Abstract
Mycobacterium ulcerans is found in aquatic ecosystems and causes Buruli ulcer in humans, a neglected but devastating necrotic disease of subcutaneous tissue that is rampant throughout West and Central Africa. Here, we report the complete 5.8-Mb genome sequence of M. ulcerans and show that it comprises two circular replicons, a chromosome of 5632 kb and a virulence plasmid of 174 kb. The plasmid is required for production of the polyketide toxin mycolactone, which provokes necrosis. Comparisons with the recently completed 6.6-Mb genome of Mycobacterium marinum revealed >98% nucleotide sequence identity and genome-wide synteny. However, as well as the plasmid, M. ulcerans has accumulated 213 copies of the insertion sequence IS2404, 91 copies of IS2606, 771 pseudogenes, two bacteriophages, and multiple DNA deletions and rearrangements. These data indicate that M. ulcerans has recently evolved via lateral gene transfer and reductive evolution from the generalist, more rapid-growing environmental species M. marinum to become a niche-adapted specialist. Predictions based on genome inspection for the production of modified mycobacterial virulence factors, such as the highly abundant phthiodiolone lipids, were confirmed by structural analyses. Similarly, 11 protein-coding sequences identified as M. ulcerans-specific by comparative genomics were verified as such by PCR screening a diverse collection of 33 strains of M. ulcerans and M. marinum. This work offers significant insight into the biology and evolution of mycobacterial pathogens and is an important component of international efforts to counter Buruli ulcer.
In 1935, two rural medical practitioners reported a series of unusual indolent skin ulcers from patients within a remote farming community in the Bairnsdale district of Southeastern Australia (Alsop 1972). Thirteen years later, a team of Australian researchers discovered the etiologic agent of “Bairnsdale ulcer,” a hitherto unknown Mycobacterium that they named Mycobacterium ulcerans (MacCallum et al. 1948). During the 1960s, many cases were reported from the Buruli County in Uganda; hence, the disease became more generally known as Buruli ulcer. The disease occurs in other parts of the world, but impoverished rural communities of West and Central Africa are worst affected. Since 1989, the disease burden has steadily increased, and the prevalence of Buruli ulcer now exceeds that of leprosy and, in some instances, tuberculosis (Johnson et al. 2005).
The epidemiology of Buruli ulcer is poorly understood, and outbreaks are sporadic and unpredictable; however, proximity to stagnant or slow-flowing watercourses is a recognized risk factor. M. ulcerans is associated with aquatic vegetation such as algae (Marsollier et al. 2004b); snails and other organisms that feed on algae may serve as passive hosts (Marsollier et al. 2004a). One report has shown that M. ulcerans can multiply in the salivary glands of carnivorous water bugs, such as Naucoris cimicoides (Marsollier et al. 2002). It is conceivable that humans become infected through contact with contaminated Naucoridae or bites, as this route of infection has been demonstrated in a murine model of the disease (Marsollier et al. 2002).
The extensive subcutaneous necrosis that results in the typical pathology of Buruli ulcer stems from the multiple actions of a lipophilic macrolide toxin named mycolactone. Recently, a virulence plasmid, pMUM001, was discovered in M. ulcerans (Stinear et al. 2004, 2005b), and this encodes three giant polyketide synthases (PKS) required for mycolactone synthesis. Injection of purified mycolactone replicates the human disease in rodent and cellular models. Mycolactone is cytotoxic, has anti-phagocytic activity, induces apoptosis of antigen-presenting cells, and inhibits the pro-inflammatory cytokine response (Coutanceau et al. 2005). As a consequence, from a relatively early stage in disease M. ulcerans is found primarily in an extracellular location in mammalian tissues, in contrast to other pathogenic mycobacteria (Coutanceau et al. 2005). Intriguingly, mycolactone is required for lysis of the plasmatocytes that transport the bacterium away from the coelomic cavity in N. cimicoides (Marsollier et al. 2005), suggesting that mycolactone may have an important role in the adaptation of M. ulcerans to an arthropod niche.
Phylogenetic relationships inferred from limited multi-locus sequence comparisons suggest that M. ulcerans has recently diverged from a Mycobacterium marinum progenitor (Stinear et al. 2000). M. marinum does not contain pMUM001 and makes no mycolactone, but causes a tuberculoid-like disease in fish and frogs and, occasionally, a limited cutaneous infection in humans characterized by intracellular multiplication and a granulomatous host response. Unlike M. ulcerans, M. marinum replicates in 4 not 50 h, produces light-inducible carotenoid pigments, and can utilize glucose, acetate, succinate, and pyruvate as sole carbon sources.
Currently, there is no vaccine to prevent Buruli ulcer and, owing to the painless nature of the lesions, patients often refer to clinicians so late that surgical excision followed by skin grafts is the only effective recourse. Treatment with rifampin and streptomycin shows some efficacy but has not yet been validated by a controlled clinical trial (Etuaful et al. 2005). A recent resolution adopted by the World Health Assembly called for intensified research to develop tools to diagnose, treat, and prevent Buruli ulcer (World Health Organization, http://www.who.int/buruli/en/). The genome sequence of M. ulcerans described herein provides an important resource for addressing these aims.
Results
General features
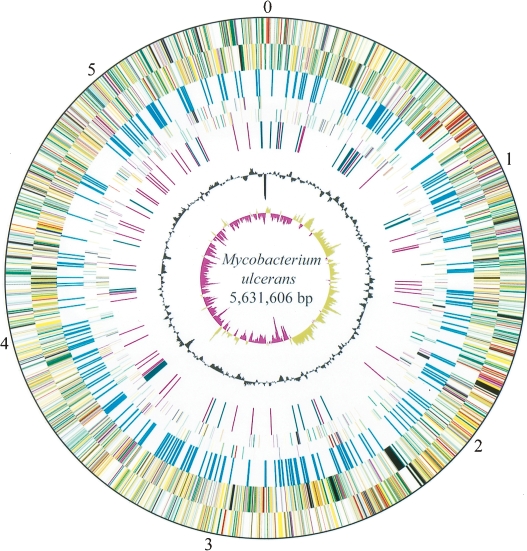
The genome comprises 5,805,761 bp and is composed of two circular replicons, a 5,631,606-bp chromosome (Fig. 1) and a 174,155-bp plasmid, pMUM001. The plasmid, which has an average G+C content of 62.5%, contains 81 protein-coding sequences (CDS) and encodes the polyketide synthases and polyketide-modifying enzymes that are required to produce mycolactone (Stinear et al. 2004, 2005a, b).
Figure 1.
Circular representation of the Mycobacterium ulcerans chromosome. The scale is shown in megabases in the outer black circle. Moving inward, the next two circles show forward and reverse strand CDS, respectively, with colors representing the functional classification (red, replication; light blue, regulation; light green, hypothetical protein; dark green, cell wall and cell processes; orange, conserved hypothetical protein; cyan, IS elements; yellow, intermediate metabolism; gray, lipid metabolism; purple, PE/PPE). The location of each copy of IS2404 and IS2606 is then shown (cyan). The following two circles show forward and reverse strand pseudogenes (colors represent the functional classification), followed by the G+C content and finally the GC skew (G−C)/(G+C) using a 20-kb window.
The chromosome contains 4160 CDS and 771 pseudogenes, and functional information could be attributed to nearly 70% of these. There are 45 genes encoding tRNA and a single rRNA operon located 1.2 Mb from oriC. The chromosome harbors two prophage, phiMU01 and phiMU02, and 302 insertion sequence elements (ISE), including 209 complete or partial copies of IS2404 and 83 copies of IS2606 (Table 1; Fig. 1; Stinear et al. 1999). Cumulative GC skew analysis demonstrated a bias in G+C content between the leading and lagging strands, indicating a probable origin of replication around dnaA; however, deviations in this bias were also evident (Fig. 1), suggesting the recent occurrence of several large chromosomal rearrangements.
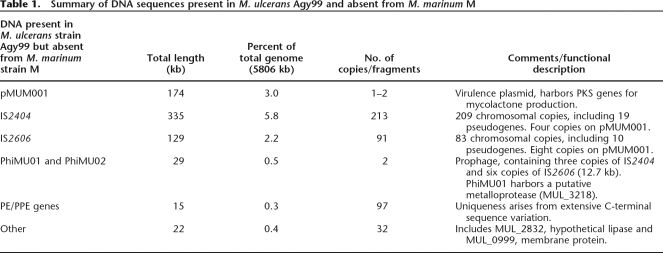
Table 1.
Summary of DNA sequences present in M. ulcerans Agy99 and absent from M. marinum M
Genome comparisons reveal the origin of M. ulcerans
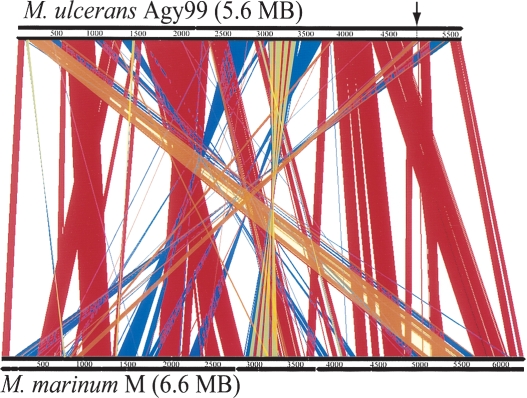
To gain insight into these rearrangements, the recently completed 6,636,827-bp genome sequence of M. marinum, with its 5426 CDS, was compared with that of M. ulcerans (Fig. 2). This revealed the existence of many deletions, accounting for 1064 kb, and many DNA rearrangements in M. ulcerans; some of these are asymmetrical around the origin and terminus of replication (Figs. 1, 2). Symmetrical chromosome rearrangements are commonly seen in bacterial comparisons (Eisen et al. 2000); asymmetric rearrangements that switch genes from the leading to the lagging strands are uncommon, but have been reported in bacteria with large numbers of ISE. Close inspection reveals that in M. ulcerans at least 12 of the rearrangements shown in Figure 2 have resulted in transfer to the lagging strand of 273 CDS with leading strand orthologs in M. marinum and M. tuberculosis, and some of these CDS are predicted to have key roles in cellular metabolism. For example, a 71-kb fragment spanning the region 5,032,986 to 5,104,623 in M. ulcerans has been relocated from a region around 945,816 to 1,012,490 in M. marinum (Fig. 2). CDS now on the lagging strand in this 71-kb region include (1) murB (MUL_4552), encoding a reductase required for peptidoglycan biosynthesis; (2) icl (MUL_4536), encoding isocitrate lyase, a key enzyme of the glyoxylate shunt; (3) proC (MUL_4570), encoding a carboxylate reductase that is an essential gene in M. tuberculosis required for proline biosynthesis; and (4) the hemACDB operon (MUL_4598 to MUL_4561), a group of essential genes in M. tuberculosis required for porphyrin biosynthesis (Sassetti et al. 2001). Such exchanges of CDS involved in cell wall biosynthesis and carbon, amino acid, and cofactor metabolism to the lagging strand might explain in part the slow growth of M. ulcerans, as transcription of these (and other) potentially essential genes could be slowed or disrupted by competition between DNA and RNA polymerases on the lagging strand at the replication forks (Rocha and Danchin 2003). However, it is important to note that chromosome remodeling in M. ulcerans has not resulted in a significant net change in strand coding bias as 239 CDS have moved from lagging to leading strand.
Figure 2.
Diagram depicting the rearrangements in, and deletions from, the genome of M. ulcerans compared with M. marinum. The figure shows a linear genomic comparison generated with ACT (Carver et al. 2005). (Red lines) Regions of DNA:DNA identity, (blue lines) inverted regions, (orange) strand-displacing rearrangements, (arrow) position of the displaced 71-kb fragment.
Further comparisons between the genomes of M. ulcerans and M. marinum confirmed a very close relationship between these two species and show that M. ulcerans has recently evolved from M. marinum. Approximately 90% of the M. ulcerans CDS have syntenic orthologs in M. marinum with an average DNA identity level of 98.3% (excluding ISE and PE/PPE genes). This compares with only 60% of M. ulcerans CDS that have an M. tuberculosis ortholog and an average DNA identity of 78.5%.
Identification of M. ulcerans-specific DNA
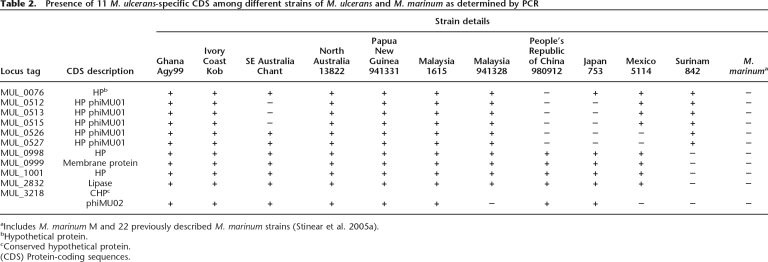
Many regions of difference are due to insertions and deletions in both species (Supplemental Table S1). In addition to pMUM001, 475 kb of DNA was identified in M. ulcerans that is absent from M. marinum, and this is distributed throughout the chromosome (Fig. 1; Table 1). Most of this is accounted for by IS2404 and IS2606. The remaining 79 kb includes the prophages and extensive DNA sequence variation among the PE/PPE genes (Table 1). There are 11 predicted CDS that present a diagnostic interest (Table 1). PCR screening for these 11 genes among a diverse panel of M. ulcerans and M. marinum strains confirmed that they are all M. ulcerans-specific and fully conserved (Table 2).
Table 2.
Presence of 11 M. ulcerans-specific CDS among different strains of M. ulcerans and M. marinum as determined by PCR
aIncludes M. marinum M and 22 previously described M. marinum strains (Stinear et al. 2005a).
bHypothetical protein.
cConserved hypothetical protein.
(CDS) Protein-coding sequences.
Insertion sequences and phages
Sequencing revealed many more copies of IS2404 and IS2606 (Table 1) than were first estimated (Stinear et al. 1999). These elements have profoundly affected genome plasticity, flanking many of the chromosome inversions, marking the site of DNA deletions, and causing 111 of the 743 predicted pseudogenes by inserting within genes. The M. marinum M strain has only 64 pseudogenes and possesses neither IS2404 nor IS2606. There are 10 other putative ISE belonging to various families, including one single-copy element (MUL_2378) belonging to the IS3 family that is present in nine copies in M. marinum. The two prophages named phiMU01 (18 kb, 18 CDS) and phiMU02 (24 kb, 17 CDS) resemble other mycobacteriophages in overall structure, integrating near tRNA genes and containing CDS associated with replication functions. However, phiMU02 may be non-functional, as several of its genes have been inactivated by multiple IS2606 insertions.
Pseudogene accumulation
Foremost among pseudogenes in M. ulcerans are members of the PE and PPE multigene families encoding the Gly-Ala-rich cell envelope proteins confined to mycobacteria. M. marinum has 170 PE and 105 PPE, respectively, compared with M. ulcerans that has retained only 70 intact PE and 46 PPE, with the remaining orthologs showing widespread sequence variation (Fig. 1; Table 1). These changes suggest a decreased requirement for PE/PPE proteins. The degeneration appears symptomatic of an intermediate stage of reductive evolution with M. ulcerans heading toward the highly contracted genome state of M. leprae, which is nearly devoid of these proteins (Cole et al. 2001).
The loss of paralogous genes and subsequent decrease in genetic redundancy may also contribute to slow growth via a diminished gene dose effect. For example, the disaccharide trehalose is essential for mycobacterial cell wall biogenesis and can be synthesized by three potential pathways (OtsAB, TreYZ, and TreS). As in M. tuberculosis, M. ulcerans has two otsB genes, otsB1 and otsB2; however, otsB1 has been disrupted by IS2404. Similarly, one of the three gltA genes coding for glutamine synthetase, a central enzyme in the nitrogen regulon, is a pseudogene in M. ulcerans, as are three of four phospholipase C paralogs required for phospholipolysis.
Other gene inactivations suggest divergence of the ecological niches of M. ulcerans and M. marinum. The crtB locus in M. marinum is responsible for the production of light-inducible carotenoids that protect the bacterium from incident sunlight. M. ulcerans does not produce these pigments, suggesting it is not exposed to direct sunlight in its habitat, yet it contains an identical crtB region. One explanation for the lack of photochromogenicity may be the introduction of a premature stop codon in crtI, encoding phytoene dehydrogenase, the second committed step in pigment synthesis.
Protein families
To better understand the biology of M. ulcerans, the predicted proteins were classified into families and compared with those found in other mycobacteria. Only one protein family (N > 5), comprising eight metal-dependent hydrolases, was found that had no counterpart in tubercle and leprosy bacilli. Otherwise, the same themes were observed (Tekaia et al. 1999), namely an abundance of proteins involved in lipid metabolism and prominent PE and PPE families. However, the number of potential transport proteins and enzyme systems predicted to be involved in carbon and energy metabolism was higher. M. marinum has 192 CDS associated with substrate transport while M. ulcerans has only 128.
If M. ulcerans occupies several distinct niches, including plant biofilms, insects, and humans, then its metabolism may be versatile. In an aquatic environment, M. ulcerans might obtain its energy and carbon from degradation of plant saccharides, as these stimulate growth in vitro (Marsollier et al. 2004b). Little is known, however, about its metabolism in insect tissues, although it may be significant that both M. marinum and M. ulcerans encode four potential chitinases/transglycosidases (BuruList/mast/P4.27.html), one of which is attached to a PE domain and thus is expected to be in the cell envelope. These enzymes might mediate attachment to, or degradation of, the N-acetyl-d-glucosamine polymers that comprise chitin, a major component of the exoskeletons of insects and crustaceans.
Primary metabolism and respiration
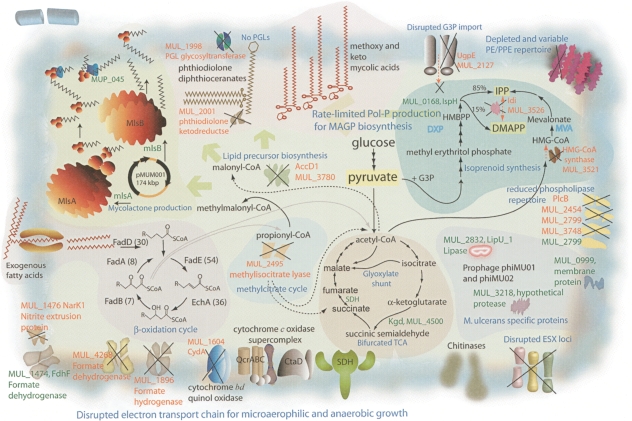
Metabolic pathway reconstruction supported the importance of lipid degradation for central carbon metabolism and uncovered intact glycolysis and pentose phosphate pathways, but no Entner-Doudoroff pathway. As in M. tuberculosis, M. ulcerans has a bifurcated TCA cycle, as it lacks α-ketoglutarate dehydrogenase (Fig. 3) and thus probably converts α-ketoglutarate to succinate via α-ketoglutarate decarboxylase (Tian et al. 2005). The TCA cycle incorporates the glyoxylate shunt, with two copies of the gene encoding isocitrate lyase. Thus, like M. tuberculosis, M. ulcerans may direct the two-carbon degradation products (acetyl-coA) from β-oxidation of host fatty acids into the TCA cycle operating in biosynthetic mode.
Figure 3.
Some important biochemical pathways. The main biochemical activities of M. ulcerans discussed in the text are depicted; these are all present in M. marinum except for mycolactone biosynthesis encoded by pMUM001. Some key genes and pathways that have been inactivated in M. ulcerans are indicated.
The β-oxidation of fatty acids also generates propionyl-CoA, which can be metabolized in several ways including via the methylcitrate pathway (Fig. 3). M. ulcerans appears to have all the methylcitrate enzymes except methylisocitrate lyase, whose putative gene (MUL_2495) has a frameshift mutation. A reduced capacity to metabolize propionic acid might be detrimental for growth; however, in M. tuberculosis the combination of the two isocitrate lyases can complement the absence of methylisocitrate lyase (Munoz-Elias and McKinney 2005).
Analysis of potential electron transport chains suggests M. ulcerans is capable of growth under aerobic but not anaerobic conditions as it lacks the nitrate and fumarate reductase systems. Based on gene orthology with M. tuberculosis, complete NADH oxidase, ubiquinol cyctochrome c oxidase, and ATP synthase complexes indicate ATP generation by oxidative phosphorylation during aerobic growth. M. marinum contains a potential alternative anaerobic respiratory pathway involving nitrite reductase, encoded by nrfD and formate dehydrogenase, where formate oxidation is coupled to nitrite reduction. M. ulcerans contains an intact nitrite reductase (nrfD), but the linked formate dehydrogenase system has been inactivated by multiple mutations (Fig. 3). Loss of this enzyme, a known selenoprotein, is consistent with deletion of the selenocysteine locus from M. ulcerans. M. marinum may be capable of nitrite reduction via the nirBD nitrite reductase and NarK transporter, but while M. ulcerans also contains these loci, nirB and narK are pseudogenes.
Under microaerophilic conditions, mycobacteria up-regulate expression of a high oxygen affinity cytochrome bd oxidase (cydABCD). This locus is present in both M. ulcerans and M. marinum, but in M. ulcerans the cydA ortholog is a pseudogene. A knockout mutation of cydA in Mycobacterium smegmatis leads to a competitive growth disadvantage under microaerophilic conditions (Kana et al. 2001). The above observations suggest that M. ulcerans has depleted respiratory versatility. Nevertheless, it has maintained >400 putative oxidoreductases, dehydrogenases, and mono- and dioxygenases (M. marinum has >590), suggesting that a robust and complex respiratory potential remains under aerobic conditions. As reported in other bacteria undergoing reductive evolution, the cytochrome P450 repertoire was particularly prone to pseudogene formation (Cole et al. 2001).
Cell wall lipids and secondary metabolism
The cell envelope of M. ulcerans and the secondary metabolites therein have been less well studied than those of other mycobacteria. From the genome sequence several predictions can be made, as reduction of secondary metabolite loci is widespread, notably through depletion of PKS genes. M. marinum has 27 PKS genes, but M. ulcerans has reduced its complement to 12. Ten of these PKS produce important cell-wall-associated lipids including mycolic acids (pks13), phthiodiolones (ppsA-E), phenol phthiodiolones (pks15/1), mannosyl-β1-phopholipids (pks12), and mycobactins (mbtC,D) (Matsunaga et al. 2004; Portevin et al. 2004). Neither M. ulcerans nor M. marinum contains the pks2 locus, required for the production of the sulfolipids. A larger PKS operon of unknown function harbors pks9 and pks11, but this operon may be inactive in M. ulcerans, as its other PKS genes (pks7, pks8, pks10) have become pseudogenes. One predicted consequence for M. ulcerans of downsizing its PKS complement would be liberation of energy (NADP) and substrate (malonyl-CoA and methylmalonyl-CoA) for the production of mycolactone. Contraction of secondary metabolism is also reflected in reduction of the MmpL family of lipid and polyketide transporters from 25 in M. marinum to only six in M. ulcerans.
Despite the loss of many pks, M. ulcerans has retained a significant anabolic lipid potential highlighting the central importance of particular lipids to the preservation of the mycobacterial cell wall. Structural analyses of some of the signature lipid species were undertaken and then correlated with the genome findings. M. ulcerans and M. marinum have been previously shown to produce diunsaturated, methoxy, and ketomycolic acids (Daffe et al. 1991). Central to synthesis of these mycolates and other lipids are the fatty acid synthases I and II. All the principal components of these enzymes are present in M. ulcerans, and the key proteins share >90% identity with those of M. marinum and M. tuberculosis.
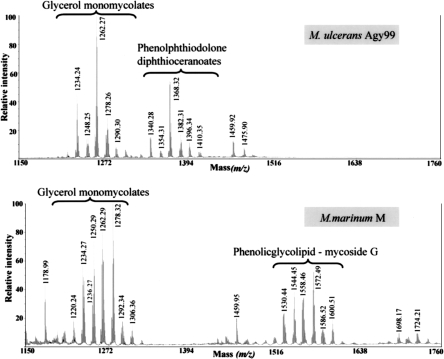
M. ulcerans also produces a highly abundant and apolar cell wall lipid consisting of diesters of phthiodiolone (Daffe et al. 1984; Onwueme et al. 2005). The common β-diol backbone of phthiodiolones in M. ulcerans is produced from C16–C18 fatty acids by five type I PKS encoded by the genes ppsA–E. Processing of p-hydroxyphenylalkanoate rather than fatty acids by these PKS results in the production of phenolphthiodiolones, which can be further modified by glycosylation to form the virulence factors and immune modulators, the phenolic glycolipids (PGLs) (Daffe et al. 1992; Reed et al. 2004). In M. tuberculosis, Rv2962 encodes the glycosyl transferase that adds the first sugar residue to the highly related lipid, phenolphthiocerol (Perez et al. 2004). The absence of PGLs in M. ulcerans Agy99 is readily explained, as MUL_1998, the Rv2962 ortholog, is a pseudogene. This observation was supported by MALDI-TOF MS detection of the substrate for the MUL_1998 gene product, phenolphthiodiolone diphthioceranoate, in M. ulcerans but not in its glycosylated form, which was detected in M. marinum (Figs. 3, 4).
Figure 4.
Partial matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectra of lipids from M. ulcerans and M. marinum. These spectra show the presence of diesters of phthiocerol and phthiodiolone, and of phenolic glycolipids in M. marinum, whereas only diesters of phthiodiolone and phenolphthiodiolone occur in M. ulcerans.
M. ulcerans and M. marinum contain both a mevalonate (MVA) and non-mevalonate (DXP) pathway for polyprenoid synthesis, the first example of such in the mycobacteria and a rare occurrence among bacteria (Rohdich et al. 2004). Polyprenyl phosphate (Pol-P) forms lipid-linked sugar intermediates that are required by bacteria during cell wall biosynthesis. They are formed by sequential condensation of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). In other mycobacteria these molecules are synthesized via the DXP pathway; the same appears true for M. ulcerans, as the MVA pathway is probably inactive because one of the genes in the MVA operon, MUL_3521, encoding a putative hydroxymethylglutaryl-coenzyme A synthase, has been disrupted (Fig. 3).
Implications for pathogenesis from genome reduction
Comparative genomics also shows that the M. ulcerans genome is contracting and that extensive pseudogene formation has occurred. There are 157 regions, accounting for 1232 kb, present in M. marinum but absent from M. ulcerans. These were assigned a M. ulcerans Region of Difference (MURD) number (Supplemental Table S1). While some of these represent insertions in M. marinum, including seven prophage and 13 ISE, totaling 168 kb, the majority result from the accumulated deletion of 1064 kb of DNA from M. ulcerans. In this way many paralogous gene families have been lost in a situation reminiscent of M. leprae where considerable genetic downsizing has occurred (Cole et al. 2001). Most notable among these are several secondary metabolism loci and the PE/PPE genes, the latter accounting for 45% of the MURDs. Other deletions affect genes involved in intermediary and energy metabolism such as potassium transport (MURD9), hydrogenase (MURD17), and production of selenocysteine (MURD142), a rare amino acid found in the active sites of formate dehydrogenase and many bacterial antioxidant proteins.
Noteworthy deletions in M. ulcerans include the ESX loci. While there are five complete ESX systems in M. marinum, only three remain in M. ulcerans as the other two have incurred deletions. In M. tuberculosis, the esx1 locus encodes a novel protein secretion apparatus, which contributes to its virulence, intercellular spread, and immunogenicity (Brodin et al. 2004). In M. ulcerans, esx1 has been disrupted by two deletions (MURD151 and MURD152). Effector proteins secreted by ESX systems belong to the ESAT-6 or the EspA families (Fortune et al. 2005), and 13 ESAT-6 proteins exist in M. ulcerans. Interestingly, the espA gene, encoding a putative substrate of ESX-1, has been deleted (MURD111). Loss of these systems, which trigger granuloma formation by M. marinum and M. tuberculosis, may contribute to the predominantly extracellular location of M. ulcerans in infected tissue. Supplemental Table S1 also lists 102 orthologous M. tuberculosis genes that M. ulcerans has lost by deletion.
Discussion
Bacteria that have recently passed an evolutionary bottleneck and are now adapting to a new, more stable environment, such as Mycobacterium leprae, Yersinia pestis, and Bordetella pertussis (Parkhill et al. 2003), have some of the following genomic signatures: proliferation of ISE, accumulation of pseudogenes, chromosomal rearrangements, genome downsizing, and acquisition of foreign genes, often via plasmids or bacteriophage, that confer a fitness advantage in the new environment. They also show a high degree of genetic relatedness or clonality. All six genomic signatures are shared by M. ulcerans, which appears to be undergoing reductive evolution and, as such, may represent a good model for studying this phenomenon and the response to changing from an environmental to host-adapted, possibly arthropod niche.
Our study provides compelling evidence for M. ulcerans being a descendant of an M. marinum progenitor strain that acquired the virulence plasmid, pMUM001, from another actinobacterium. This event then led to divergence of the ecological niches of M. ulcerans and M. marinum, although other factors may have contributed. For instance, the crtB locus is responsible for the production of light-inducible carotenoids that protect M. marinum from incident sunlight (Ramakrishnan et al. 1997). M. ulcerans does not produce these pigments, due to a lesion in crtI, so its impaired ability to withstand exposure to direct sunlight would lead to selection for progeny capable of adapting to, and settling in, a more protected niche. The strong selective pressure exerted by mycolactone could have enabled M. ulcerans to colonize aquatic insects or other animals sharing its ecosystem.
Once within a more stable and protected habitat, loss of gene function and slower growth would be readily tolerated. Genomic analysis suggests possible mechanisms that have impacted on growth rate including the loss, or reduction in number, of genes for transport of potassium and other solutes; enzymes such as hydrogenase, glutamine synthase, and phospholipase C; and for trehalose and isoprenoid production, among others; in turn, the capacity for free-living would diminish.
Unlike other pathogenic mycobacteria, M. ulcerans is found predominantly in extracellular form in infected human tissue, although it passes via a transient intracellular stage (Coutanceau et al. 2005). In part, this is due to the antiphagocytic properties of mycolactone, but loss of the protein secretion system, ESX-1, that mediates the export of the proteins ESAT-6 and EspA may also contribute. Inactivation of ESX-1 in M. marinum or M. tuberculosis results in reduced uptake by phagocytes and greatly diminished intercellular spread (Brodin et al. 2004). It is also conceivable that some of the functions encoded by the M. ulcerans-restricted CDS could contribute to the pathology associated with Buruli ulcer. These genes or their products may also find application in the development of new diagnostic tests, which are required to help control the spread of Buruli ulcer, and their conservation in all isolates of M. ulcerans examined is encouraging in this respect.
Methods
M. ulcerans Agy99
M. ulcerans Agy99 was isolated from an ulcerative lesion on the right elbow of a female patient from the Ga district of Ghana in 1999. Multi-locus sequence typing and inter-IS PCR analysis confirmed that this strain belonged to the African epidemic genotype. DNA was prepared for sequencing four passages after primary isolation.
Whole-genome sequencing
A whole-genome shotgun library was prepared in the vector pCDNA2.1 using Escherichia coli strain XL2-blue (Invitrogen). Genomic DNA was sheared by sonication and size fractionated to produce plasmids with inserts in the size ranges of 2–3 kb, 3–5 kb, and 5–10 kb. These were used as template in DNA cycle sequencing reactions to obtain 75,415 end-sequence reads using an ABI 3700 DNA sequencer (Applied Biosystems), providing theoretical 8× coverage. Sequence assembly was performed by PHRAP and GAP4 as described (Cole et al. 1998). Gap closure was facilitated by scaffolding using a large insert BAC library (Stinear et al. 2005a) augmented with primer walking and PCR.
Genome annotation and comparative genomics
Annotation and database construction was performed with Wasabi, an in-house, Web-interfaced MySQL database for management of genome annotation, Artemis, and GenoList. A complete database is available at http://genolist.pasteur.fr/BuruList/. CDS were predicted using GenemarkS and GenoStar, and compared with public sequence databases using the BLAST suite of algorithms. Pseudogenes had one or more mutations that would prevent correct translation. Mutations were checked against original sequence data to exclude sequencing errors. The Artemis Comparison Tool (Carver et al. 2005) was used for comparative genome analysis.
Lipid analysis
Lipids were extracted with CHCl3/CH3OH (1:1 v/v) and subjected to MALDI-TOF mass spectrometry using a Voyager DE-STR MALDI-TOF instrument (PerSeptive Biosystems) equipped with a pulse nitrogen laser emitting at 337 nm and analyzed in the reflector mode as previously described (Perez et al. 2004).
Acknowledgments
We thank Paul Harrison, Ivan Moszer, and Roland Brosch for sharing expertise and helpful discussions. We gratefully acknowledge the financial support of the Génopole program, the Association Française Raoul Follereau, the National Health and Medical Research Council of Australia, and the World Health Organization through grants provided by the Nippon Foundation, Tokyo, Japan.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to Genbank under accession number CP000325.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5942807
References
- Alsop D. The Bairnsdale ulcer. Aust. N. Z. J. Surg. 1972;41:317–319. [PubMed] [Google Scholar]
- Brodin P., Rosenkrands I., Andersen P., Cole S.T., Brosch R., Rosenkrands I., Andersen P., Cole S.T., Brosch R., Andersen P., Cole S.T., Brosch R., Cole S.T., Brosch R., Brosch R. ESAT-6 proteins: Protective antigens and virulence factors? Trends Microbiol. 2004;12:500–508. doi: 10.1016/j.tim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rajandream M.A., Barrell B.G., Parkhill J., Barrell B.G., Parkhill J., Parkhill J. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Eiglmeier K., Gas S., Barry C.E., III, Gas S., Barry C.E., III, Barry C.E., III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cole S.T., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Honore N., Garnier T., Churcher C., Harris D., Garnier T., Churcher C., Harris D., Churcher C., Harris D., Harris D., et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Coutanceau E., Marsollier L., Brosch R., Perret E., Goossens P., Tanguy M., Cole S.T., Small P.L., Demangel C., Marsollier L., Brosch R., Perret E., Goossens P., Tanguy M., Cole S.T., Small P.L., Demangel C., Brosch R., Perret E., Goossens P., Tanguy M., Cole S.T., Small P.L., Demangel C., Perret E., Goossens P., Tanguy M., Cole S.T., Small P.L., Demangel C., Goossens P., Tanguy M., Cole S.T., Small P.L., Demangel C., Tanguy M., Cole S.T., Small P.L., Demangel C., Cole S.T., Small P.L., Demangel C., Small P.L., Demangel C., Demangel C. Modulation of the host immune response by a transient intracellular stage of Mycobacterium ulcerans: The contribution of endogenous mycolactone toxin. Cell. Microbiol. 2005;7:1187–1196. doi: 10.1111/j.1462-5822.2005.00546.x. [DOI] [PubMed] [Google Scholar]
- Daffe M., Laneelle M.A., Roussel J., Asselineau C., Laneelle M.A., Roussel J., Asselineau C., Roussel J., Asselineau C., Asselineau C. Specific lipids from Mycobacterium ulcerans. Ann. Microbiol. (Paris) 1984;135A:191–201. [PubMed] [Google Scholar]
- Daffe M., Laneelle M.A., Lacave C., Laneelle M.A., Lacave C., Lacave C. Structure and stereochemistry of mycolic acids of Mycobacterium marinum and Mycobacterium ulcerans. Res. Microbiol. 1991;142:397–403. doi: 10.1016/0923-2508(91)90109-n. [DOI] [PubMed] [Google Scholar]
- Daffe M., Varnerot A., Levy-Frebault V.V., Varnerot A., Levy-Frebault V.V., Levy-Frebault V.V. The phenolic mycoside of Mycobacterium ulcerans: Structure and taxonomic implications. J. Gen. Microbiol. 1992;138:131–137. doi: 10.1099/00221287-138-1-131. [DOI] [PubMed] [Google Scholar]
- Eisen J.A., Heidelberg J.F., White O., Salzberg S.L., Heidelberg J.F., White O., Salzberg S.L., White O., Salzberg S.L., Salzberg S.L. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-research0011. research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etuaful S., Carbonnelle B., Grosset J., Lucas S., Horsfield C., Phillips R., Evans M., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Carbonnelle B., Grosset J., Lucas S., Horsfield C., Phillips R., Evans M., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Grosset J., Lucas S., Horsfield C., Phillips R., Evans M., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Lucas S., Horsfield C., Phillips R., Evans M., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Horsfield C., Phillips R., Evans M., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Phillips R., Evans M., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Evans M., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Ofori-Adjei D., Klustse E., Owusu-Boateng J., Klustse E., Owusu-Boateng J., Owusu-Boateng J., et al. Efficacy of the combination rifampin–streptomycin in preventing growth of Mycobacterium ulcerans in early lesions of Buruli ulcer in humans. Antimicrob. Agents Chemother. 2005;49:3182–3186. doi: 10.1128/AAC.49.8.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune S.M., Jaeger A., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Jaeger A., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Sarracino D.A., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Chase M.R., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Sassetti C.M., Sherman D.R., Bloom B.R., Rubin E.J., Sherman D.R., Bloom B.R., Rubin E.J., Bloom B.R., Rubin E.J., Rubin E.J. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.D., Stinear T., Small P.L., Pluschke G., Merritt R.W., Portaels F., Huygen K., Hayman J.A., Asiedu K., Stinear T., Small P.L., Pluschke G., Merritt R.W., Portaels F., Huygen K., Hayman J.A., Asiedu K., Small P.L., Pluschke G., Merritt R.W., Portaels F., Huygen K., Hayman J.A., Asiedu K., Pluschke G., Merritt R.W., Portaels F., Huygen K., Hayman J.A., Asiedu K., Merritt R.W., Portaels F., Huygen K., Hayman J.A., Asiedu K., Portaels F., Huygen K., Hayman J.A., Asiedu K., Huygen K., Hayman J.A., Asiedu K., Hayman J.A., Asiedu K., Asiedu K. Buruli ulcer (M. ulcerans infection): New insights, new hope for disease control. PLoS Med. 2005;2:e108. doi: 10.1371/journal.pmed.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana B.D., Weinstein E.A., Avarbock D., Dawes S.S., Rubin H., Mizrahi V., Weinstein E.A., Avarbock D., Dawes S.S., Rubin H., Mizrahi V., Avarbock D., Dawes S.S., Rubin H., Mizrahi V., Dawes S.S., Rubin H., Mizrahi V., Rubin H., Mizrahi V., Mizrahi V. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 2001;183:7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum P., Tolhurst J., Buckle G., Ha S., Tolhurst J., Buckle G., Ha S., Buckle G., Ha S., Ha S. A new mycobacterial infection in man. J. Pathol. Bacteriol. 1948;60:93–122. [PubMed] [Google Scholar]
- Marsollier L., Robert R., Aubry J., Saint Andre J.P., Kouakou H., Legras P., Manceau A.L., Mahaza C., Carbonnelle B., Robert R., Aubry J., Saint Andre J.P., Kouakou H., Legras P., Manceau A.L., Mahaza C., Carbonnelle B., Aubry J., Saint Andre J.P., Kouakou H., Legras P., Manceau A.L., Mahaza C., Carbonnelle B., Saint Andre J.P., Kouakou H., Legras P., Manceau A.L., Mahaza C., Carbonnelle B., Kouakou H., Legras P., Manceau A.L., Mahaza C., Carbonnelle B., Legras P., Manceau A.L., Mahaza C., Carbonnelle B., Manceau A.L., Mahaza C., Carbonnelle B., Mahaza C., Carbonnelle B., Carbonnelle B. Aquatic insects as a vector for Mycobacterium ulcerans. Appl. Environ. Microbiol. 2002;68:4623–4628. doi: 10.1128/AEM.68.9.4623-4628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsollier L., Severin T., Aubry J., Merritt R.W., Saint Andre J.P., Legras P., Manceau A.L., Chauty A., Carbonnelle B., Cole S.T., Severin T., Aubry J., Merritt R.W., Saint Andre J.P., Legras P., Manceau A.L., Chauty A., Carbonnelle B., Cole S.T., Aubry J., Merritt R.W., Saint Andre J.P., Legras P., Manceau A.L., Chauty A., Carbonnelle B., Cole S.T., Merritt R.W., Saint Andre J.P., Legras P., Manceau A.L., Chauty A., Carbonnelle B., Cole S.T., Saint Andre J.P., Legras P., Manceau A.L., Chauty A., Carbonnelle B., Cole S.T., Legras P., Manceau A.L., Chauty A., Carbonnelle B., Cole S.T., Manceau A.L., Chauty A., Carbonnelle B., Cole S.T., Chauty A., Carbonnelle B., Cole S.T., Carbonnelle B., Cole S.T., Cole S.T. Aquatic snails, passive hosts of Mycobacterium ulcerans. Appl. Environ. Microbiol. 2004a;70:6296–6298. doi: 10.1128/AEM.70.10.6296-6298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsollier L., Stinear T., Aubry J., Saint Andre J.P., Robert R., Legras P., Manceau A.L., Audrain C., Bourdon S., Kouakou H., Stinear T., Aubry J., Saint Andre J.P., Robert R., Legras P., Manceau A.L., Audrain C., Bourdon S., Kouakou H., Aubry J., Saint Andre J.P., Robert R., Legras P., Manceau A.L., Audrain C., Bourdon S., Kouakou H., Saint Andre J.P., Robert R., Legras P., Manceau A.L., Audrain C., Bourdon S., Kouakou H., Robert R., Legras P., Manceau A.L., Audrain C., Bourdon S., Kouakou H., Legras P., Manceau A.L., Audrain C., Bourdon S., Kouakou H., Manceau A.L., Audrain C., Bourdon S., Kouakou H., Audrain C., Bourdon S., Kouakou H., Bourdon S., Kouakou H., Kouakou H., et al. Aquatic plants stimulate the growth of and biofilm formation by Mycobacterium ulcerans in axenic culture and harbor these bacteria in the environment. Appl. Environ. Microbiol. 2004b;70:1097–1103. doi: 10.1128/AEM.70.2.1097-1103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsollier L., Aubry J., Coutanceau E., Andre J.P., Small P.L., Milon G., Legras P., Guadagnini S., Carbonnelle B., Cole S.T., Aubry J., Coutanceau E., Andre J.P., Small P.L., Milon G., Legras P., Guadagnini S., Carbonnelle B., Cole S.T., Coutanceau E., Andre J.P., Small P.L., Milon G., Legras P., Guadagnini S., Carbonnelle B., Cole S.T., Andre J.P., Small P.L., Milon G., Legras P., Guadagnini S., Carbonnelle B., Cole S.T., Small P.L., Milon G., Legras P., Guadagnini S., Carbonnelle B., Cole S.T., Milon G., Legras P., Guadagnini S., Carbonnelle B., Cole S.T., Legras P., Guadagnini S., Carbonnelle B., Cole S.T., Guadagnini S., Carbonnelle B., Cole S.T., Carbonnelle B., Cole S.T., Cole S.T. Colonization of the salivary glands of Naucoris cimicoides by Mycobacterium ulcerans requires host plasmatocytes and a macrolide toxin, mycolactone. Cell. Microbiol. 2005;7:935–943. doi: 10.1111/j.1462-5822.2005.00521.x. [DOI] [PubMed] [Google Scholar]
- Matsunaga I., Bhatt A., Young D.C., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Bhatt A., Young D.C., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Young D.C., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Cheng T.Y., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Eyles S.J., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Besra G.S., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Briken V., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Porcelli S.A., Costello C.E., Jacobs W.R., Jr., Costello C.E., Jacobs W.R., Jr., Jacobs W.R., Jr., et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J. Exp. Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias E.J., McKinney J.D., McKinney J.D. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwueme K.C., Vos C.J., Zurita J., Soll C.E., Quadri L.E., Vos C.J., Zurita J., Soll C.E., Quadri L.E., Zurita J., Soll C.E., Quadri L.E., Soll C.E., Quadri L.E., Quadri L.E. Identification of phthiodiolone ketoreductase, an enzyme required for production of mycobacterial diacyl phthiocerol virulence factors. J. Bacteriol. 2005;187:4760–4766. doi: 10.1128/JB.187.14.4760-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Sebaihia M., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Sebaihia M., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Preston A., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Murphy L.D., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Thomson N., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Harris D.E., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Holden M.T., Churcher C.M., Bentley S.D., Mungall K.L., Churcher C.M., Bentley S.D., Mungall K.L., Bentley S.D., Mungall K.L., Mungall K.L., et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Perez E., Constant P., Lemassu A., Laval F., Daffe M., Guilhot C., Constant P., Lemassu A., Laval F., Daffe M., Guilhot C., Lemassu A., Laval F., Daffe M., Guilhot C., Laval F., Daffe M., Guilhot C., Daffe M., Guilhot C., Guilhot C. Characterization of three glycosyltransferases involved in the biosynthesis of the phenolic glycolipid antigens from the Mycobacterium tuberculosis complex. J. Biol. Chem. 2004;279:42574–42583. doi: 10.1074/jbc.M406246200. [DOI] [PubMed] [Google Scholar]
- Portevin D., De Sousa-D’Auria C., Houssin C., Grimaldi C., Chami M., Daffe M., Guilhot C., De Sousa-D’Auria C., Houssin C., Grimaldi C., Chami M., Daffe M., Guilhot C., Houssin C., Grimaldi C., Chami M., Daffe M., Guilhot C., Grimaldi C., Chami M., Daffe M., Guilhot C., Chami M., Daffe M., Guilhot C., Daffe M., Guilhot C., Guilhot C. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. 2004;101:314–319. doi: 10.1073/pnas.0305439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan L., Tran H.T., Federspiel N.A., Falkow S., Tran H.T., Federspiel N.A., Falkow S., Federspiel N.A., Falkow S., Falkow S. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: Isolation, characterization, and gene disruption via homologous recombination. J. Bacteriol. 1997;179:5862–5868. doi: 10.1128/jb.179.18.5862-5868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed M.B., Domenech P., Manca C., Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., III, Domenech P., Manca C., Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., III, Manca C., Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., III, Su H., Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., III, Barczak A.K., Kreiswirth B.N., Kaplan G., Barry C.E., III, Kreiswirth B.N., Kaplan G., Barry C.E., III, Kaplan G., Barry C.E., III, Barry C.E., III A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- Rocha E.P., Danchin A., Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- Rohdich F., Bacher A., Eisenreich W., Bacher A., Eisenreich W., Eisenreich W. Perspectives in anti-infective drug design. The late steps in the biosynthesis of the universal terpenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate. Bioorg. Chem. 2004;32:292–308. doi: 10.1016/j.bioorg.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Sassetti C.M., Boyd D.H., Rubin E.J., Boyd D.H., Rubin E.J., Rubin E.J. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T., Ross B.C., Davies J.K., Marino L., Robins-Browne R.M., Oppedisano F., Sievers A., Johnson P.D., Ross B.C., Davies J.K., Marino L., Robins-Browne R.M., Oppedisano F., Sievers A., Johnson P.D., Davies J.K., Marino L., Robins-Browne R.M., Oppedisano F., Sievers A., Johnson P.D., Marino L., Robins-Browne R.M., Oppedisano F., Sievers A., Johnson P.D., Robins-Browne R.M., Oppedisano F., Sievers A., Johnson P.D., Oppedisano F., Sievers A., Johnson P.D., Sievers A., Johnson P.D., Johnson P.D. Identification and characterization of IS2404 and IS2606: Two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 1999;37:1018–1023. doi: 10.1128/jcm.37.4.1018-1023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T.P., Jenkin G.A., Johnson P.D.R., Davies J.K., Jenkin G.A., Johnson P.D.R., Davies J.K., Johnson P.D.R., Davies J.K., Davies J.K. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 2000;182:6322–6330. doi: 10.1128/jb.182.22.6322-6330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T.P., Mve-Obiang A., Small P.L., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Mve-Obiang A., Small P.L., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Small P.L., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Frigui W., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Pryor M.J., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Brosch R., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Jenkin G.A., Johnson P.D., Davies J.K., Lee R.E., Johnson P.D., Davies J.K., Lee R.E., Davies J.K., Lee R.E., Lee R.E., et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T.P., Hong H., Frigui W., Pryor M.J., Brosch R., Garnier T., Leadlay P.F., Cole S.T., Hong H., Frigui W., Pryor M.J., Brosch R., Garnier T., Leadlay P.F., Cole S.T., Frigui W., Pryor M.J., Brosch R., Garnier T., Leadlay P.F., Cole S.T., Pryor M.J., Brosch R., Garnier T., Leadlay P.F., Cole S.T., Brosch R., Garnier T., Leadlay P.F., Cole S.T., Garnier T., Leadlay P.F., Cole S.T., Leadlay P.F., Cole S.T., Cole S.T. Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J. Bacteriol. 2005a;187:1668–1676. doi: 10.1128/JB.187.5.1668-1676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear T.P., Pryor M.J., Porter J.L., Cole S.T., Pryor M.J., Porter J.L., Cole S.T., Porter J.L., Cole S.T., Cole S.T. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiol. 2005b;151:683–692. doi: 10.1099/mic.0.27674-0. [DOI] [PubMed] [Google Scholar]
- Tekaia F., Gordon S.V., Garnier T., Brosch R., Barrell B.G., Cole S.T., Gordon S.V., Garnier T., Brosch R., Barrell B.G., Cole S.T., Garnier T., Brosch R., Barrell B.G., Cole S.T., Brosch R., Barrell B.G., Cole S.T., Barrell B.G., Cole S.T., Cole S.T. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber. Lung Dis. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- Tian J., Bryk R., Itoh M., Suematsu M., Nathan C., Bryk R., Itoh M., Suematsu M., Nathan C., Itoh M., Suematsu M., Nathan C., Suematsu M., Nathan C., Nathan C. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: Identification of α-ketoglutarate decarboxylase. Proc. Natl. Acad. Sci. 2005;102:10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]