Abstract
Determining how transcriptional regulatory signals are encoded in vertebrate genomes is essential for understanding the origins of multicellular complexity; yet the genetic code of vertebrate gene regulation remains poorly understood. In an attempt to elucidate this code, we synergistically combined genome-wide gene-expression profiling, vertebrate genome comparisons, and transcription factor binding-site analysis to define sequence signatures characteristic of candidate tissue-specific enhancers in the human genome. We applied this strategy to microarray-based gene expression profiles from 79 human tissues and identified 7187 candidate enhancers that defined their flanking gene expression, the majority of which were located outside of known promoters. We cross-validated this method for its ability to de novo predict tissue-specific gene expression and confirmed its reliability in 57 of the 79 available human tissues, with an average precision in enhancer recognition ranging from 32% to 63% and a sensitivity of 47%. We used the sequence signatures identified by this approach to successfully assign tissue-specific predictions to ∼328,000 human–mouse conserved noncoding elements in the human genome. By overlapping these genome-wide predictions with a data set of enhancers validated in vivo, in transgenic mice, we were able to confirm our results with a 28% sensitivity and 50% precision. These results indicate the power of combining complementary genomic data sets as an initial computational foray into a global view of tissue-specific gene regulation in vertebrates.
Increasing lines of evidence support the notion that the majority of functional elements in the human genome do not code for proteins (Waterston et al. 2002), yet our ability to systematically categorize and predict their function remains limited. For instance, most progress in elucidating transcriptional regulatory mechanisms has stemmed from computational and experimental analyses of transcription factors (TFs) acting within promoter regions of functionally related cohorts of genes. While informative (Bajic et al. 2004; Sharan et al. 2004; Kim et al. 2005; Xie et al. 2005), these studies did not assess distant-acting regulatory elements and thereby only sampled a limited portion of the vertebrate gene regulatory network (Levine and Tjian 2003; Cawley et al. 2004). Several recent studies have provided conclusive evidence that the complex transcriptional expression pattern of human genes is mediated through multiple discrete sequences, often located hundreds of kilobases (kb) away from their core promoters (Lettice et al. 2003; Nobrega et al. 2003). In these studies, evolutionary sequence conservation has served as a reliable indicator of biological activity, with an increasing number of distant noncoding evolutionarily conserved regions (ECRs) validated as tissue-specific enhancers during development (Loots et al. 2000; Lettice et al. 2003; Nobrega et al. 2003; de la Calle-Mustienes et al. 2005; Dermitzakis et al. 2005; Woolfe et al. 2005; Pennacchio et al. 2006). Although genome comparisons have provided a powerful approach for identifying noncoding ECRs that are under selective pressure, we have yet to develop reliable high-throughput computational methods for the discovery of distant regulatory elements with predetermined functional specificity. Here we explore a strategy for translating noncoding sequence data into transcriptional regulatory information that ultimately serves two vital purposes: to define the genetic vocabulary of tissue-specific gene regulation and to use this information to predict tissue-specific enhancers in the entire human genome, de novo. This approach combines genome-wide tissue-specific gene expression profiling data (Su et al. 2002), vertebrate genome comparisons, and pattern analysis of transcription factor binding sites (TFBS). Our results support the notion of an existing underlying genetic code of gene regulation in mammals, and provide an initial foundation for deciphering this code using primarily a computational approach.
Results
Predicting candidate regulatory elements for tissue-specific genes
As a first step toward directly relating gene expression to comparative sequence data, we clustered overlapping gene transcripts in the human genome and identified 18,504 unique protein-coding loci (the boundaries of each locus were defined by the neighboring genes, independent of the absolute size of the locus; see Methods). We next assigned transcriptional information obtained from the GNF Atlas2 gene expression database (gnfAtlas2) (Su et al. 2002) to these genomic loci. This included 79 human tissues with the majority of human loci (85%) successfully linked to their corresponding gene-expression pattern. For each represented tissue, we defined two sets of genes: high expressors and low expressors. Our goal was to compare the genomic loci containing these two contrasting gene sets (across available tissues) to search for shared noncoding DNA sequence features in the vicinity of genes highly and concordantly expressed in a given tissue, while simultaneously minimizing the presence of these features in loci of genes with low expression within the tissue of investigation. Thus, this approach sought to identify primary candidate sequences responsible for tissue-specific enhancers that control high gene expression within a given tissue. To focus our search on a uniform cohort of highly expressed genes, we selected the top 300 most highly expressed genes (about 2% of the total number of genes in gnfAtlas2) for each tissue. To establish a large background data set, in which the probability of finding a tissue-specific enhancer might be predicted to be the lowest, we selected approximately one-third of all genes in gnfAtlas2—comprising 5000 genes with the lowest level of expression in the corresponding tissue. The same number of genes in the high and low expression groups for different tissues allowed an unbiased analysis of the method’s performance across the panel of all studied tissues.
We initially observed a strong correlation between the tissue specificity of a gene and the size of the locus, such that loci of highly expressed genes in the central nervous system (CNS) were, on average, significantly larger than the global median locus length. In contrast, loci corresponding to highly expressed genes in the immune system or various tumor tissues were significantly shorter (Supplemental Fig. S1). For example, the median locus length of a human gene highly expressed in fetal brain was 245 kb, while genes highly expressed in testis were on average 3.6 times shorter (68 kb) (Supplemental Fig. S1). We also found that 10% of the brain and CNS loci coincided with vast noncoding regions termed gene deserts (Nobrega et al. 2003) in the human genome (a twofold increase over the expected value; P < 10−7), consistent with the observation that most enhancers identified within gene deserts, to date, are biased toward brain and/or CNS expression during vertebrate development (Nobrega et al. 2003; Uchikawa et al. 2004; de la Calle-Mustienes et al. 2005; Pennacchio et al. 2006). Finally, we observed a linear correlation between locus length and the number of human/mouse noncoding ECRs, regardless of the tissue under investigation (Supplemental Fig. S1E).
Recent studies suggest that the most highly conserved noncoding ECRs within a locus commonly possess gene regulatory function (Nobrega et al. 2003; Ovcharenko et al. 2004b; Prabhakar et al. 2006). Therefore, we selected the three most conserved human/mouse noncoding ECRs for each of the 18,504 human genes in our study, as well as noncoding ECRs overlapping with the gene’s promoter region (see Methods for selection procedure details). This selection generated a data set of 60 thousand (k) candidate regulatory elements in the human genome, averaging 4.2 candidate regulatory elements per locus. Classification of these elements based on their genomic location annotated 31% as intergenic, 28% as promoter, 20% as intronic, 13% as 3′ UTR, and 8% as 5′ UTR. Approximately 24 k of these elements flanked 6059 genes with the highest gene expression in at least one of the 79 tissues, while ∼55 k of these elements flanked 15,632 genes with the lowest gene expression (serving as a negative control data set).
To explore the sequence motifs of these noncoding ECRs linked to genes displaying high versus low expression in the same tissue, we used a previously described motif-identification strategy (Loots and Ovcharenko 2004) and identified 1.8 million (M) evolutionarily conserved putative TFBS within this data set (see Methods). We found that several individual motifs were significantly enriched in 43 human tissues (Supplemental Table S1). For example, we observed a strong association among NRF1, POU2F1, MEF2A, and CREB1, transcription factors known to play key roles in brain and neural development (Ilia et al. 2003; Okuda et al. 2004; Chang et al. 2005; Shalizi and Bonni 2005; Riccio et al. 2006) in candidate regulatory elements from loci highly expressed in human fetal brain (Supplemental Table S1A). However, as described in further detail below, no single TF by itself was sufficient to predict where a candidate enhancer will drive gene expression.
Determining sequence signatures of candidate tissue-specific enhancers
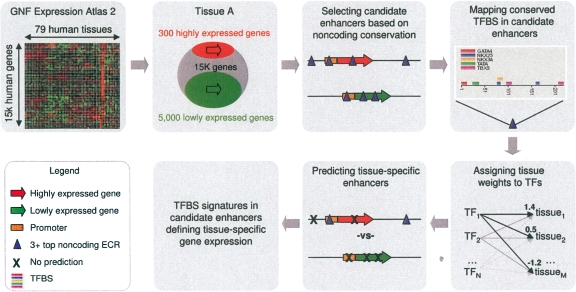
Based on the presumed combinatorial nature of multiple TFs to mediate a given enhancer’s activity, we used an analysis strategy that simultaneously scored the impact of multiple TFBS motifs in an attempt to classify candidate enhancers based on sequence signatures that define gene expression in a particular tissue. This was accomplished by assigning a weight to each TF that quantifies its association with a given tissue. By summing these TFBS motif weights, we were thus able to generate a regulatory potential tissue-specificity score for each of the 24 k candidate enhancers of highly expressed genes as well as 55 k background elements of the low expressed genes. This scoring scheme provided the means to optimize TF weights in an effort to enrich for positively scoring candidate enhancers in tissue-specific loci of highly expressed genes while simultaneously minimizing their presence in loci of genes with low expression (independently performed for each tissue; see Methods). We named this approach Enhancer Identification (or EI) and its application allowed us to select candidate tissue-specific enhancers from the pool of conserved noncoding elements in loci of genes highly expressed in a given tissue (Fig. 1). We performed EI analysis independently on both human and mouse gene expression data, and while we primarily utilized human statistics in our discussion, mouse data analysis is provided in the Supplemental materials (Supplemental Figs. S1, S2; Tables S2, S7).
Figure 1.
Schematic of the general EI strategy for defining signatures of tissue-specific enhancers.
The EI scoring optimization allowed us to maximize our resolving power to the point where 60% (±5%) of genes highly expressed in a tissue group contain signatures that are present in <15% of the low expressed genes for any given tissue (Fig. 2B). For example, EI identified at least one fetal lung candidate enhancer for 65% of genes with high fetal lung expression, while no such candidates were identified in the non-intergenic regions (promoter, UTR, or intronic) of >86% of genes with low fetal lung expression (intergenic regions were excluded from the negative control group to prevent potential associations with neighboring genes’ regulation [see Methods]). Of the original 24 k candidate regulatory elements linked to genes highly expressed in one or more of the 79 available tissues, EI optimization identified 7187 candidate enhancers with signatures that define tissue-specific expression. The database that summarizes these candidate tissue-specific enhancers is available at http://www.dcode.org/EI. Through this consolidation of the data set we found that 47% of human noncoding ECRs defined as candidate enhancers were predictive of expression in more than one tissue, consistent with our finding that 66% of the human genes in this study are highly expressed in multiple tissues. Since these candidate enhancers were mainly assigned to different tissues that are functionally related (e.g., CD4 and CD8 T-cells) (Supplemental Table S6), it is possible that the transcriptional regulation of genes expressed in similar tissues could be achieved through shared gene-regulatory mechanisms. These findings are consistent with in vivo expression data derived from enhancer scans in transgenic mice, indicating that one-third of embryonic enhancers active during a single time-point in development drive expression in more than one tissue type (Nobrega et al. 2003; de la Calle-Mustienes et al. 2005; Woolfe et al. 2005; Pennacchio et al. 2006). Finally, we also found that 20% of highly expressed genes within our data set harbor more than one distinct candidate enhancer predicted to be active in the same tissue, supporting the hypothesis that certain genes contain multiple discrete regulatory elements that overlap in their enhancer activity (Frazer et al. 2004; de la Calle-Mustienes et al. 2005).
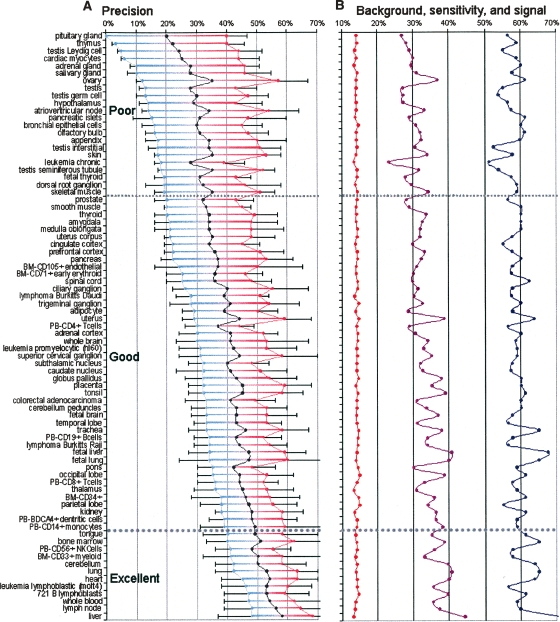
Figure 2.
Precision (A) and sensitivity (B) of the EI method of recognizing human tissue-specific enhancers. Lower- and upper-bound estimates of precision along with their average are given in red, blue, and black on precision plots (A), respectively. Standard deviation is also depicted for each lower- and upper-bound estimate. Tissues are split into poor, good, and excellent groups based on the lower-bound estimate of the precision. See Supplemental Figure S2 for corresponding mouse data. Navy and red curves on sensitivity plots (B) measure the percentage of high- and low-expressed gene loci with tissue-specific enhancers, respectively; while the middle purple curve estimates EI sensitivity for de novo enhancer recognition.
Since the EI method is based on the weighting of multiple TFs for their association with tissue-specific expression, we sought to further explore the nature of this combinatorial TF-scoring scheme. We found that in no case was a single TF sufficient to predict tissue-specific gene expression, supporting the notion that tissue-specific gene regulation is a direct result of interplay among multiple TFs. To quantify the impact of an individual ith TF on predicting gene expression in a particular tissue t, we calculated the TF importance parameter (Iit) defined as the product of the TF occurrence (percentage of tissue-specific candidate enhancers with a particular conserved TFBS) and its weight in a tissue-specific group of candidate enhancers (Supplemental Table S2). Since TF importance compounds the effects of TF occurrence and weight, it presents an integrative measure of the TF’s role in generating high positive scores of tissue-specific candidate regulatory elements. At the same time, it minimizes the impact of TFs that are rare or have small weights and thus do not contribute significantly to establishing either a positive or a negative tissue-specificity score. This quantification allowed for the identification of cohorts of TFs in candidate enhancers potentially involved in tissue-specific regulatory networks, i.e., those TFs both with high weights and high occurrences (see Supplemental Materials). As an example of a high TF impact on tissue-specific regulation, the photoreceptor-specific CRX TF has the highest importance parameter value in eye development (Supplemental Table S2) consistent with the known function of this regulatory protein in Cone-Rod Dystrophy (CRD), an inherited progressive disease that causes deterioration of the cone and rod photoreceptor cells and leads to blindness (Itabashi et al. 2004).
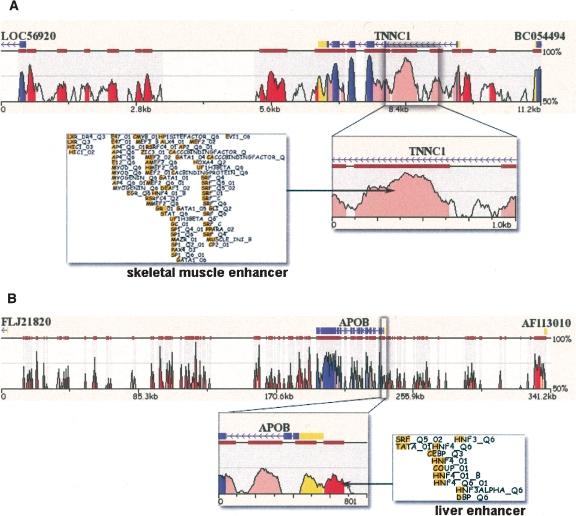
To illustrate this method’s ability to predict functional enhancers, we examined two well-characterized enhancers, one for skeletal muscle and one for liver, flanking the human cardiac/slow skeletal muscle troponin C (TNNC1) and the apolipoprotein B (APOB) genes, respectively (Fig. 3). An EI scan of the TNNC1 locus first identified four noncoding ECRs (of 12 total) as candidate regulatory elements (two intergenic, one intronic, and one promoter element). Subsequent EI optimization then correctly predicted the noncoding ECR in intron 1 as a skeletal muscle enhancer in precise agreement with the previously defined TNNC1 skeletal muscle enhancer (Christensen et al. 1993; Parmacek et al. 1994). In a second example, EI correctly identified the APOB promoter element as a fetal liver (and adult liver) enhancer and predicted transcription factors HNF4 and C/EBP to be activating APOB expression, in concordance with previous experimental studies (Novak et al. 1998).
Figure 3.
EI annotation of TNNC1 skeletal muscle (A) and APOB (B) liver enhancer. Zoomed-in view of Mulan (Ovcharenko et al. 2005) human/mouse evolutionary conservation profiles for these loci depicts candidate enhancer elements, followed by profile of conserved TFBS present within.
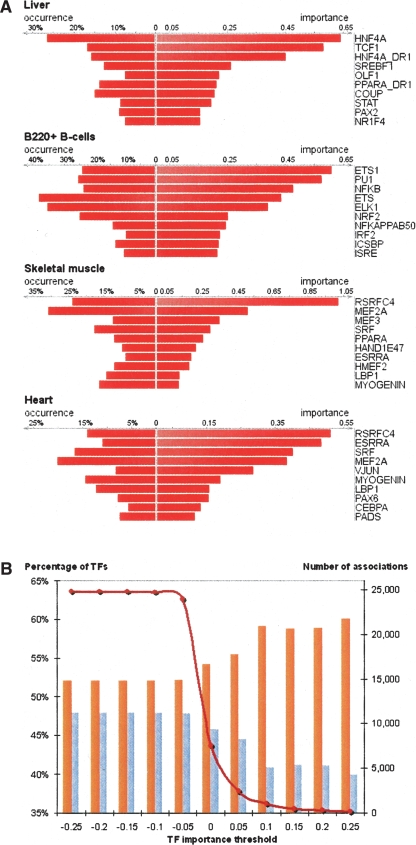
To explore the possibility of synergistic TF linkage that may be biologically required for directing tissue-specific gene expression, we extracted the top 10 scoring TFs for each tissue based on their importance in predicting tissue-specific expression. As an example, we focused on the TF characteristics of two similar tissue types: heart and skeletal muscle (Fig. 4A) (a complete list of the top TF for each tissue is provided in Supplemental Table S2). We observed that five of the top 10 TF predictions for both these muscle types are shared, four of which (MEF2, SRF, myogenin, and ESRRA) are strongly linked to transcriptional regulation in muscle tissue and associated with various human cardiac myopathies (Sakuma et al. 2003; Huss et al. 2004; Kadi et al. 2004; Parlakian et al. 2005). As a second example, the top 10 TF predictors of liver expression included Hepatocyte Nuclear Factor 1 (TCF1), HNF4A, PPARA, SREBF1, HNF4-DR1, NR2F2, and NR1H4 (Fig. 4A), all of which are known regulatory proteins important in liver function (Shih et al. 2001; Zannis et al. 2001; Cheng et al. 2006). These two examples highlight the biological plausibility of the EI method to predict tissue-specific gene expression.
Figure 4.
Importance and occurrence of individual TFs in candidate enhancers corresponding to mouse liver, B-cells, heart, and skeletal muscle (A). Binning of 25 k predicted TF-tissue associations by a minimal TF importance threshold (B). The number of TF-tissue associations almost does not change in the area of negative TF importance thresholds and rapidly decreases in the area of positive TF importance thresholds (dark red graph; right y-axis) indicative of a small number of TFs with large positive importance values and an even smaller number of TFs with large negative importance values. The percentage of TF-tissue associations that are confirmed by an increase in TF gene expression (orange bars) increases with the increase of minimal TF importance (followed by the corresponding decrease in the number of nonconfirmed associations—blue bars). As ∼60% of predicted TF-tissue associations with a minimal TF importance of 0.1 are supported by an increased level of TF gene expression in the corresponding tissue, this threshold could serve as a cut-off of reliability in TF-tissue association predictions.
To globally address the power of the predicted TF-tissue associations in addition to the support gained from the above selected examples, we mapped TFs to the human genome and determined the tissue gnfAtlas2 expression profile for each TF gene. Our rationale was that if tissue-specific gene expression predictiveness is based on TFBS density in candidate enhancer sequences, then the TF required for this function should be expressed in the tissue of activity. Thus, we attempted to correlate positive TF importance with the level of TF gene expression in the available 79 human tissues. This was accomplished by adjusting the minimal TF importance threshold increasingly from −0.25 to +0.25 (thus gradually increasing the ratio of TFs with positive importance values in the group) to determine whether TF expression and enhancer predictiveness were positively correlated (Fig. 4B). Indeed, we observed that ∼60% of predicted positive TF-tissue associations corresponding to TF importance thresholds of ≥0.1 were supported by an increased level of gene expression in the associated tissue (Fig. 4B). One possible explanation for the lack of total concordance between the predicted TF-tissue associations and tissue specificity is the ubiquitous nature of TF gene expression that often leads to ambiguous definitions of tissue specificity with increased and decreased level of gene expression in gnfAtlas2. Manual curation of these interactions revealed that 90% (142/158) of predicted TF-tissue associations with ≥0.25 TF importance threshold are supported by published literature or alternative sources of experimental evidence (see Supplemental materials; Table S3).
Since any parametric optimization approach could potentially introduce “overfitting”—the identification of random profiles that separate genes with high versus low expression purely by chance—we attempted to cross-validate our results. This was accomplished by characterizing the ability of the EI method to annotate tissue-specific enhancers in loci of highly expressed genes without any a priori knowledge of tissue specificity of gene expression (i.e., these genes were excluded from the training set; see Methods). This approach allowed us to quantify both the method’s precision (defined as the proportion of predicted elements that act as tissue-specific enhancers) and sensitivity for each tissue (Fig. 2). Through this analysis, we observed a high variability in EI precision across the 79 sampled human tissues, and hence, these tissues were classified into three quality groups (Fig. 2A): (1) poor (lower-bound precision, P↓ < 20%), (2) good (lower-bound precision, P↓ between 20% and 40%), and (3) excellent (lower-bound precision, P↓ > 40%) (Fig. 2). Next, we grouped lower- and upper-bound precision values to use their average as an estimate for the true precision and found that 72% (57/79) of human tissues have an average precision of 40%. These data allowed us to conclude that overfitting did not account for the majority of signals obtained from the EI predictive method. In contrast, EI was suboptimal for the remaining 22 human tissues, which fell into the poor category where the average precision was below 30%, indicating that overfitting likely explained a significant fraction of the signature derived for these tissues. Tissues comprised within this category mainly consisted of multiple gland and germ tissues, as well as structures such as the appendix and olfactory bulb. Based on these observations, we placed low significance values on the predictions derived for these tissues, and their enhancer predictions should be treated cautiously as they are likely to represent false-positives. In contrast, the average precision of the excellent group was above 50% for 12 tissues, including heart, liver, tongue, blood, and several immune tissues (Fig. 2) Thus, these tissue types bear the highest confidence of EI predictions.
Assigning tissue-specific predictions to conserved noncoding sequences in the human genome
Since the EI method can generate tissue-specific predictions for any conserved element, we used this approach to score 364 k previously reported candidate enhancers in the human genome (Prabhakar et al. 2006) (see Methods). In total, EI was able to assign a tissue specificity to 90% (328 k) of these elements, covering 4.0% of the human genome. This large data set comprises tissue-specificity predictions for the majority (86%) of genes in the human genome. It also represents an important resource for prioritizing tissue-specific enhancers in loci of genes with known functions when one is interested in sifting through multiple evolutionarily conserved elements and prioritizing only those that correspond to candidate enhancers with matching tissue specificity. We should note that we observed an overlap in tissue-specificity predictions as a result of several related tissues having similar EI recognition profiles (Supplemental Tables S6, S7). For example, 24% of CD4+ T-cell-predicted elements were also classified as CD8+ T-cell, while 14% of liver-predicted elements were simultaneously classified as fetal liver. In contrast, only 0.8% of CD4+ T-cell-predicted elements were simultaneously classified as fetal liver predictions. This suggests that the direct EI tissue-specificity annotation of conserved elements may fail to distinguish between closely related tissues, but can possibly distinguish between major tissue categories or different organs. However, since tissue specificity of these 364 k predictions could not always be supported by high expression of flanking genes, we anticipate this data set to feature a relatively high rate of false-positive predictions, warranting further validation of our predictions in elements that had been characterized using sophisticated in vivo approaches.
Experimental validation of tissue-specific enhancer predictions
Based on our genome-wide predictions of tissue-specific activities for all noncoding ECRs, we sought to determine their performance against existing enhancer data of gene expression derived from transgenic mouse studies. As a test bed, we examined the EI tissue-specific predictions for five previously characterized enhancers expressed in the brain and nervous system in the 1-Mb region upstream of the DACH1 gene (Nobrega et al. 2003). We found that three of these elements were predicted to have enhancer activity limited to brain tissues, while the two remaining elements were not assigned to any tissue (Supplemental Table S4). While these initial correlations were based on a small sample set, the statistical significance of this match is supported by a P-value of 0.004 (see Supplemental materials).
To expand these data beyond the limited published in vivo data for distant-acting enhancer elements, we next performed a large-scale analysis of our whole-genome predictions against a publicly available data set of 106 elements that have been shown to act as tissue-specific enhancers in the mouse at embryonic day 11.5 of development (E11.5) (data available at http://enhancer.lbl.gov) (Pennacchio et al. 2006) (Supplemental Table S5). In this data set, we found 71 (67%) enhancers to dictate expression in forebrain, midbrain, hindbrain, and/or neural tube. We thus assessed whether whole-genome EI tissue-specific predictions overlap with these in vivo characterized enhancers. Indeed, 28% (20/71) of these elements were selectively predicted as enhancers active in the brain and/or the nervous system. In addition, another 7% (5/71) of these validated CNS-specific enhancers had EI predictions with a mixed annotation of brain/CNS and another organ/tissue, suggesting that these elements are possibly multifunctional. We also observed 21% (15/71) of predictions provided tissue annotations inconsistent with the experimental data, while the remaining 44% (31/71) elements had no tissue-specific prediction(s) (Supplemental Table S5). This corresponded to 28% sensitivity and 50% precision in recognition of brain and CNS-specific enhancers using the EI method, de novo. By calculating the distribution of brain/CNS predictions in a large random data set (see Supplemental materials) we found the overlap of this analysis with experimental data to be 2.5-fold greater than what would be expected by chance, corresponding to a P-value of 0.0001.
To further explore the relationship between the 20 concordant EI whole-genome predictions and the existing in vivo nervous system data set described above, we examined the distribution of the predictions within the 18 different brain tissues present in the gnfAtlas2 database. While we found four or less of these, in vivo-defined CNS enhancers were predicted to be expressed in each of the 17 adult brain tissues present in the expression annotation, 11 of them were annotated to the fetal brain category in the gnfAtlas2 (the probability of this observation being random is <10−7 [see Supplemental materials]). This high ratio of fetal-brain predictions is consistent with the entire in vivo expression data set that corresponds to a single time point of enhancer analysis during embryonic development at E11.5. This suggests that the fetal brain-enhancer recognition profile of EI is a specific signature of in vivo embryonic brain enhancers, in contrast to enhancers active in specialized compartments of the adult brain. It is unclear, however, whether these enhancers are exclusively active during embryonic time points and not during adult stages. Additional in vivo data sets based on nonembryonic time points will further aid in assessing the ability of this approach to predict enhancer elements active in adult tissues.
Discussion
Deciphering the genetic code of gene regulation in vertebrate genomes remains a significant challenge that has been partially aided by the availability of the human and other vertebrate genome sequences. However, while techniques such as comparative genomics can enrich for putative enhancer sequences based on evolutionary conservation, predicting their tissue specificity has been difficult. Nevertheless, several proof-of-principle studies have demonstrated that there is a vaguely defined, but computationally recognizable genetic code of gene regulatory elements corresponding to selected biological functions (Thompson et al. 2004; Hallikas et al. 2006; Sun et al. 2006). Additional studies have also revealed the power of microarray expression data to correlate the distribution of evolutionarily conserved putative TFBS in the promoters of coexpressed human (and mouse) genes with the level and dynamics of gene expression (Sharan et al. 2003; Das et al. 2006). These early focused studies suggest that computationally predicting enhancer function is a solvable problem. We therefore developed a multifaceted approach coupling TF-binding specificities, comparative genomics, and microarray expression data in an attempt to recognize sequence signatures within putative enhancer elements in the human genome. Through these efforts, we show that it is possible to identify tissue-specific enhancers for 72% of human sampled tissues by constructing sequence-recognition profiles based on the distribution of TFBS in noncoding ECRs linked to genes expressed in similar tissues.
One of the inferences we can formulate based on the results of the EI method introduced here is the proportion of enhancer activity assigned to promoters versus more distant-acting sequences. This measurement was possible since the EI approach utilizes the three most highly conserved human–mouse elements neighboring the gene under investigation and thus goes beyond promoter only exploration of cis-regulatory features, the dominant method currently used in regulatory genomics. Through the comparison of the EI signal strength in promoter versus nonpromoter conserved elements, we found that only 23% of EI candidate enhancers map to promoter regions of corresponding genes. While a caveat to this analysis is the incomplete status of precisely defined promoter boundaries, this result is consistent with ChIP-chip and in vivo enhancer studies, which also suggest that more than half of human genes potentially rely on distant mechanisms of gene regulation (Lettice et al. 2003; Levine and Tjian 2003; Nobrega et al. 2003; Cawley et al. 2004).
Since this method can be applied to the analysis of any set of coexpressed genes, this provides a rapid and efficient approach for translating gene-expression data into function-specific gene regulatory principles. Thereby, it should be straightforward to extend this method to other tissues, developmental time-points, or functional gene categories (such as Gene Ontology and KEGG data sets [Kanehisa and Goto 2000; Harris et al. 2004], for example). In addition, the elements identified in this study represent a data set of tissue-specific candidate enhancers that could be used to guide the ongoing large-scale experimental efforts aimed at exploring transcriptional regulatory function in human, mouse, and other vertebrate genomes. Since the backbone of the EI optimization method is the association of TFs with tissue-specificities, we were able to predict over 7 k such associations and retrieve experimental evidence for 90% of them in a selected group of 158 TF-tissue associations (at a TF importance threshold of 0.25). Furthermore, characterization of TF spacing in predicted tissue-specific enhancers allowed us to extract ∼1 k TF pairs significantly enriched as putative synergistic activators in a given tissue (see Supplemental materials). While we were able to bring forth published evidence for several predicted TF co-occurrences, the vast majority of TF-tissue linkages and their potential interactions represent novel regulatory associations that could be used in facilitating future studies of the complexities of gene-regulatory pathways.
It is likely that computational approaches that identify gene-regulatory elements and assign tissue specificity to enhancer function will greatly improve over time. Current challenges include the varying quality and the limited number of tissues (and primarily adult origin) uniformly profiled in humans and mice by microarray analysis. Further difficulties arise from the small size of available in vivo spatial and temporal enhancer data to further serve as training sets, as well as our incomplete knowledge of TFs and their precise sequence-based binding specificities currently available in the TRANSFAC database (Wingender et al. 2000). The current method specifically targets identification of noncoding signatures specific to the highest tissue-specific expression and thus leads to the identification of tissue-specific enhancers. Therefore, studies aimed at identification of tissue-specific repressors would need to use a modified version of this approach with a different selection of genes in the signal and background data sets. In addition, the comparative analysis exploited here was limited to human–mouse genome alignments under one alignment and conservation scoring method. Nevertheless, despite these limitations, our finding of EI’s ability to identify tissue-specific enhancers with the available data sets is encouraging, and represents a platform for further efforts in this area.
In summary, the data presented here provide further support for the notion that sequence-based features in vertebrate cis-regulatory elements are computationally recognizable, similar to previous successes in the inference of coding, intron–exon, core promoter, and repetitive DNA sequence signatures. Even though our study is limited by the availability and reliability of position weight matrices (PWM) of known TFs, the methods introduced here present a universal framework for the de novo prediction of regulatory elements with shared biological function, as well as for defining novel interactions among transcription factors that can explain tissue-specific function of enhancer elements. Future computational efforts linked to topics such as human disease and vertebrate phenotypic diversity are likely to refine the predictive ability of our strategy and provide insights into gene regulatory mechanisms of unexplained biological phenomena.
Methods
Gene annotation and expression data integration
The UCSC Genome Browser (Kent et al. 2002) database was used to extract genes and link them to their physical chromosome location. Human and mouse “knownGene” transcripts (Karolchik et al. 2003) were mapped to the NCBI Build 35 of the human (hg17) and mouse (mm7) genomes and grouped into 18,504 and 17,636 nonoverlapping loci, respectively. GNF Novartis Atlas2 tissue-specific gene expression (Su et al. 2002) was extracted from the gnfAtlas2 table and mapped to their respective genes using the knownToGnfAtlas2 table (both tables are available in the UCSC Genome Browser database). At least one tissue-expression profile was available for each of 15,690 human and 14,303 mouse genes.
Identification of noncoding ECRs and candidate regulatory elements
Human–mouse alignments generated by the ECR Browser (Ovcharenko et al. 2004a) were extracted for all ECRs that satisfy the threshold of 100 bp in length and 70% identity as a basis for the study. These ECRs were subsequently filtered out for overlapping coding exons of “knownGene” genes, resulting in a data set of 1.4 M noncoding ECRs, from which 60 k candidate regulatory elements were selected by identifying the three most conserved ECRs for each locus as well as ECRs overlapping the 1.5-kb promoter region of each gene. The conservation level was quantified as a product of each element’s percent identity and its length. Lower to upper quartile distribution in the length of candidate regulatory elements spanned from 206 to 780 bp with the median of the distribution being 420 bp. While our selection of the three top ECRs per gene was somewhat arbitrary, two factors weighed in on this choice. First, by performing our analysis on a gene-by-gene basis, we selected an equal number of candidate enhancers for each gene. For example, this generally resulted in higher conservation thresholds for well-conserved loci of developmental genes and lower thresholds for rapidly diverging loci of immune system genes. Second, by choosing only the three top ECRs per gene, we limited our analysis to a tractable group of the most highly conserved elements while still maintaining the ability to capture several discrete enhancers that may individually direct expression to different tissues. Furthermore, the selection of the top most conserved elements has been previously linked to their increased likelihood of being biologically functional (Ovcharenko et al. 2004b; Prabhakar et al. 2006).
Profiling putative TFBS in candidate gene regulatory elements
Human–mouse ECR Brower genome alignments (Ovcharenko et al. 2004a) were processed by rVista 2.0 (Loots and Ovcharenko 2004) to identify evolutionarily conserved putative TFBS in the human and mouse genomes. A previously described optimized PWM threshold (Cartharius et al. 2005; Ovcharenko et al. 2005) was used to limit the appearance of predictions to five TFBS per 10 kb of random sequence. In total, 13.4 M conserved putative TFBS were identified using 554 TRANSFAC 9.4 PWMs (Wingender et al. 2000) and three manually curated PWMs for TBX5, NKX2.5, and GLI TFs (B. Bruneau and J. Aronowicz, pers. comm.). These putative TFBS were then grouped into 364 separate TF families (as several TFs have multiple overlapping definitions in the TRANSFAC database). These TF families are simply referred to as TFs in the text. The identification of evolutionarily conserved TFBS was performed independently of the identification of ECRs and candidate regulatory elements. Therefore, these TFs were superimposed with the 60 k candidate regulatory elements to construct a data set of 1.8 M putative TFBS in candidate regulatory elements as the basis for the study. On average, there were 29.4 TFBS identified in a candidate regulatory element. The number of TFBS in an average candidate regulatory element varied across the panel of sampled tissues in concordance with the varying length of these elements (Supplemental Table S8). Thus, some tissues (such as CNS tissues, for example) contained a much larger number of ECRs in their loci than average, while other loci (such as liver or bone marrow, for example) contained a significantly lower number of ECRs in their loci. As a result of the positive correlation between the ECR size and the number of putative TFBS within them (figure insert in Supplemental Table S8), more TFBS were observed per candidate regulatory element in genes displaying higher versus lower levels of overall DNA conservation. Finally, 52% of candidate regulatory elements contained multiple occurrences of the same TFBS in a single candidate regulatory element; though an individual TF was found to cluster in only ∼0.6% of candidate regulatory elements. Tissue-specific enrichment of putative TFBS in candidate enhancers was calculated as the ratio of their occurrence in loci of genes with high versus low expression in a given tissue for each TF. To correct for multiple hypothesis testing, the hypergeometric distribution with Bonferroni correction was used (Supplemental Table S1).
Assigning tissue specificity scores to candidate enhancers
The distribution of putative TFBS inside a candidate enhancer (or noncoding ECR) was utilized to assign a tissue-specificity score to that element. First, a tissue-specificity weight 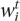 was assigned to each ith TF as a measure of its association with the tissue t. Next, the distribution of putative TFBS in the kth candidate enhancer was scored to define candidate enhancer tissue specificity:
was assigned to each ith TF as a measure of its association with the tissue t. Next, the distribution of putative TFBS in the kth candidate enhancer was scored to define candidate enhancer tissue specificity:
where 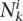 is the number of ith TF putative TFBS located in the kth candidate enhancer and the summation was performed over all nTF TFs. TF weights were allowed to vary from −1 to 10. Large positive weights
is the number of ith TF putative TFBS located in the kth candidate enhancer and the summation was performed over all nTF TFs. TF weights were allowed to vary from −1 to 10. Large positive weights 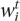 indicate a strong correlation between the ith TF and the tth tissue-specificity, while large negative weights indicate the unlikely presence of the ith TF in a candidate enhancer that is active in the tissue t.
indicate a strong correlation between the ith TF and the tth tissue-specificity, while large negative weights indicate the unlikely presence of the ith TF in a candidate enhancer that is active in the tissue t.
EI optimization to define TF tissue-specific weights
To identify tissue-specific candidate enhancers, the Brent’s method optimization was performed on TF weights 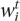 that maximizes the number of positively scoring candidate enhancers in loci of genes with high expression in a given tissue (L+) and simultaneously minimizes the number of positively scoring candidate enhancers in loci of genes with low expression in that tissue (L−). Optimization was performed independently for each different tissue. To ensure a reliable and specific identification of noncoding features in loci of highly expressed tissue-specific genes, a large background data set was included comprising 5 k loci of genes with low expression assigned to each tissue. A scoring function Ft,
that maximizes the number of positively scoring candidate enhancers in loci of genes with high expression in a given tissue (L+) and simultaneously minimizes the number of positively scoring candidate enhancers in loci of genes with low expression in that tissue (L−). Optimization was performed independently for each different tissue. To ensure a reliable and specific identification of noncoding features in loci of highly expressed tissue-specific genes, a large background data set was included comprising 5 k loci of genes with low expression assigned to each tissue. A scoring function Ft,
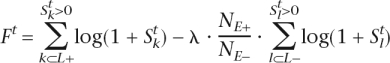 |
containing summations over all positively scoring candidate enhancers associated with L+ (k⊂L+) and L− (l⊂L−) was maximized to perform the optimization of weights (the distribution of positively scoring candidate enhancers in L+ and L− was allowed to change dynamically following the change in TF weights). The ratio of the total number of candidate enhancers in L+ (NE+) to the total number of candidate enhancers in L− (NE−) was introduced to the scoring function to account for differences in the number of genes with high versus low gene expression and the number of corresponding candidate enhancers. λ, or the signal enrichment coefficient served to increase the negative impact of positively scoring noncoding ECRs in L−. λ was selected as 1 during the initial optimization step and then gradually increased to 10,000 to achieve the greatest separation between loci of highly and lowly expressed genes. Optimization was initialized with TF weights estimated using the density of putative TFBS in L+ and L− as
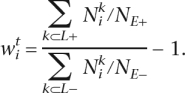 |
Initial TF weights were upper-bounded by 1 and the optimization was performed contiguously and recursively for each ith TF. It was interrupted after achieving an increase l < 0.1 in the scoring function during a cycle of TF weights optimizations across all TFs. An important property of this optimization is the dynamic selection of the positively scoring subset of tissue-specific candidate enhancers from the original set of candidate enhancers.
Cross-validation
For cross-validation experiments, we expanded the data set of highly expressed genes to 400 and subdivided this set into two groups consisting of (1) 300 genes for EI optimization, and (2)100 genes for testing the signal recognition. None of the 100 test genes were included in the set used for the optimization of the algorithm. Cross-validation was repeated four times to estimate the statistical error in precision and sensitivity. The four cross-validation replicas of 100 test genes did not overlap with each other to ensure that four independent quantifications are carried out. Similarly, in each case a different group of 500 genes was removed from the background data set for each cycle of EI optimization. Using this approach, four independent rounds of EI optimization were performed with 300 signal (highly expressed) and 4500 background (lowly expressed) genes and subsequently applied the generated TF profiles to independently calculate the percentage of tissue-specific candidate enhancers from the 100 test (R+) and 500 control (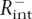 ) data sets. Optimization and testing of control genes was restricted to nonintergenic regions to avoid potential cross-talk with tissue-specific enhancers controlling the expression of neighboring genes. Therefore, EI precision in recognizing tissue-specific enhancers (which measures the ratio of true positive tissue-specific enhancers in the full data set of predicted elements)
) data sets. Optimization and testing of control genes was restricted to nonintergenic regions to avoid potential cross-talk with tissue-specific enhancers controlling the expression of neighboring genes. Therefore, EI precision in recognizing tissue-specific enhancers (which measures the ratio of true positive tissue-specific enhancers in the full data set of predicted elements)
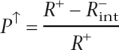 |
represents the upper-bound estimate of the precision. By excluding the nonintergenic component of loci of test highly expressed genes from the quantification, after that, one decreases the percentage of recognized test genes to 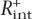 and the corresponding precision
and the corresponding precision
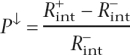 |
then represents the lower bound of the precision of the method. By averaging these two values, an estimate of the real precision of the method (P) was obtained. Also, R+ served as an estimate for the lower-bound sensitivity of the method (Sn↓) in the de novo recognition of tissue-specific enhancers (which measures the probability of a tissue-specific enhancer to be detected by EI) in cases where the corresponding gene does not belong to a specific group of highly expressed genes.
Mapping TFs to known transcripts
TRANSFAC names of sampled TFs were used for automated (and manually curated after that) GenBank queries to identify the name and chromosomal location of the human gene best matching each TF. For example, we were able to map the AML1 TF matrix to the human runt-related transcription factor 1 (RUNX1) residing at chr21 (q22.12). In total, 314 of 364 utilized TRANSFAC TFs were successfully mapped to human genes. In several instances, a TF mapped to more than one gene locus (in the case of E2F1DP2 heterodimer, the TF complex mapped to E2F1 and TFDP2 genes; similarly, the SREBP TF mapped to both SREBF1 and SREBF2 genes); in such cases, the expression profiles were averaged across all genes corresponding to the TF or TF complex.
Permutation analysis to identify significant tissue-specific inter-TF interactions
The distribution of positively scoring TFBS was analyzed in tissue-specific candidate enhancers independently for each tissue. Only TFs with individual TF occurrence ≥5% or TF importance ≥0.05 were subselected for the analysis. The number of TF–TF pairs with the minimal and maximal inter-TF distances of five and 100, respectively, was calculated for each pair of TFs. A total of 10 k permutations randomizing the distribution of TF name labels among different TFBS were performed. The total number of TFBS for each TF as well as positions of individual TFBS was kept intact during the randomization. Next, a subset of TF pairs was extracted that occur less frequently in 95% of permutation tests than in the original distribution (corresponding to a P-value <0.05 to observe the original distribution by chance) and that corresponded to at least a twofold increase in their density in the original distribution as compared with an average pair density in permutation tests.
Assigning tissue-specific enhancer predictions to a whole-genome data set of human–mouse noncoding ECRs
TFBS distributions were scored for 364 k previously cataloged human/mouse conserved noncoding sequences (Prabhakar et al. 2006), and a comprehensive 1.4-M noncoding ECRs set for the entire human genome, to identify 328 and 588 k elements, respectively, that have a positive tissue-specificity score according to EI tissue-specificity profiles. A P-value 0.05 cut-off was used for the 364 k set that corresponds to an estimate of 0.05 false-positive enhancer predictions per 10 kb of random sequence (Prabhakar et al. 2006). In cases of multiple tissue associations assigned to an element, up to three top-scoring associations were selected with the score of at least 50% of the most top-scoring tissue association (data available at http://www.dcode.org/EI). The same score selection procedure was applied for the analysis of organ specificities.
Acknowledgments
We thank Shyam Prabhakar and Alex Poliakov for providing Gumby enhancer predictions in the human genome. G.G.L. and I.O. were supported by an LLNL LDRD-04-ERD-052 grant; and I.O. was in part supported by an LLNL LDRD-06-ERD-004 grant. The work was performed under the auspices of the United States Department of Energy by the University of California, Lawrence Livermore National Laboratory Contract W-7405-Eng-48. L.A.P. was supported by the Grant HL066681, Berkeley-PGA, under the Programs for Genomic Application, funded by National Heart, Lung, & Blood Institute, and HG003988 funded by National Human Genome Research Institute and performed under Department of Energy Contract DE-AC02-05CH11231, University of California, E.O. Lawrence Berkeley National Laboratory.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5972507
References
- Bajic V.B., Tan S.L., Suzuki Y., Sugano S., Tan S.L., Suzuki Y., Sugano S., Suzuki Y., Sugano S., Sugano S. Promoter prediction analysis on the whole human genome. Nat. Biotechnol. 2004;22:1467–1473. doi: 10.1038/nbt1032. [DOI] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T., Klingenhoff A., Frisch M., Bayerlein M., Werner T., Frisch M., Bayerlein M., Werner T., Bayerlein M., Werner T., Werner T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chang W.T., Chen H.I., Chiou R.J., Chen C.Y., Huang A.M., Chen H.I., Chiou R.J., Chen C.Y., Huang A.M., Chiou R.J., Chen C.Y., Huang A.M., Chen C.Y., Huang A.M., Huang A.M. A novel function of transcription factor alpha-Pal/NRF-1: Increasing neurite outgrowth. Biochem. Biophys. Res. Commun. 2005;334:199–206. doi: 10.1016/j.bbrc.2005.06.079. [DOI] [PubMed] [Google Scholar]
- Cheng W., Guo L., Zhang Z., Soo H.M., Wen C., Wu W., Peng J., Guo L., Zhang Z., Soo H.M., Wen C., Wu W., Peng J., Zhang Z., Soo H.M., Wen C., Wu W., Peng J., Soo H.M., Wen C., Wu W., Peng J., Wen C., Wu W., Peng J., Wu W., Peng J., Peng J. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev. Biol. 2006;294:482–496. doi: 10.1016/j.ydbio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Christensen T.H., Prentice H., Gahlmann R., Kedes L., Prentice H., Gahlmann R., Kedes L., Gahlmann R., Kedes L., Kedes L. Regulation of the human cardiac/slow-twitch troponin C gene by multiple, cooperative, cell-type-specific, and MyoD-responsive elements. Mol. Cell. Biol. 1993;13:6752–6765. doi: 10.1128/mcb.13.11.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D., Nahle Z., Zhang M.Q., Nahle Z., Zhang M.Q., Zhang M.Q. Adaptively inferring human transcriptional subnetworks. Mol. Syst. Biol. 2006;2:2006.0029. doi: 10.1038/msb4100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Allende M.L., Gomez-Skarmeta J.L., Gomez-Skarmeta J.L. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis E.T., Reymond A., Antonarakis S.E., Reymond A., Antonarakis S.E., Antonarakis S.E. Conserved non-genic sequences—An unexpected feature of mammalian genomes. Nat. Rev. Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- Frazer K.A., Tao H., Osoegawa K., de Jong P.J., Chen X., Doherty M.F., Cox D.R., Tao H., Osoegawa K., de Jong P.J., Chen X., Doherty M.F., Cox D.R., Osoegawa K., de Jong P.J., Chen X., Doherty M.F., Cox D.R., de Jong P.J., Chen X., Doherty M.F., Cox D.R., Chen X., Doherty M.F., Cox D.R., Doherty M.F., Cox D.R., Cox D.R. Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res. 2004;14:367–372. doi: 10.1101/gr.1961204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas O., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Rautiainen R., Partanen J., Ukkonen E., Taipale J., Partanen J., Ukkonen E., Taipale J., Ukkonen E., Taipale J., Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Eilbeck K., Lewis S., Marshall B., Mungall C., Lewis S., Marshall B., Mungall C., Marshall B., Mungall C., Mungall C., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss J.M., Torra I.P., Staels B., Giguere V., Kelly D.P., Torra I.P., Staels B., Giguere V., Kelly D.P., Staels B., Giguere V., Kelly D.P., Giguere V., Kelly D.P., Kelly D.P. Estrogen-related receptor α directs peroxisome proliferator-activated receptor α signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilia M., Sugiyama Y., Price J., Sugiyama Y., Price J., Price J. Gender and age related expression of Oct-6–a POU III domain transcription factor, in the adult mouse brain. Neurosci. Lett. 2003;344:138–140. doi: 10.1016/s0304-3940(03)00445-2. [DOI] [PubMed] [Google Scholar]
- Itabashi T., Wada Y., Sato H., Kawamura M., Shiono T., Tamai M., Wada Y., Sato H., Kawamura M., Shiono T., Tamai M., Sato H., Kawamura M., Shiono T., Tamai M., Kawamura M., Shiono T., Tamai M., Shiono T., Tamai M., Tamai M. Novel 615delC mutation in the CRX gene in a Japanese family with cone-rod dystrophy. Am. J. Ophthalmol. 2004;138:876–877. doi: 10.1016/j.ajo.2004.05.067. [DOI] [PubMed] [Google Scholar]
- Kadi F., Johansson F., Johansson R., Sjostrom M., Henriksson J., Johansson F., Johansson R., Sjostrom M., Henriksson J., Johansson R., Sjostrom M., Henriksson J., Sjostrom M., Henriksson J., Henriksson J. Effects of one bout of endurance exercise on the expression of myogenin in human quadriceps muscle. Histochem. Cell Biol. 2004;121:329–334. doi: 10.1007/s00418-004-0630-z. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B., Richmond T.A., Wu Y., Green R.D., Ren B., Wu Y., Green R.D., Ren B., Green R.D., Ren B., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice L.A., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Heaney S.J., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E., Goode D., Elgar G., Hill R.E., de Graaff E., Elgar G., Hill R.E., de Graaff E., Hill R.E., de Graaff E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Levine M., Tjian R., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., III, Zody M.C., Chang J.L., Kulbokas E.J., III, Zody M.C., Kulbokas E.J., III, Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Loots G.G., Ovcharenko I., Ovcharenko I. rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Miller W., Rubin E.M., Frazer K.A., Rubin E.M., Frazer K.A., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M., Ovcharenko I., Afzal V., Rubin E.M., Afzal V., Rubin E.M., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Novak E.M., Dantas K.C., Charbel C.E., Bydlowski S.P., Dantas K.C., Charbel C.E., Bydlowski S.P., Charbel C.E., Bydlowski S.P., Bydlowski S.P. Association of hepatic nuclear factor-4 in the apolipoprotein B promoter: A preliminary report. Braz. J. Med. Biol. Res. 1998;31:1405–1408. doi: 10.1590/s0100-879x1998001100006. [DOI] [PubMed] [Google Scholar]
- Okuda T., Tagawa K., Qi M.L., Hoshio M., Ueda H., Kawano H., Kanazawa I., Muramatsu M., Okazawa H., Tagawa K., Qi M.L., Hoshio M., Ueda H., Kawano H., Kanazawa I., Muramatsu M., Okazawa H., Qi M.L., Hoshio M., Ueda H., Kawano H., Kanazawa I., Muramatsu M., Okazawa H., Hoshio M., Ueda H., Kawano H., Kanazawa I., Muramatsu M., Okazawa H., Ueda H., Kawano H., Kanazawa I., Muramatsu M., Okazawa H., Kawano H., Kanazawa I., Muramatsu M., Okazawa H., Kanazawa I., Muramatsu M., Okazawa H., Muramatsu M., Okazawa H., Okazawa H. Oct-3/4 repression accelerates differentiation of neural progenitor cells in vitro and in vivo. Brain Res. Mol. Brain Res. 2004;132:18–30. doi: 10.1016/j.molbrainres.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I., Nobrega M.A., Loots G.G., Stubbs L., Nobrega M.A., Loots G.G., Stubbs L., Loots G.G., Stubbs L., Stubbs L. ECR Browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004a;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I., Stubbs L., Loots G.G., Stubbs L., Loots G.G., Loots G.G. Interpreting mammalian evolution using Fugu genome comparisons. Genomics. 2004b;84:890–895. doi: 10.1016/j.ygeno.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I., Loots G.G., Giardine B.M., Hou M., Ma J., Hardison R.C., Stubbs L., Miller W., Loots G.G., Giardine B.M., Hou M., Ma J., Hardison R.C., Stubbs L., Miller W., Giardine B.M., Hou M., Ma J., Hardison R.C., Stubbs L., Miller W., Hou M., Ma J., Hardison R.C., Stubbs L., Miller W., Ma J., Hardison R.C., Stubbs L., Miller W., Hardison R.C., Stubbs L., Miller W., Stubbs L., Miller W., Miller W. Mulan: Multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlakian A., Charvet C., Escoubet B., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Charvet C., Escoubet B., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Escoubet B., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Mericskay M., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Molkentin J.D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Gary-Bobo G., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., De Windt L.J., Ludosky M.A., Paulin D., Daegelen D., Ludosky M.A., Paulin D., Daegelen D., Paulin D., Daegelen D., Daegelen D., et al. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation. 2005;112:2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778. [DOI] [PubMed] [Google Scholar]
- Parmacek M.S., Ip H.S., Jung F., Shen T., Martin J.F., Vora A.J., Olson E.N., Leiden J.M., Ip H.S., Jung F., Shen T., Martin J.F., Vora A.J., Olson E.N., Leiden J.M., Jung F., Shen T., Martin J.F., Vora A.J., Olson E.N., Leiden J.M., Shen T., Martin J.F., Vora A.J., Olson E.N., Leiden J.M., Martin J.F., Vora A.J., Olson E.N., Leiden J.M., Vora A.J., Olson E.N., Leiden J.M., Olson E.N., Leiden J.M., Leiden J.M. A novel myogenic regulatory circuit controls slow/cardiac troponin C gene transcription in skeletal muscle. Mol. Cell. Biol. 1994;14:1870–1885. doi: 10.1128/mcb.14.3.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Dubchak I., Holt A., Lewis K.D., Holt A., Lewis K.D., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Prabhakar S., Poulin F., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Poulin F., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Shoukry M., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Afzal V., Rubin E.M., Couronne O., Pennacchio L.A., Rubin E.M., Couronne O., Pennacchio L.A., Couronne O., Pennacchio L.A., Pennacchio L.A. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A., Alvania R.S., Lonze B.E., Ramanan N., Kim T., Huang Y., Dawson T.M., Snyder S.H., Ginty D.D., Alvania R.S., Lonze B.E., Ramanan N., Kim T., Huang Y., Dawson T.M., Snyder S.H., Ginty D.D., Lonze B.E., Ramanan N., Kim T., Huang Y., Dawson T.M., Snyder S.H., Ginty D.D., Ramanan N., Kim T., Huang Y., Dawson T.M., Snyder S.H., Ginty D.D., Kim T., Huang Y., Dawson T.M., Snyder S.H., Ginty D.D., Huang Y., Dawson T.M., Snyder S.H., Ginty D.D., Dawson T.M., Snyder S.H., Ginty D.D., Snyder S.H., Ginty D.D., Ginty D.D. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol. Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Sakuma K., Nishikawa J., Nakao R., Nakano H., Sano M., Yasuhara M., Nishikawa J., Nakao R., Nakano H., Sano M., Yasuhara M., Nakao R., Nakano H., Sano M., Yasuhara M., Nakano H., Sano M., Yasuhara M., Sano M., Yasuhara M., Yasuhara M. Serum response factor plays an important role in the mechanically overloaded plantaris muscle of rats. Histochem. Cell Biol. 2003;119:149–160. doi: 10.1007/s00418-003-0499-2. [DOI] [PubMed] [Google Scholar]
- Shalizi A.K., Bonni A., Bonni A. Brawn for Brains: The role of MEF2 proteins in the developing nervous system. Curr. Top. Dev. Biol. 2005;69:239–266. doi: 10.1016/S0070-2153(05)69009-6. [DOI] [PubMed] [Google Scholar]
- Sharan R., Ovcharenko I., Ben-Hur A., Karp R.M., Ovcharenko I., Ben-Hur A., Karp R.M., Ben-Hur A., Karp R.M., Karp R.M. CREME: A framework for identifying cis-regulatory modules in human-mouse conserved segments. Bioinformatics. 2003;19:i283–i291. doi: 10.1093/bioinformatics/btg1039. [DOI] [PubMed] [Google Scholar]
- Sharan R., Ben-Hur A., Loots G.G., Ovcharenko I., Ben-Hur A., Loots G.G., Ovcharenko I., Loots G.G., Ovcharenko I., Ovcharenko I. CREME: Cis-Regulatory Module Explorer for the human genome. Nucleic Acids Res. 2004;32:W253–W256. doi: 10.1093/nar/gkh385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih D.Q., Bussen M., Sehayek E., Ananthanarayanan M., Shneider B.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., Bussen M., Sehayek E., Ananthanarayanan M., Shneider B.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., Sehayek E., Ananthanarayanan M., Shneider B.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., Ananthanarayanan M., Shneider B.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., Shneider B.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L., Bollileni J.S., Gonzalez F.J., Breslow J.L., Gonzalez F.J., Breslow J.L., Breslow J.L., et al. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., Vega R.G., Sapinoso L.M., Moqrich A., Sapinoso L.M., Moqrich A., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Chen G., Streb J.W., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Chen G., Streb J.W., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Streb J.W., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Long X., Yang Y., Stoeckert C.J., Jr., Miano J.M., Yang Y., Stoeckert C.J., Jr., Miano J.M., Stoeckert C.J., Jr., Miano J.M., Miano J.M. Defining the mammalian CArGome. Genome Res. 2006;16:197–207. doi: 10.1101/gr.4108706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W., Palumbo M.J., Wasserman W.W., Liu J.S., Lawrence C.E., Palumbo M.J., Wasserman W.W., Liu J.S., Lawrence C.E., Wasserman W.W., Liu J.S., Lawrence C.E., Liu J.S., Lawrence C.E., Lawrence C.E. Decoding human regulatory circuits. Genome Res. 2004;14:1967–1974. doi: 10.1101/gr.2589004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa M., Takemoto T., Kamachi Y., Kondoh H., Takemoto T., Kamachi Y., Kondoh H., Kamachi Y., Kondoh H., Kondoh H. Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech. Dev. 2004;121:1145–1158. doi: 10.1016/j.mod.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F., Meinhardt T., Pruss M., Reuter I., Schacherer F., Pruss M., Reuter I., Schacherer F., Reuter I., Schacherer F., Schacherer F. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis V.I., Kan H.Y., Kritis A., Zanni E., Kardassis D., Kan H.Y., Kritis A., Zanni E., Kardassis D., Kritis A., Zanni E., Kardassis D., Zanni E., Kardassis D., Kardassis D. Transcriptional regulation of the human apolipoprotein genes. Front. Biosci. 2001;6:D456–D504. doi: 10.2741/zannis. [DOI] [PubMed] [Google Scholar]