3402 proteins) (Fig. 4). However, we could not rule out the possibility that these lineage-specific proteins were produced by false predictions of ORFs. Many of the lineage-specific proteins of rice could only be classified into Category V (Supplemental Fig. 6). The skewed length distribution of the Oryzeae-specific proteins (Supplemental Fig. 7) supported the hypothesis that there may be several bogus ORFs included in the Category V set, as noted in “Curation of ORF Functions.” The rice genome might contain a large number of species-specific short proteins, but it seems also possible that many of the transcripts unique to rice are non-protein-coding or are experimental artifacts. In addition, only a few monocotyledon- or eudicotyledon-specific proteins were detected (Fig. 4), suggesting that investigations into plant species other than O. sativa and A. thaliana, at the molecular level, may not have been as detailed as they could have been. Further DNA sequencing in a variety of plant species may reduce the number of apparent lineage-specific protein-coding genes found in this study.
Figure 4.
Result of similarity search of the O. sativa and A. thaliana proteins against the protein databases. The proteins were classified into five groups: homologous to nonplant, (nonflowering) plant, (nonmonocot/eudicot) flowering plant, (non-Oryzeae/Arabidopsis) monocot/eudicot, and Oryzeae/Arabidopsis-specific proteins.
Discussion
The curated annotation presented and described in this study revealed that the functions of 19,969 (70.0%) ORFs could be inferred by either sequence similarity or motif searches (Categories I, II, and III) (Table 2). Since we aimed to provide basic annotation only in this study, further functional assignment will be assisted in the future by sophisticated methods such as a tertiary structure-based approach (e.g., see McDermott and Samudrala 2003). Moreover, the insertional mutants that have been produced by several groups (Hirochika et al. 2004) will likely serve as an essential resource for the experimental validation of biological sequence annotations. We found that >4000 protein-coding regions were disrupted by Tos17 (Table 2), for which mutant lines are available (Miyao et al. 2003). Additionally, 402 T-DNA and 173 Ds insertion lines, respectively, were found to have disrupted protein-coding regions (Table 2). Those of the Category II genes will be the first targets in further functional analysis for experimental annotations. It is anticipated that mutant strains will also help us to investigate the functions of hypothetical proteins.
Most ORFs were predicted computationally. However, we could confirm the ORFs for 834 transcripts by comparison with the proteome data (Table 2). As the number of proteins directly determined by protein sequencing increases over time, we expect to be able to filter out a greater percentage of bogus ORFs from our data set. The proteome data will also provide experimentally validated evidence of any post-translational modifications, tissue-specificity, and cellular localization (Komatsu et al. 2004; Komatsu and Tanaka 2005), which will permit us to infer the functions of what we currently refer to as hypothetical proteins.
Since we focused on those genes that were validated by the cDNAs currently available and since the cDNA data set is incomplete, the estimated gene number may be regarded as a lower estimate. The presence of a transcript may not necessarily be used as the only criterion for identifying genes. In particular, there may exist a substantial number of rare transcripts or non-protein-coding genes that are currently undetected in rice. The experience in mice has shown that as more cDNA sequences were obtained, an increasingly large number of novel genes with no coding potential could be detected (Carninci et al. 2005). Future estimates could be validated by further experiments such as genome-wide tiling microarrays (Li et al. 2006).
Although we detected 5663 lineage-specific gene candidates in rice (Fig. 4), it is unlikely that all of them were newly derived from nonfunctional DNA sequences. There are several possibilities that could account for those genes that appear to be unique to rice. First, these genes may have diverged to such an extent that their homologs could no longer be detected by sequence similarity search. This is a probable scenario among the duplicated genes for which purifying selection is not strong. Second, independent gene deletions and insufficient data sampling could have led to an apparent uniqueness of genes (Salzberg et al. 2001; Stanhope et al. 2001). Indeed, we found 609 O. sativa ORFs that showed similarity to protein(s) in the protein databases, but that did not have any homolog(s) to gene products in the reported A. thaliana transcriptome. This finding suggests that these genes had been deleted during the evolutionary process leading to the creation of the A. thaliana we see today. A combination of multiple factors complicates efforts to trace back the genes’ ancestry. Finally, many of the “unique” genes seem to be non-protein-coding, as previously mentioned (Supplemental Fig. 7). Additionally, there may be other factors at work of which we are not aware, that may lead to the true number of functional RNAs being much higher than we estimated in this study (Table 3).
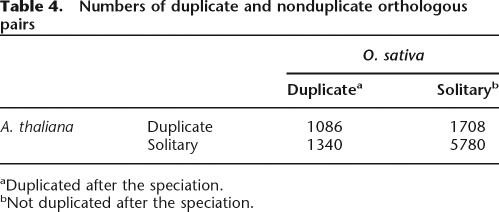
Since the distributions of gene duplicates were quite similar between O. sativa and A. thaliana (Fig. 2; Supplemental Fig. 6), there may be some common factor that accounts for this observed similarity. A probable candidate is natural selection enforcing limitations on the number of duplicate genes. If the duplication was selectively neutral, genes would be duplicated or remain as single copies at random in both species. In order to assess whether duplication in the two species was random, we calculated the ratio of those orthologs that have undergone intraspecific duplication events to those that have not, for both species (Table 4). We found that the numbers obtained were different from those that would be expected if duplication and deletion events had been random (P < 10−90, Fisher’s exact test). It seems that duplication of some genes may have been neutral or beneficial, while others were so deleterious that, if the gene was retained at all, it only remained as a single copy. Thus, the current gene composition of both O. sativa and A. thaliana seems to be partly due to natural selection, which shaped the similar genetic makeup of the genomes of these representative flowering plants.
Table 4.
Numbers of duplicate and nonduplicate orthologous pairs
aDuplicated after the speciation.
bNot duplicated after the speciation.
The nucleotide sequence of the genome is so vast as to make it unreadable to human eyes alone. A representation of the underlying biology that is comprehensible to humans can only be inferred through analytical programs. The high-quality automated annotation polished by extensive manual curation provides a more sharply focused view of the genome that we hope will allow more accurately targeted experimental work and comparative analysis.
Methods
Automated annotation
We used the genome sequence assembled by the International Rice Genome Sequencing Project (2005) and cDNAs registered in the International Nucleotide Sequence Databases. Genes and ORFs were identified or predicted by our custom-made annotation pipeline. For details, see the Supplemental Methods.
Curating ORF functional assignment
The ORFs were classified into five categories. When we discovered an ORF with a translated amino acid sequence identical to a protein from UniProtKB/SWISS-PROT, RefSeq, or Rice Proteome Database entry, this ORF was classified into the “known” protein group (Category I). If the sequence was ≥50% identical to a protein in the databases and the function of the protein was confirmed experimentally, the sequence was classified into the “similar” protein group (Category II). Sequences that contained conserved motifs in InterProScan hits were “functional-domain-containing” proteins (Category III). If the function of an amino acid sequence could not be inferred but the sequence showed ≥50% identity and ≥50% coverage of a hypothetical protein, the ORF was classified as a “conserved hypothetical” protein (Category IV). The others were “hypothetical” proteins (Category V).
If a locus contained more than one gene structure, curators selected one of them as a representative transcript by examining exon numbers and some other features (for details, see Imanishi et al. 2004). Next, curators thoroughly examined the automated functional annotations, focusing on descriptions and cited literature pertaining to homologs in the protein databases so that computationally predicted proteins were discarded for functional inference. The descriptions were only used for our annotation if the protein had been examined experimentally. Manual curation was assisted by our in-house system developed for the RAP.
All information regarding the ORF functions and gene positions in the genome can be downloaded at http://rapdownload.lab.nig.ac.jp/ (Ohyanagi et al. 2006). Locus IDs were designed as described in the Supplemental Methods. The annotations are also available under accession numbers AP008207–AP008218 in the International Nucleotide Sequence Databases (http://www.insdc.org/).
Comparison of the O. sativa and A. thaliana protein data sets
The O. sativa proteins determined in this paper were compared with the A. thaliana protein set of MIPS (http://mips.gsf.de/proj/plant/jsf/athal/download/index.jsp), which contains 26,521 ORFs deduced from the nuclear genome (Schoof et al. 2004). Each protein was subjected to BLASTP search against the combined protein data set of O. sativa and A. thaliana, and reciprocal best hits with E-values of <10−5 were taken as possible orthologs between the two species. Paralogs derived in each lineage before or after their divergence were determined by the following method. A paralog of O. sativa detected by BLASTP was aligned together with the ortholog pair by using CLUSTALW (Thompson et al. 1994). The evolutionary distance between each pairing of these three sequences was estimated by the Poisson-γ correction with the shape parameter of 2.25, which approximately corresponds to Dayhoff’s correction (Nei and Kumar 2000). Let dOA be the distance between the orthologs of O. sativa and A. thaliana and dOOp be the distance between the O. sativa paralogs. If the molecular clock hypothesis holds true and the paralog was derived after the two species diverged, the branch leading to the A. thaliana ortholog should be longer than the others. The difference, D, between dOOp and dOA is defined by:
The variance of D, V(D), can be computed as follows:
For the methods of estimating the variance and covariance of the Poisson-γ correction, see Ota and Nei (1994). If D is significantly smaller than 0 by the Z test, the paralog was derived after divergence from the common ancestor (Nei and Kumar 2000), and thus was assigned as a lineage-specific duplicate.
Acknowledgments
We are grateful to C. Robin Buell, Hisakazu Iwama, Satoshi Fukuchi, Craig Gough, Kumiko Suzuki, Junko Sugiyama, Emiko Saito, Masato Kawabata, Chikatada Satoh, Shigetoyo Furukawa, Satoshi Nobushima, Ryo Aono, Tomohiro Endo, and Michitoshi Nagamochi for their support. We thank all the participants of the First Rice Annotation Project Meeting (RAP1). We also thank the Computer Center for Agriculture, Forestry and Fisheries Research for assisting RAP1. This work was supported by a grant from the Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5509507
References
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L., Coleman C., Liu R., Ma J., Ramakrishna W., Coleman C., Liu R., Ma J., Ramakrishna W., Liu R., Ma J., Ramakrishna W., Ma J., Ramakrishna W., Ramakrishna W. Consistent over-estimation of gene number in complex plant genomes. Curr. Opin. Plant Biol. 2004;7:732–736. doi: 10.1016/j.pbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Camon E., Magrane M., Barrell D., Binns D., Fleischmann W., Kersey P., Mulder N., Oinn T., Maslen J., Cox A., Magrane M., Barrell D., Binns D., Fleischmann W., Kersey P., Mulder N., Oinn T., Maslen J., Cox A., Barrell D., Binns D., Fleischmann W., Kersey P., Mulder N., Oinn T., Maslen J., Cox A., Binns D., Fleischmann W., Kersey P., Mulder N., Oinn T., Maslen J., Cox A., Fleischmann W., Kersey P., Mulder N., Oinn T., Maslen J., Cox A., Kersey P., Mulder N., Oinn T., Maslen J., Cox A., Mulder N., Oinn T., Maslen J., Cox A., Oinn T., Maslen J., Cox A., Maslen J., Cox A., Cox A., et al. The Gene Ontology Annotation (GOA) Project: Implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 2003;13:662–672. doi: 10.1101/gr.461403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chaw S.M., Chang C.C., Chen H.L., Li W.H., Chang C.C., Chen H.L., Li W.H., Chen H.L., Li W.H., Li W.H. Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- Das S., Yu L., Gaitatzes C., Rogers R., Freeman J., Bienkowska J., Adams R.M., Smith T.F., Lindelien J., Yu L., Gaitatzes C., Rogers R., Freeman J., Bienkowska J., Adams R.M., Smith T.F., Lindelien J., Gaitatzes C., Rogers R., Freeman J., Bienkowska J., Adams R.M., Smith T.F., Lindelien J., Rogers R., Freeman J., Bienkowska J., Adams R.M., Smith T.F., Lindelien J., Freeman J., Bienkowska J., Adams R.M., Smith T.F., Lindelien J., Bienkowska J., Adams R.M., Smith T.F., Lindelien J., Adams R.M., Smith T.F., Lindelien J., Smith T.F., Lindelien J., Lindelien J. Biology’s new Rosetta stone. Nature. 1997;385:29–30. doi: 10.1038/385029a0. [DOI] [PubMed] [Google Scholar]
- Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- Fernandes J., Brendel V., Gai X., Lal S., Chandler V.L., Elumalai R.P., Galbraith D.W., Pierson E.A., Walbot V., Brendel V., Gai X., Lal S., Chandler V.L., Elumalai R.P., Galbraith D.W., Pierson E.A., Walbot V., Gai X., Lal S., Chandler V.L., Elumalai R.P., Galbraith D.W., Pierson E.A., Walbot V., Lal S., Chandler V.L., Elumalai R.P., Galbraith D.W., Pierson E.A., Walbot V., Chandler V.L., Elumalai R.P., Galbraith D.W., Pierson E.A., Walbot V., Elumalai R.P., Galbraith D.W., Pierson E.A., Walbot V., Galbraith D.W., Pierson E.A., Walbot V., Pierson E.A., Walbot V., Walbot V. Comparison of RNA expression profiles based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Physiol. 2002;128:896–910. doi: 10.1104/pp.010681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner J., Schroeder S., Polacco M.L., Sanchez-Villeda H., Fang Z., Morgante M., Landewe T., Fengler K., Useche F., Hanafey M., Schroeder S., Polacco M.L., Sanchez-Villeda H., Fang Z., Morgante M., Landewe T., Fengler K., Useche F., Hanafey M., Polacco M.L., Sanchez-Villeda H., Fang Z., Morgante M., Landewe T., Fengler K., Useche F., Hanafey M., Sanchez-Villeda H., Fang Z., Morgante M., Landewe T., Fengler K., Useche F., Hanafey M., Fang Z., Morgante M., Landewe T., Fengler K., Useche F., Hanafey M., Morgante M., Landewe T., Fengler K., Useche F., Hanafey M., Landewe T., Fengler K., Useche F., Hanafey M., Fengler K., Useche F., Hanafey M., Useche F., Hanafey M., Hanafey M., et al. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 2004;134:1317–1326. doi: 10.1104/pp.103.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., Glazebrook J., Sessions A., Oeller P., Varma H., Sessions A., Oeller P., Varma H., Oeller P., Varma H., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pave A., Gautier C., Gouy M., Mercier R., Pave A., Gouy M., Mercier R., Pave A., Mercier R., Pave A., Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H.S., Xie Q., Fei J.F., Chua N.H., Xie Q., Fei J.F., Chua N.H., Fei J.F., Chua N.H., Chua N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., Guiderdoni E., An G., Hsing Y.I., Eun M.Y., Han C.D., Upadhyaya N., Ramachandran S., Zhang Q., Pereira A., Guiderdoni E., An G., Hsing Y.I., Eun M.Y., Han C.D., Upadhyaya N., Ramachandran S., Zhang Q., Pereira A., An G., Hsing Y.I., Eun M.Y., Han C.D., Upadhyaya N., Ramachandran S., Zhang Q., Pereira A., Hsing Y.I., Eun M.Y., Han C.D., Upadhyaya N., Ramachandran S., Zhang Q., Pereira A., Eun M.Y., Han C.D., Upadhyaya N., Ramachandran S., Zhang Q., Pereira A., Han C.D., Upadhyaya N., Ramachandran S., Zhang Q., Pereira A., Upadhyaya N., Ramachandran S., Zhang Q., Pereira A., Ramachandran S., Zhang Q., Pereira A., Zhang Q., Pereira A., Pereira A., et al. Rice mutant resources for gene discovery. Plant Mol. Biol. 2004;54:325–334. doi: 10.1023/B:PLAN.0000036368.74758.66. [DOI] [PubMed] [Google Scholar]
- Huttenhofer A., Schattner P., Polacek N., Schattner P., Polacek N., Polacek N. Non-coding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Imanishi T., Itoh T., Suzuki Y., O’Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Itoh T., Suzuki Y., O’Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Suzuki Y., O’Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., O’Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., Tamura T., Yamaguchi-Kabata Y., Tanino M., Yamaguchi-Kabata Y., Tanino M., Tanino M., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:856–875. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Izawa T., Takahashi Y., Yano M., Takahashi Y., Yano M., Yano M. Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr. Opin. Plant Biol. 2003;6:113–120. doi: 10.1016/s1369-5266(03)00014-1. [DOI] [PubMed] [Google Scholar]
- Jabbari K., Cruveiller S., Clay O., Le Saux J., Bernardi G., Cruveiller S., Clay O., Le Saux J., Bernardi G., Clay O., Le Saux J., Bernardi G., Le Saux J., Bernardi G., Bernardi G. The new genes of rice: A closer look. Trends Plant Sci. 2004;9:281–285. doi: 10.1016/j.tplants.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Jantasuriyarat C., Gowda M., Haller K., Hatfield J., Lu G., Stahlberg E., Zhou B., Li H., Kim H., Yu Y., Gowda M., Haller K., Hatfield J., Lu G., Stahlberg E., Zhou B., Li H., Kim H., Yu Y., Haller K., Hatfield J., Lu G., Stahlberg E., Zhou B., Li H., Kim H., Yu Y., Hatfield J., Lu G., Stahlberg E., Zhou B., Li H., Kim H., Yu Y., Lu G., Stahlberg E., Zhou B., Li H., Kim H., Yu Y., Stahlberg E., Zhou B., Li H., Kim H., Yu Y., Zhou B., Li H., Kim H., Yu Y., Li H., Kim H., Yu Y., Kim H., Yu Y., Yu Y., et al. Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction. Plant Physiol. 2005;138:105–115. doi: 10.1104/pp.104.055624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., Yazaki J., Ishikawa M., Yamada H., Ooka H., Ishikawa M., Yamada H., Ooka H., Yamada H., Ooka H., Ooka H., et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Komatsu S., Tanaka N., Tanaka N. Rice proteome analysis: A step toward functional analysis of the rice genome. Proteomics. 2005;5:938–949. doi: 10.1002/pmic.200401040. [DOI] [PubMed] [Google Scholar]
- Komatsu S., Kojima K., Suzuki K., Ozaki K., Higo K., Kojima K., Suzuki K., Ozaki K., Higo K., Suzuki K., Ozaki K., Higo K., Ozaki K., Higo K., Higo K. Rice Proteome Database based on two-dimensional polyacrylamide gel electrophoresis: Its status in 2003. Nucleic Acids Res. 2004;32:D388–D392. doi: 10.1093/nar/gkh020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J., Dey N., Kim C.-S., Bharti A.K., Rudd S., Mayer K.F.X., Larkins B.A., Becraft P., Messing J., Dey N., Kim C.-S., Bharti A.K., Rudd S., Mayer K.F.X., Larkins B.A., Becraft P., Messing J., Kim C.-S., Bharti A.K., Rudd S., Mayer K.F.X., Larkins B.A., Becraft P., Messing J., Bharti A.K., Rudd S., Mayer K.F.X., Larkins B.A., Becraft P., Messing J., Rudd S., Mayer K.F.X., Larkins B.A., Becraft P., Messing J., Mayer K.F.X., Larkins B.A., Becraft P., Messing J., Larkins B.A., Becraft P., Messing J., Becraft P., Messing J., Messing J. Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Res. 2004;14:1932–1937. doi: 10.1101/gr.2780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li L., Wang X., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Wang X., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Stolc V., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Li X., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Zhang D., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Su N., Tongprasit W., Li S., Cheng Z., Wang J., Tongprasit W., Li S., Cheng Z., Wang J., Li S., Cheng Z., Wang J., Cheng Z., Wang J., Wang J., et al. Genome-wide transcription analyses in rice using tiling microarrays. Nat. Genet. 2006;38:124–129. doi: 10.1038/ng1704. [DOI] [PubMed] [Google Scholar]
- Liu Q., Feng Y., Zhao X.A., Dong H., Xue Q., Feng Y., Zhao X.A., Dong H., Xue Q., Zhao X.A., Dong H., Xue Q., Dong H., Xue Q., Xue Q. Synonymous codon usage bias in Oryza sativa. Plant Sci. 2004;167:101–105. [Google Scholar]
- Lynch M., Conery J.S., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- MacIntosh G.C., Wilkerson C., Green P.J., Wilkerson C., Green P.J., Green P.J. Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol. 2001;127:765–776. [PMC free article] [PubMed] [Google Scholar]
- McDermott J., Samudrala R., Samudrala R. Bioverse: Functional, structural and contextual annotation of proteins and proteomes. Nucleic Acids Res. 2003;31:3736–3737. doi: 10.1093/nar/gkg550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S., Crosby M., Mungall C., Matthews B., Campbell K., Hradecky P., Huang Y., Kaminker J., Millburn G., Prochnik S., Crosby M., Mungall C., Matthews B., Campbell K., Hradecky P., Huang Y., Kaminker J., Millburn G., Prochnik S., Mungall C., Matthews B., Campbell K., Hradecky P., Huang Y., Kaminker J., Millburn G., Prochnik S., Matthews B., Campbell K., Hradecky P., Huang Y., Kaminker J., Millburn G., Prochnik S., Campbell K., Hradecky P., Huang Y., Kaminker J., Millburn G., Prochnik S., Hradecky P., Huang Y., Kaminker J., Millburn G., Prochnik S., Huang Y., Kaminker J., Millburn G., Prochnik S., Kaminker J., Millburn G., Prochnik S., Millburn G., Prochnik S., Prochnik S., et al. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0083. research0083.1–0083.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A., Tanaka K., Murata K., Sawaki H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H., Tanaka K., Murata K., Sawaki H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H., Murata K., Sawaki H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H., Sawaki H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H., Takeda S., Abe K., Shinozuka Y., Onosato K., Hirochika H., Abe K., Shinozuka Y., Onosato K., Hirochika H., Shinozuka Y., Onosato K., Hirochika H., Onosato K., Hirochika H., Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Kumar S., Kumar S. Molecular evolution and phylogenetics. Oxford University Press; Oxford: 2000. [Google Scholar]
- Ohyanagi H., Tanaka T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Tanaka T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., Antonio B.A., Nagamura Y., Imanishi T., Nagamura Y., Imanishi T., Imanishi T., et al. The Rice Annotation Project Database (RAP-DB): Hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., Nei M., Nei M. Estimation of the number of amino-acid substitutions per site when the substitution rate varies among sites. J. Mol. Evol. 1994;38:642–643. doi: 10.1007/BF00175826. [DOI] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R., Harte N., Mulder N., Apweiler R., Lopez R., Mulder N., Apweiler R., Lopez R., Apweiler R., Lopez R., Lopez R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S.L., White O., Peterson J., Eisen J.A., White O., Peterson J., Eisen J.A., Peterson J., Eisen J.A., Eisen J.A. Microbial genes in the human genome: Lateral transfer or gene loss? Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Matsumoto T., Yamamoto K., Sakata K., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Matsumoto T., Yamamoto K., Sakata K., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Yamamoto K., Sakata K., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Sakata K., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., Niimura Y., Cheng Z., Nagamura Y., Cheng Z., Nagamura Y., Nagamura Y., et al. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–316. doi: 10.1038/nature01184. [DOI] [PubMed] [Google Scholar]
- Schoof H., Karlowski W.M., Karlowski W.M. Comparison of rice and Arabidopsis annotation. Curr. Opin. Plant Biol. 2003;6:106–112. doi: 10.1016/s1369-5266(03)00003-7. [DOI] [PubMed] [Google Scholar]
- Schoof H., Ernst R., Nazarov V., Pfeifer L., Mewes H.W., Mayer K.F., Ernst R., Nazarov V., Pfeifer L., Mewes H.W., Mayer K.F., Nazarov V., Pfeifer L., Mewes H.W., Mayer K.F., Pfeifer L., Mewes H.W., Mayer K.F., Mewes H.W., Mayer K.F., Mayer K.F. MIPS Arabidopsis thaliana Database (MAtDB): An integrated biological knowledge resource for plant genomics. Nucleic Acids Res. 2004;32:D373–D376. doi: 10.1093/nar/gkh068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope M.J., Lupas A., Italia M.J., Koretke K.K., Volker C., Brown J.R., Lupas A., Italia M.J., Koretke K.K., Volker C., Brown J.R., Italia M.J., Koretke K.K., Volker C., Brown J.R., Koretke K.K., Volker C., Brown J.R., Volker C., Brown J.R., Brown J.R. Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature. 2001;411:940–944. doi: 10.1038/35082058. [DOI] [PubMed] [Google Scholar]
- Sunkar R., Girke T., Jain P.K., Zhu J.K., Girke T., Jain P.K., Zhu J.K., Jain P.K., Zhu J.K., Zhu J.K. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J., Higgins D.G., Gibson T.J., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranquilla T.A., Cortese R., Melton D., Smith J.D., Cortese R., Melton D., Smith J.D., Melton D., Smith J.D., Smith J.D. Sequences of four tRNA genes from Caenorhabditis elegans and the expression of C. elegans tRNALeu (anticodon IAG) in Xenopus oocytes. Nucleic Acids Res. 1982;10:7919–7934. doi: 10.1093/nar/10.24.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Maehara T., Shimokawa T., Yamamoto S., Harada C., Takazaki Y., Ono N., Mukai Y., Koike K., Yazaki J., Maehara T., Shimokawa T., Yamamoto S., Harada C., Takazaki Y., Ono N., Mukai Y., Koike K., Yazaki J., Shimokawa T., Yamamoto S., Harada C., Takazaki Y., Ono N., Mukai Y., Koike K., Yazaki J., Yamamoto S., Harada C., Takazaki Y., Ono N., Mukai Y., Koike K., Yazaki J., Harada C., Takazaki Y., Ono N., Mukai Y., Koike K., Yazaki J., Takazaki Y., Ono N., Mukai Y., Koike K., Yazaki J., Ono N., Mukai Y., Koike K., Yazaki J., Mukai Y., Koike K., Yazaki J., Koike K., Yazaki J., Yazaki J., et al. A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell. 2002;14:525–535. doi: 10.1105/tpc.010274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Lee D.Y., Miyao A., Hirochika H., An G., Hirano H.-Y., Lee D.Y., Miyao A., Hirochika H., An G., Hirano H.-Y., Miyao A., Hirochika H., An G., Hirano H.-Y., Hirochika H., An G., Hirano H.-Y., An G., Hirano H.-Y., Hirano H.-Y. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell. 2006;18:15–28. doi: 10.1105/tpc.105.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Guo L., Fu Y., Borsuk L.A., Wen T.J., Skibbe D.S., Cui X., Scheffler B.E., Cao J., Emrich S.J., Guo L., Fu Y., Borsuk L.A., Wen T.J., Skibbe D.S., Cui X., Scheffler B.E., Cao J., Emrich S.J., Fu Y., Borsuk L.A., Wen T.J., Skibbe D.S., Cui X., Scheffler B.E., Cao J., Emrich S.J., Borsuk L.A., Wen T.J., Skibbe D.S., Cui X., Scheffler B.E., Cao J., Emrich S.J., Wen T.J., Skibbe D.S., Cui X., Scheffler B.E., Cao J., Emrich S.J., Skibbe D.S., Cui X., Scheffler B.E., Cao J., Emrich S.J., Cui X., Scheffler B.E., Cao J., Emrich S.J., Scheffler B.E., Cao J., Emrich S.J., Cao J., Emrich S.J., Emrich S.J., et al. Evaluation of five ab initio gene prediction programs for the discovery of maize genes. Plant Mol. Biol. 2005;57:445–460. doi: 10.1007/s11103-005-0271-1. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Deng Y., Dai L., Zhou Y., Zhang X., Dai L., Zhou Y., Zhang X., Zhou Y., Zhang X., Zhang X., et al. 2002A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science 29679 –92. [DOI] [PubMed] [Google Scholar]
- Yu J., Wang J., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Wang J., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., Zhou J., Ni P., Dong W., Hu S., Zeng C., Ni P., Dong W., Hu S., Zeng C., Dong W., Hu S., Zeng C., Hu S., Zeng C., Zeng C., et al. The genomes of Oryza sativa: A history of duplications. PLoS. Biol. 2005;3:266–281. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R., Apweiler R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zhang D., Choi D.W., Wanamaker S., Fenton R.D., Chin A., Malatrasi M., Turuspekov Y., Walia H., Akhunov E.D., Kianian P., Choi D.W., Wanamaker S., Fenton R.D., Chin A., Malatrasi M., Turuspekov Y., Walia H., Akhunov E.D., Kianian P., Wanamaker S., Fenton R.D., Chin A., Malatrasi M., Turuspekov Y., Walia H., Akhunov E.D., Kianian P., Fenton R.D., Chin A., Malatrasi M., Turuspekov Y., Walia H., Akhunov E.D., Kianian P., Chin A., Malatrasi M., Turuspekov Y., Walia H., Akhunov E.D., Kianian P., Malatrasi M., Turuspekov Y., Walia H., Akhunov E.D., Kianian P., Turuspekov Y., Walia H., Akhunov E.D., Kianian P., Walia H., Akhunov E.D., Kianian P., Akhunov E.D., Kianian P., Kianian P., et al. Construction and evaluation of cDNA libraries for large-scale expressed sequence tag sequencing in wheat (Triticum aestivum L.) Genetics. 2004;168:595–608. doi: 10.1534/genetics.104.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]