Abstract
Restriction site associated DNA (RAD) tags are a genome-wide representation of every site of a particular restriction enzyme by short DNA tags. Most organisms segregate large numbers of DNA sequence polymorphisms that disrupt restriction sites, which allows RAD tags to serve as genetic markers spread at a high density throughout the genome. Here, we demonstrate the applicability of RAD markers for both individual and bulk-segregant genotyping. First, we show that these markers can be identified and typed on pre-existing microarray formats. Second, we present a method that uses RAD marker DNA to rapidly produce a low-cost microarray genotyping resource that can be used to efficiently identify and type thousands of RAD markers. We demonstrate the utility of the former approach by using a tiling path array for the fruit fly to map a recombination breakpoint, and the latter approach by creating and using an enriched RAD marker array for the threespine stickleback. The high number of RAD markers enabled localization of a previously identified region, as well as a second region also associated with the lateral plate phenotype. Taken together, our results demonstrate that RAD markers, and the method to develop a RAD marker microarray resource, allow high-throughput, high-resolution genotyping in both model and nonmodel systems.
Over the past century, much has been learned about the genetic bases of developmental and physiological processes through intensive genetic screens in a handful of model organisms. Because of their power, forward genetic approaches in these models will continue to play a vital role in understanding genetic networks (Davidson 2001). In addition, a large pool of phenotypic variation exists throughout the amazing diversity of the millions of species outside of the laboratory, the genetic basis of which has been relatively unexplored (Carroll et al. 2005). Methods that facilitate the rapid identification of the genes that underlie both mutagen-induced and natural phenotypic variation will help to provide a much deeper understanding of many biological processes (Fisher 1930; Orr and Presgraves 2000).
Genetic mapping is a critical tool for deciphering the molecular underpinnings of phenotypic variation. Recombination mapping has long been used to correlate genetic markers with the presence or absence of Mendelian traits. The same fundamental approach allows a statistical association between a genomic region and a proportion of the phenotypic variance for quantitative traits (Lynch and Walsh 1998; Mackay 2001; Doerge 2002). Furthermore, the potential of association mapping, which takes advantage of numerous generations of ancestral recombination, is beginning to be recognized as a way to directly map variation in natural populations without the need to first produce laboratory crosses (Luikart et al. 2003; Storz 2005; Breseghello and Sorrells 2006). All of these approaches have in common the need for high-throughput, cost-effective methods to identify and genotype polymorphisms.
Genotyping methods have traditionally been expensive and labor intensive, but new high-throughput technologies hold promise. Allele-specific oligonucleotide microarrays allow many thousands of single nucleotide polymorphisms (SNPs) to be scored in a single hybridization (Wang et al. 1998; Matsuzaki et al. 2004). Other methods have been developed that use direct hybridization of labeled genomic DNA to short oligonucleotide arrays (Winzeler et al. 1998; Borevitz et al. 2003). Like SNPs, single feature polymorphisms (SFPs) can be discovered and typed at a high density given the availability of a short oligonucleotide array. However, the barriers to their widespread use are substantial. Some combination of prior sequencing, resequencing for marker identification, oligonucleotide synthesis, and other expensive production costs is required to create the physical resources necessary for these methods. For nonmodel systems, such high-resolution mapping resources are not feasible with current methods. Other microarray-based techniques have recently been developed that do not require significant prior investment (Jaccoud et al. 2001; Wenzl et al. 2004). While these procedures are an attractive option for use in nonmodel organisms without extensive resources, the low number of markers limits their applications.
Here we demonstrate restriction site associated DNA (RAD) marker genotyping, a method that can identify and type a large number of markers on a resource that is easy to produce for both model and nonmodel organisms. First, we apply these markers to rapidly map a recombination breakpoint in the model organism Drosophila melanogaster using a pre-existing genomic tiling path microarray. Second, we demonstrate a more powerful aspect of these markers by presenting a method that uses RAD marker DNA to rapidly produce a low-cost microarray genotyping resource. We produce a RAD marker microarray for threespine stickleback (Gasterosteus aculeatus) that allows for the typing of thousands of markers in parallel for both bulk and individual samples. The approach of using RAD markers, and the method to develop a RAD marker microarray resource, can greatly increase the rate at which the genetic bases of traits are identified in model and nonmodel systems.
Results
Restriction site associated DNA (RAD) markers
SNPs that disrupt restriction sites have long been assayed by a variety of genetic markers (e.g., RFLP, AFLP) (Botstein et al. 1980; Vos et al. 1995). A drawback of present techniques based on restriction site polymorphisms is that they screen only a small subset of available restriction sites for any particular enzyme. We have developed an approach that allows for nearly every restriction site of a particular enzyme to be screened in parallel (Fig. 1). Genomic DNA is digested with a particular restriction enzyme and subsequently ligated to biotinylated linkers. The DNA is then randomly sheared into fragments much smaller than the average distance between restriction sites, leaving attached to the linkers only the fragments that were directly flanking a restriction site. Streptavidin beads are used to immobilize these fragments while the rest of the DNA is removed. Digestion at the original restriction sites is used to release the fragments from the beads. This process specifically isolates DNA tags directly flanking the restriction sites of a particular restriction enzyme throughout the genome. What we have termed restriction site associated DNA (RAD) markers can be identified and/or typed by detecting differential hybridization patterns of RAD-tag samples on a microarray (Fig. 1).
Figure 1.
Restriction site associated DNA (RAD) markers can be identified and typed by detecting differential hybridization patterns of RAD tags on a microarray. Genomic DNA samples S1 and S2 contain the recognition sequence for various restriction enzymes at locations throughout the genome. Dark blue triangles represent restriction sites of a particular enzyme. Some of these restriction sites are only present in one sample because of polymorphisms that disrupt the recognition sequence (red asterisks). The two samples are separately digested with a particular restriction enzyme and then ligated to biotinylated linkers (light blue ellipses). The DNA is randomly sheared leaving only the fragments that were directly flanking a restriction site attached to biotin linkers. These fragments are purified using streptavidin beads and released by digestion at the original restriction site. Loci containing polymorphisms, such as the left locus of S2 or the right locus of S1, will not contain tags for that locus in the purified RAD-tag sample, thus resulting in differential hybridization patterns of RAD tags on a microarray.
RAD markers rapidly localized a breakpoint to a 150-kb region in Drosophila
We first confirmed the feasibility of using RAD markers for genotyping by identifying a recombination breakpoint in D. melanogaster, a model organism with a rich set of genomic resources. We had previously designed a spotted, tiling path microarray made up of 3-kb clones used in sequencing the Drosophila genome (Hollich et al. 2004; Sandmann et al. 2006), which made it possible to test the behavior of RAD markers between distinct fly lines. RAD tags were isolated separately from EcoRI-digested genomic DNA of two polymorphic strains (Oregon-R and Canton-S). The two RAD-tag samples were competitively hybridized directly against each other to the genomic tiling array to identify polymorphic markers. Two independent experimental replicates had a correlation coefficient of r = 0.94. By using a twofold cutoff of the averaged ratios, we identified 1117 polymorphic array elements between the two lines.
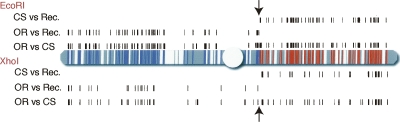
A fly stock carrying a homozygous, recombinant second chromosome was created with genetic material from the Oregon-R and Canton-S parents. We compared RAD tags from the recombinant line to each of the parents, allowing us to infer patterns of chromosomal inheritance (Fig. 2). A similar pattern of markers on the right part of chromosome 2 results from both the Oregon-R versus Canton-S hybridization and the recombinant line versus Canton-S hybridization, indicating that the chromosomal material in the recombinant came from the Oregon-R parent. The left part of the second chromosome showed the opposite pattern, indicating that the genetic material came from the Canton-S parent. The polymorphism pattern exhibited a clear switch between RAD markers at positions 3,320,000 and 3,470,000, indicating that the recombination breakpoint was in that 150-kb region. The experiment was repeated using XhoI-digested genomic DNA. As predicted, a different set of markers was identified, but the recombination breakpoint was mapped to the same region. Thus, RAD markers were easily identified by hybridization to a tiling path microarray and used to rapidly and precisely localize a genetic breakpoint.
Figure 2.
Recombination breakpoint mapping using RAD markers in Drosophila. Polymorphic RAD markers between Oregon-R (OR) and Canton-S (CS) flies were identified on a genomic tiling path microarray. Black tick marks above the diagrammed second chromosome represent the genomic locations of EcoRI RAD tags with an average differential hybridization greater than twofold. Tick marks below the chromosome represent XhoI RAD tags with a differential hybridization >2.3-fold. Polymorphic markers are found when the recombinant line (Rec.) is compared to the OR parental line on the left arm of the chromosome and when compared to the CS parental line on most of the right arm of the chromosome. Marker patterns indicative of CS inheritance are shown as blue regions of the chromosome, and OR inheritance as red. Arrows mark the recombination breakpoint. Large gaps in the tiling array are shown as white regions of the chromosome.
RAD marker microarrays are an inexpensive resource for identifying and typing RAD markers
In the absence of genomic tiling path microarrays, as was used in the previously described Drosophila experiment, RAD markers could be identified by hybridization to cDNA or oligonucleotide microarrays. However, the reduced sequence representation of these microarrays decreases the number of identifiable RAD markers. In many cases, there is no suitable pre-existing array on which to hybridize RAD tags. In order to mitigate this problem, we developed a new type of array for optimal identification and typing of RAD markers. This microarray is composed of RAD tags themselves, preferentially those tags that are informative RAD markers (Fig. 3). We tested this array production method in the threespine stickleback, G. aculeatus, an emerging model organism lacking the genomic resources of Drosophila, but possessing a diverse array of phenotypes (Bell and Foster 1994; Peichel 2005; Cresko et al. 2006).
Figure 3.
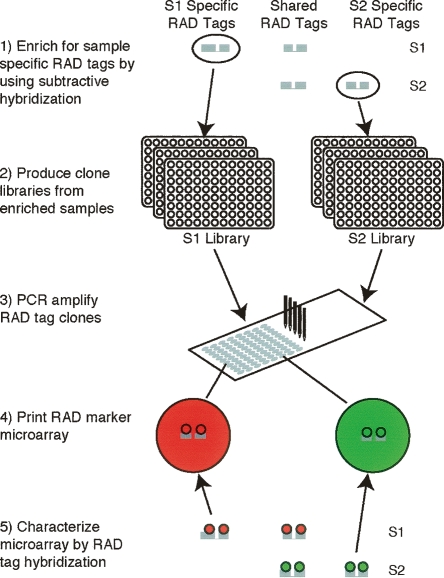
Enriched RAD marker microarray production and characterization. RAD-tag samples S1 and S2 contain polymorphic sets of RAD tags. RAD tags that are present in both individuals will not serve as informative markers. In order to produce an array that types a large number of informative markers, subtractive hybridization is used to enrich for sample-specific RAD tags. RAD-tag clone libraries are generated from these enriched samples. These clone libraries are used as templates for PCR, the products of which are spotted to produce RAD marker microarrays. To identify informative markers, RAD-tag samples S1 and S2 are fluorescently labeled and competitively hybridized directly against each other to the array.
We chose two polymorphic stickleback individuals, one from an ancestral oceanic population (Rabbit Slough, RS) and another from a derived freshwater population (Bear Paw, BP; see Methods). Individuals from both populations had been used previously for experimental mapping studies of bony armor variation (Cresko et al. 2004), and the BP population provided the source material for the stickleback genome project. EcoRI RAD-tag samples were isolated from these individuals, and subtractive hybridizations between the samples were performed in order to enrich for informative RAD tags. BP tags were used as the driver in one subtraction in order to enrich for tags specific to the RS sample, and the reciprocal subtraction was also performed in order to enrich for BP-specific tags. RAD-tag libraries of 9600 clones were made from the enriched samples of each individual. These clone sets were used as templates for PCR, the products of which were spotted as 19,200 elements to create an enriched RAD-tag microarray.
RAD marker microarrays allow for the identification of thousands of polymorphic markers
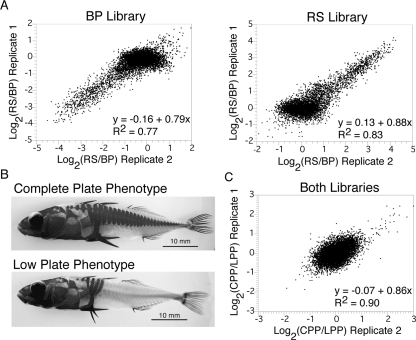
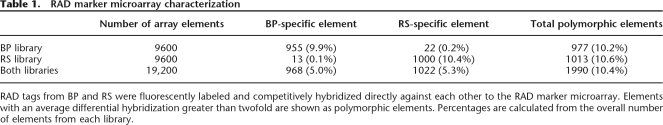
We determined the number of informative markers typed by this enriched RAD marker array by competitively hybridizing directly against each other the RAD-tag samples from the same BP and RS individuals that were used to produce the RAD array. Experimental replicates were highly reproducible (Fig. 4A). A twofold cutoff of the average of two experimental replicates resulted in the identification of ∼2000 array elements with differential hybridization between the RS and BP samples. As predicted, array elements for which the increased hybridization was associated with the BP-tag sample were isolated specifically from the BP individual. A similar specificity occurred for the RS-tag sample and the RS-derived array elements (Fig. 4A; Table 1). As a control, we investigated if the hybridization differences we observed between the RS and BP RAD-tag samples were due to the differential presence of individual RAD tags, or merely reflected intrinsic differences in genomic content. Genomic DNA from these individuals was sheared, labeled, and hybridized together to the array without RAD-tag isolation. Of the 1990 elements with hybridization differences between the RS and BP RAD-tag samples, only 112 had hybridization differences between the sheared genomic samples, indicating that the arrays were specifically detecting the presence and absence of the RAD tags.
Figure 4.
RAD marker microarray characterization and bulk mapping experiments. (A) RAD tags from BP (Bear Paw individual) and RS (Rabbit Slough individual) were fluorescently labeled and competitively hybridized directly against each other to the RAD marker microarray. Scatterplots are of two experimental replicates. Array elements from the BP library are shown on the left, RS library elements on the right. Elements having high ratios are specifically from the RS library; elements having low ratios are from the BP library. The least-squares, best-fit regression equations (bottom right of each plot) were calculated for elements with an average hybridization difference greater than twofold. (B) The complete and low lateral plate phenotypes. Lateral plate reduction behaves as a recessive Mendelian locus. (C) Bulk-segregant analysis of the lateral plate phenotype. RAD tags isolated from a complete plate pool (CPP) and a low plate pool (LPP) were fluorescently labeled and competitively hybridized directly against each other to the RAD array. Elements from both libraries are shown together. Notice the larger hybridization differences seen from RAD markers that are linked to the complete plate allele than to the low plate allele. This is expected because of the recessive nature of the low plate phenotype.
Table 1.
RAD marker microarray characterization
RAD tags from BP and RS were fluorescently labeled and competitively hybridized directly against each other to the RAD marker microarray. Elements with an average differential hybridization greater than twofold are shown as polymorphic elements. Percentages are calculated from the overall number of elements from each library.
The RAD genotyping approach efficiently localized the stickleback major lateral plate locus through bulk segregant analysis
We tested if the RAD marker microarray could be used for mapping the genetic basis of phenotypic differences among natural populations of stickleback. The reduction of bony lateral plates behaves as a recessive Mendelian locus that maps to linkage group (LG) IV and has been repeatedly fixed in multiple freshwater populations (Fig. 4B; Colosimo et al. 2004, 2005; Cresko et al. 2004). Because the phenotype is easily distinguished and has been previously mapped, we used it as a test case for genetic mapping using the newly created enriched RAD array.
We analyzed the RAD markers from a stickleback F2 mapping family that was segregating the lateral plate phenotype. This mapping family was produced by crossing F1 individuals that were the product of a mating between a RS oceanic fish with a fish from Mud Lake (MD). Similar to the BP population, MD is a derived freshwater population that has evolved a low lateral plate phenotype. Two genomic DNA pools were made, one from 16 individuals sharing the complete lateral plate phenotype, and the other from an equal number of fish with the low lateral plate phenotype. RAD-tag samples were isolated from each of these pools, differentially labeled, and competitively hybridized against each other to the RAD-tag microarray. As expected for a bulk segregant analysis, only a small subset of the 1990 markers identified in the initial RS versus BP hybridization experiments exhibited an association with the lateral plate phenotype (Fig. 4C).
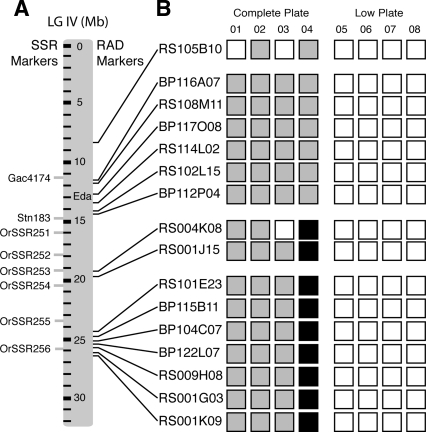
We identified the genomic positions of the RAD markers with the greatest hybridization differences in the bulk experiments (see Methods) (Birney et al. 2006). As expected, a set of six markers clustered in a 3-Mb region of LG IV very near Stn183, the marker initially linked to the lateral plate phenotype in Alaskan populations (Cresko et al. 2004), and centered only 200 kb away from Eda, the previously identified lateral plate locus (Fig. 5A; Colosimo et al. 2005). Surprisingly, a second cluster of seven RAD markers was also identified in a 2-Mb region nearly 13 Mb away from Eda. Three additional markers were located away from the two clusters. In order to validate the association of the second cluster, we designed primers for a microsatellite marker near the RAD marker RS009H08 (OrSSR256), and typed the 76 individual F2 progeny from the RS × MD family used for the RAD bulk mapping. All 18 low plate individuals were homozygous for the low plate allele, and all 58 complete plate individuals had at least one copy of the complete plate allele. No individuals were homozygous for the low plate allele at only one of the two regions. These data are consistent with either two distinct regions that are both associated with the lateral plate phenotype, or one large block of association that exhibits a very nonrandom distribution of RAD markers.
Figure 5.
Bulk segregant analysis of RAD markers identifies two regions associated with the lateral plate phenotype. Sixteen RAD markers (small black bars) with large hybridization differences in the bulk experiments of the complete plate pool versus low plate pool were sequenced and placed on linkage group IV. (A) Six markers clustered in a 3-Mb region surrounding Eda, the previously identified lateral plate locus. Seven markers also cluster in a 2-Mb region near position 25 Mb, suggesting that this region is also associated with the lateral plate phenotype. A microsatellite marker near the center of this cluster, OrSSR256, confirmed this result. (B) Individual RAD marker data confirm the bulk mapping results. The RAD-tag sample from each of eight F2 individuals was run directly against the tags from BP (Bear Paw individual) in one hybridization experiment and RS (Rabbit Slough individual) in a separate experiment. These RAD-tag hybridization patterns can be used to determine individual genotypes (open box, homozygous BP; gray box, heterozygous; black box, homozygous RS) (see Supplemental Fig. 1 for raw data). Columns represent individuals; rows represent markers. One of the four complete plate individuals (04) is heterozygous at the RAD markers around Eda (presence of both complete plate and low plate tags), but is homozygous for the complete plate allele at the 25-Mb cluster and the two markers at 19 Mb, suggesting a recombination event between the Eda cluster and the markers at 19 Mb. Individual 03 lacked the complete plate allele at RS004K08 but had a complete plate allele at RS001J15 and the cluster around Eda, suggesting both a recombination event between RS001J15 and RS004K08 and another recombination event in the region between RS004K08 and the cluster around Eda. Microsatellite markers confirmed the recombination event between RS001J15 and RS004K08 and further localized the second recombination event between OrSSR252 and OrSSR251.
RAD bulk mapping results were confirmed by analyzing recombinant individuals
RAD-tag samples were isolated from four complete and four low plate individuals of the RS × MD F2 family. The RAD-tag sample from each F2 individual was run directly against the tags from the BP individual in one hybridization experiment and the RS individuals in a separate experiment. As expected, within the two clusters (around Eda or the second cluster around 25 Mb), all of the complete plate individuals had at least one complete plate allele, while all of the low plate individuals were homozygous for the low plate allele (Fig. 5B). The individual genotypes also suggested that two out of the eight individuals had recombination events between the two clusters. One complete plate individual was homozygous for the complete plate allele at one of the clusters, but heterozygous at the other (Fig. 5B), suggesting a single recombination event. Another complete plate individual was heterozygous at one cluster, homozygous low plate at part of the central region, and heterozygous at the other cluster (Fig. 5B), consistent with either a double recombination or two single recombination events between the two clusters. We designed and scored five new microsatellite markers that confirmed these recombination patterns detected by the RAD markers. These data confirmed that the bulk analysis selectively identified two regions of complete association with the lateral plate phenotype.
Discussion
RAD markers
Current restriction site polymorphism-based genotyping techniques screen for polymorphisms in only a subset of the restriction sites of a particular enzyme. The RAD method presented here, a combination of digestion followed by random shearing, isolates DNA tags that correspond to nearly every instance of a particular restriction site throughout a genome. RAD marker profiles can be compared among individual or bulk samples by microarray hybridization.
The number of unique informative markers for a given enzyme will be determined by the SNP frequency and the size of the genome. For a restriction enzyme that recognizes a 6-bp nucleotide sequence, we estimate that an informative RAD marker will be present on average less than every 100 kb between individuals or strains with an average SNP frequency of 0.5%. For most eukaryotic genomes, several thousand polymorphic RAD markers are theoretically available for any particular enzyme. Consistent with this, the low number of repeated markers that were seen on the stickleback RAD array suggests that the nearly 2000 markers represented just a fraction of the total markers available. Furthermore, a nearly linear addition of markers is obtainable with each additional restriction enzyme used. Thus, RAD markers allow high-density polymorphism discovery in virtually any genome.
RAD marker genotyping on common microarray formats
We demonstrated that RAD markers can be rapidly identified and typed by hybridization to a pre-existing genomic tiling path microarray (Hollich et al. 2004; Sandmann et al. 2006). Using this approach, we were able to rapidly map a recombination breakpoint in D. melanogaster. RAD markers could be identified by hybridization to most kinds of microarrays. However, the marker density will decrease as sequence representation decreases on the array. The primary advantage to using expression or tiling arrays for RAD marker genotyping is that these arrays are not specific to a subset of populations or strains but will be a versatile genotyping platform for any individual. Furthermore, as high-density tilling path arrays become more widely available, they could become powerful platforms for RAD marker genotyping.
The rapid creation of a high-resolution RAD marker microarray genotyping resource
We demonstrate that RAD tags themselves can be used as a source of material for the production of specialized arrays that allow for optimal identification and typing of RAD markers. Because the RAD tags are amenable to various molecular biology manipulations, DNA subtraction techniques can be used to enrich for RAD tags present in a specific sample and not another, thus greatly increasing the number of informative markers present on the array. The production of an enriched RAD marker microarray is technically straightforward and inexpensive. The reagents needed for this process are widely available, and the resulting libraries of PCR products can be used to generate thousands of RAD arrays.
The enriched RAD marker array that we developed for threespine stickleback identified almost 2000 polymorphic RAD markers. The enrichment technique used here resulted in an informative marker rate of ∼10% of array elements, more than a threefold enrichment compared to what we have measured on an unenriched RAD marker array from these same fish (data not shown). With an optimized subtraction method, libraries could be routinely generated with much higher informative marker rates. Enriched RAD marker arrays allow high-throughput, high-density genetic marker identification and genotyping for a very low cost (less than $50 per assay) and with little initial investment.
RAD marker analysis speeds biological discovery in threespine stickleback
Threespine stickleback has long been a subject of biological research because of the presence of abundant phenotypic variation in behavioral, morphological, and life history characteristics (Wootton 1976; Bell and Foster 1994; Cresko et al. 2006). While much has been learned about the processes of phenotypic evolution in stickleback, understanding the developmental genetic basis of this evolution has been hampered by a lack of genetic and genomic tools (Kingsley et al. 2004). The recent development of some of these resources has begun to allow a wide array of evolutionary genetic questions to be addressed in this small fish (Peichel 2005; Cresko et al. 2006). To fully probe these problems requires a high-throughput, low-cost platform for genotyping in numerous stickleback crosses and populations. To address the deficit, we have developed the first microarray-based mapping resource for stickleback. The enriched RAD marker array that we produced for threespine stickleback allowed for the rapid identification of the region previously determined to be responsible for the lateral plate phenotype. More importantly, having available a large number of markers led to the identification of a previously unknown region that also appears to be associated with the lateral plate phenotype.
The genotyping of individuals revealed an added complexity; individuals that are homozygous for the low plate parental allele at only one of the two regions were not represented in our family, while all individuals that are homozygous for the low plate parental allele at both regions have the low lateral plate phenotype. These data raise two intriguing hypotheses. Epistatic interactions between loci in the two regions may be causing synthetic lethality, which cause inheritance patterns of the phenotype to appear similar to a single Mendelian locus. The presence of recombination events in the complete plate but not the low plate fish suggests that the two regions are genetically separated and that synthetic lethality is preventing the recovery of recombinants in the low plate individuals. Alternatively, these populations have a chromosomal rearrangement, and in reality the two clusters are in close proximity. While subsequent work must be done in order to further investigate the genetic nature of the newly identified region and the extent to which it is important in other crosses and populations, our results do demonstrate the ability of this technique to foment additional biological discoveries, even in apparently well-understood biological problems.
The ability to score a large number of markers from the very beginning of a mapping study provides several distinct advantages for the genetic analysis of stickleback traits. The order of magnitude greater RAD marker density, as compared to the existing microsatellite marker density, helped lead to the discovery of complicated inheritance patterns for the lateral plate region. Even if many more microsatellite markers were available throughout the genome, the need to analyze them on a marker-by-marker basis means that the initial screen would involve a subset of those markers. Subsequent microsatellite genotyping efforts would be concentrated on only one area of the genome, potentially obscuring additional genetic associations. Furthermore, the high density of markers available at the initial stages of RAD analysis opens the possibility for whole genome scans in natural populations to do such things as identify signatures of selection in recently derived populations or associate phenotypes with particular markers in populations segregating a phenotype. Lastly, the cost effectiveness of this technique will allow the genetic analysis in numerous populations of similar traits, providing important data for quantifying to what extent parallel phenotypic evolution in stickleback is based on parallel evolution in developmental genetic pathways.
Conclusions
Biologists interested in the genetic basis of traits have long been limited to the intensive study of a small set of particular organisms. There are many species about which very little is known, owing to lack of genetic tools, one of the most important of which is an efficient genotyping platform. The RAD techniques described in this paper are not specific to stickleback or even fruit fly, but can be used for genetic analysis in most model and nonmodel organisms. An enormous variety of organisms exists that could be used to address a wide array of biological questions, and this approach offers the power of being able to apply rapid, high-resolution genotyping to the organism best-suited to a particular biological question.
Methods
Restriction-site associated DNA (RAD) tag isolation
Genomic DNA (2 μg) was digested for 2 h at 37°C in a 100-μL reaction with 40 units (U) of EcoRI (New England Biolabs [NEB]). Samples were extracted with phenol/chloroform and precipitated. Sample volumes were brought to 200 μL with TE and extracted with an equal volume of phenol:choloform:isoamyl alcohol (25:24:1) (Sigma) followed by an extraction with choloform:isoamyl alcohol (24:1) (Sigma). Samples were precipitated in the presence of 40 μg of glycogen (Fermentas), 0.3 M NaOAC, and 2.5 volumes of ethanol. Pellets were resuspended in 10 μL of ligation mix 1 (1× T4 DNA ligase reaction buffer [NEB], 1 μM biotinylated EcoRI linker: 5′-Bio-TTTCGACGCTCGCATCTGGA CAGG-3′, 5′-Phos-AATTCCTGTCCAGATGCGAGCGTC-3′; Integrated DNA Technologies). Prior to the addition of enzyme, samples were heated for 2 min at 50°C and allowed to cool for 10 min at room temperature. One hundred units of T4 DNA ligase (high concentration, NEB) were then added to each sample, and samples were incubated for 1.5 h at 16°C. Samples were extracted with phenol/chloroform as described above. With the biotinylated linker now incorporated, samples were randomly sheared using a Branson sonicator 450 (Duty Cycle 80, Output 1.2) with 20 pulses followed by a quick spin, 1 min on ice, and 20 additional pulses. This sonication protocol produced DNA fragments predominantly 250–500 bp long. Samples were then precipitated as described above, and pellets were resuspended in 100 μL TE. The DNA directly flanking EcoR1 restriction sites was now purified. Fifty microliters of Dynabeads M-280 Streptavidin (Invitrogen) were prepared according to the manufacturer’s protocol. Fragmented DNA was added to the bead mix and incubated for 20 min at room temperature, with intermittent mixing. The beads were immobilized, and the supernatant was removed. The beads were washed three times with 200 μL of binding and wash buffer (5 mM Tris, 0.5 mM EDTA, 1 M NaCl) followed by one wash with 200 μL of TE. The beads were immobilized, the supernatant was removed, and the beads were resuspended in 100 μL of 1× NEBuffer EcoR1 with 40 U of EcoRI (NEB) to release the purified DNA. Samples were incubated for 1 h at 37°C, with intermittent mixing. To recover the purified DNA, the beads were immobilized, and the supernatant was transferred into a new tube. Samples were extracted with phenol/chloroform and precipitated as described above, and pellets were resuspended in 20 μL of TE. These RAD-tag samples were analyzed by gel electrophoresis. We estimate that ∼50 ng of RAD-tag DNA were typically isolated from each sample. XhoI RAD-tag isolations were done as described above with the following modifications: (1) XhoI (NEB) was used; (2) ligation mix 1 contained biotinylated XhoI linker (5′-Bio-TTTCG CTCGCATCTGGACAGC-3′, 5′-Phos-TCGAGCTGTCCAGATGCGAGCGTCG-3′) in replacement of biotinylated EcoRI linker.
Fluorescent labeling and hybridization
Prior to incorporation of fluorescent dyes, RAD-tag samples were amplified. About 5 ng of a RAD tag sample were end-repaired for 15 min at 25°C in a 50-μL reaction with 1 U of Klenow fragment (NEB). Samples were extracted with phenol/chloroform and precipitated as described above, and pellets were resuspended in 10 μL of ligation mix 2 (1× T4 DNA ligase reaction buffer [NEB], 1 μM blunt-ended linker A: 5′-GCGGTGACCCGGGAGATCT GAATTC-3′, 5′-Phos-GAATTCAGATC-3′). One hundred units of T4 DNA Ligase (high concentration, NEB) were added to each sample, and samples were incubated for 1.5 h at 16°C. Samples were heat-inactivated for 10 min at 65°C. Five microliters of the ligation samples were used as a template for PCR. PCR reactions were performed in a 100-μL volume containing the following: template as described, 1× thermopol buffer (NEB), 5 U of taq (NEB), 0.2 μM 5′-GCGGTGACCCGGGAGATCTGAATTC-3′, and 0.2 mM dNTPs (Fermentas). Thermocycling conditions consisted of the following: 55°C for 2 min, 72°C for 5 min, 94°C for 5 min, 18–22 cycles (94°C for 1 min, 55°C for 1 min, 72°C for 1 min), and 72°C for 5 min. The amplified products were purified with the PCR purification kit (Qiagen). Two hundred fifty nanograms of an amplified RAD-tag sample were used as a template for random-primer-based incorporation of fluorescent nucleotides (Alexa Fluor 555 or 647) using the Bioprime plus array CGH genomic labeling system (Invitrogen). Labeling reactions were done according to manufacturers’ protocols with the following modifications: (1) Half-sized reactions were used. (2) Incubation time was increased to >4 h at 37°C. (3) After quenching the reactions, the two labeling reactions that were to be competitively hybridized directly against each other were combined and purified through one purification column. Purified samples were dried down and resuspended in 42 μL of microarray hybridization buffer (50% formamide, 5× SSC, 1% SDS, 1 μg/μL calf thymus DNA [Invitrogen]). Hybridization samples were denatured for 5 min at 95°C and hybridized to microarrays under 24 × 60-mm m-Series LifterSlips (Erie Scientific). Microarrays were placed into microarray hybridization chambers (Corning) prepared with 20 μL of distilled H2O in each end in order to maintain constant humidity. Chambers were incubated for 12–16 h at 42°C. Following incubation, slides were washed for 5 min each in 1× SSC, 0.05% SDS, 0.2× SSC, 0.05× SSC and dried by centrifugation. The arrays were immediately scanned using a GenePix 4000a Microarray Scanner (Axon Instruments) to generate images of Alexa Fluor 555 and 647 signal intensities.
Enriched RAD library production
To prepare driver samples, ∼5 ng of RAD tag sample were end-repaired for 15 min at 25°C in a 50-μL reaction with 1 U of Klenow fragment (NEB). Samples were extracted with phenol/chloroform and precipitated as described above, and pellets were resuspended in 10 μL of ligation mix 3 (1× T4 DNA ligase reaction buffer [NEB], 1 μM blunt-ended linker B: 5′Bio-GCGGTGACCCGGGAGATCTGAATTC-3′, 5′-Phos-GAATTCAGATC-3′). One hundred units of T4 DNA ligase (high concentration, NEB) were added to each sample, and samples were incubated for 1.5 h at 16°C. Samples were heat-inactivated for 10 min at 65°C. Five microliters of the ligation samples were used as a template for PCR. PCR reactions were performed in a 100-μL volume containing the following: template as described, 1× Thermopol Buffer (NEB), 5 U of Taq (NEB) 0.2 μM 5′-Bio-GCGGTGACCCGGGAGATCTGAATTC-3′, and 0.2 mM dNTPs (Fermentas). Thermocycling conditions consisted of the following: 55°C for 2 min, 72°C for 5 min, 94°C for 5 min, 24 cycles (94°C for 1 min, 55°C for 1 min, 72°C for 1 min), 72°C for 5 min. The amplified products were purified with the PCR Purification Kit (Qiagen).
To prepare tester samples, ∼5 ng of RAD-tag sample were end-repaired for 15 min at 25°C in a 50-μL reaction with 1 U of Klenow fragment (NEB). Samples were extracted with phenol/chloroform and precipitated as described above and resuspended in 10 μL of ligation mix 4 (1× T4 DNA ligase reaction buffer [NEB], 1 μM blunt-ended linker C: 5′-CTGCTCGAA TTCAAGCTTCT-3′, 5′-Phos-AGAAGCTTGAATTCGAGCAGT CAG-3′). One hundred units of T4 DNA ligase (high concentration, NEB) were added to each sample, and samples were incubated for 1.5 h at 16°C. Samples were heat-inactivated for 10 min at 65°C. Five microliters of the ligation samples were used as a template for PCR. PCR reactions were performed in a 50-μL volume containing the following: template as described, 1× PCR buffer-Mg (Invitrogen), 1.5 mM MgCl2, 2.5 U of platinum taq (Invitrogen), 0.2 μM 5′-CTGCTCGAATTCAAGCTTCT-3′, and 0.2 mM dNTPs (Fermentas). Thermocycling conditions consisted of the following: 94°C for 2 min, 20 cycles (94°C for 1 min, 55°C for 1 min, 72°C for 1 min), 72°C for 5 min. The amplified products were purified with the PCR Purification Kit (Qiagen).
Subtractive hybridization conditions followed Sive and St John (1988) with the following modifications: (1) 100 ng of tester and 2 μg of driver were used, and 10 μg of glycogen were added prior to ethanol precipitation (Fermentas). (2) After hybridizations were transferred to HB-SDS, these samples were incubated with three changes of 100 μL of Dynabeads M-280 Streptavidin (Invitrogen) for 15 min each to remove driver and hybrid molecules. Two rounds of subtraction were done. The subtracted tester samples were precipitated, and pellets were resuspended in 20 μL of TE. Five microliters of this sample were used as template in a PCR reaction. The primer and reaction conditions used were the same as described for the preparation of tester DNA. The amplified products were purified with the PCR Purification Kit (Qiagen). Products of this subtraction method are referred to as enriched RAD-tag samples.
Microarray production
Fifty nanograms of enriched RAD tag samples were cloned using the TOPO TA Cloning kit (Invitrogen). Clones were picked into 96-well plates with 125 μL of LB + kanamycin in each well and grown for 6 h at 37°C. 15% glycerol stocks were made from these clone samples in 384-well plates. One microliter of these was used as templates for PCR. PCR reactions were performed in a 50-μL volume containing the following: template as described, 50 mM KCl, 1 mM MgCl2, 10 mM Tris (pH 8.3), 0.1% Triton X-100, 2% DMSO, 0.2 μM 5′-CTGCTCGAATTCAAGCTTCT-3′, 0.1 mM dNTPs (Fermentas), and 1.25 U of Taq (NEB). Thermocycling conditions consisted of the following: 94°C for 5 min, 44 cycles (94°C for 1 min, 62°C for 1 min, 72°C for 80 sec), and 72°C for 5 min. PCR products were transferred into 384-well microarray printing plates, dried down, and stored sealed at room temperature. Prior to printing, samples were resuspended in 10 μL of printing buffer (3× SSC, 1.5 M betaine). Samples were arrayed onto GAPSII microarray slides (Corning). Print plates were then sealed and stored at −80°C. After printing, slides were exposed to 300 mJ UV and stored at room temperature in the presence of desiccant. Immediately prior to hybridization, slides were prehybridized in 50% formamide, 5× SSC, 1% SDS, 1 mg/mL BSA for 1 h at 42°C and treated to reduce background (Raghavachari et al. 2003). Slides were placed immediately into “just off the boil” distilled H2O for 2 min and dried by centrifugation.
Fish protocols
The fish used to produce the array were from lines originally derived from collections of wild stickleback in the Matanuska-Susitna Borough of Alaska. One line was derived from a freshwater population (Bear Paw Lake; N61.6139, W149.7529) and another from Rabbit Slough, an anadromous population (N61.5595, W149.2583). A Bear Paw Lake-derived fish, BP (750.00.0001), and a Rabbit Slough-derived fish, RS (612.00.0001), were used to produce the array (see Cresko et al. 2004 for more information). The F2 fish used in the mapping analysis were offspring of a full-sib cross between two F1 individuals. These F1 parents were produced from a cross between a Rabbit Slough fish and a fish from another freshwater population, Mud Lake (N61.5625, W148.9486). Similar to those from Bear Paw Lake, stickleback from Mud Lake consistently exhibit a low lateral plate phenotype. Crosses and rearing were performed as described in Cresko et al. (2004). Genomic DNA was isolated from pectoral fin clips from live individual fish using the DNeasy Tissue Kit (Qiagen). Phenotypic analyses, including bone preparation and scoring, were performed as in Cresko et al. (2004).
Microarray analysis
GenePix Pro 3.0 image analysis software (Axon Instruments) was used to measure fluorescence signal intensities of the array elements (GEO accessions GSE5773, GPL4254). The median of the ratio of Alexa Fluor 555 and 647 signal intensities of each array element was used for the analysis. Ratios were normalized to the overall difference in signal intensities from each channel. Fifteen out of the 20 highest-ratio, complete-plate-linked elements in the bulk experiments were also identified as polymorphic elements between the BP and RS individuals. The inserts from these clones were sequenced. Nine out of the top 10 lowest-ratio, low-plate-linked elements were also identified as polymorphic elements between the BP and RS individuals. We sequenced the seven of the nine lowest-ratio elements that behaved as though they were linked to the lateral plate phenotype. The sequence analysis revealed that the 15 complete-plate-linked elements represented 14 unique markers and the seven low-plate-linked elements represented six unique markers. Ten out of the 14 complete-plate-linked markers and all six of the low-plate-linked markers were able to be placed in the pre-release stickleback genome by performing a BLASTN search (Birney et al. 2006). Presence or absence calls were made based on the assumption that the RAD-tag clones that were derived from either the RS or BP individuals were present in the homozygous state in that individual. Thus, the intensities measured from the direct hybridization of BP and RS were assumed to represent a homozygous present versus a homozygous absent RAD-tag state. F2 RAD-tag samples for which the normalized intensity was <25% of homozygous presence versus homozygous absence were called as absent.
Microsatellites
The microsatellite markers used were either previously published (Peichel et al. 2001; Gac4174 and Stn183) or created de novo (GenBank accessions BV686582–BV686587). To create new microsatellite markers, we scanned the stickleback genome assembly of LGIV for regions of simple sequence repeats and designed PCR primers to flank these regions. Standard PCR conditions and agarose gel electrophoresis were used to assess microsatellite genotypes.
Acknowledgments
We thank Tressa Atwood for help with fly breakpoint mapping; Cristin Hulslander for help with isolating colonies; Catherine Wilson for scoring microsatellites; Mark Currey for fish crosses, rearing, and phenotypic analyses; and Michael Freitag for useful comments on the manuscript. We are also in debt to the PB&J research group for conversations and discussions about this technique that increased the quality of our work. This work was supported by a National Science Foundation (NSF) grant IBN 023639 (to W.A.C.), a grant (5R01RR020833) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) (to W.A.C.), and a Research Scholar Grant #RSG-03-154-01-DDC from the American Cancer Society (to E.A.J.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR, NIH, or ACS. The material in this paper may be related to U.S. patent application U.S. Patent 60/679,693. The authors thus disclose potential competing interests.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5681207
References
- Bell M.A., Foster S.A., Foster S.A. The evolutionary biology of the threespine stickleback. Oxford University Press; Oxford, New York: 1994. [Google Scholar]
- Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Cox T., Cunningham F., Curwen V., Cutts T., Cunningham F., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Liang D., Plouffe D., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Liang D., Plouffe D., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Plouffe D., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Weigel D., Berry C.C., Winzeler E., Chory J., Berry C.C., Winzeler E., Chory J., Winzeler E., Chory J., Chory J. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 2003;13:513–523. doi: 10.1101/gr.541303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., White R.L., Skolnick M., Davis R.W., White R.L., Skolnick M., Davis R.W., Skolnick M., Davis R.W., Davis R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Breseghello F., Sorrells M.E., Sorrells M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics. 2006;172:1165–1177. doi: 10.1534/genetics.105.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B., Grenier J.K., Weatherbee S.D., Grenier J.K., Weatherbee S.D., Weatherbee S.D. From DNA to diversity. Blackwell Publishing; Malden, MA: 2005. [Google Scholar]
- Colosimo P.F., Peichel C.L., Nereng K., Blackman B.K., Shapiro M.D., Schluter D., Kingsley D.M., Peichel C.L., Nereng K., Blackman B.K., Shapiro M.D., Schluter D., Kingsley D.M., Nereng K., Blackman B.K., Shapiro M.D., Schluter D., Kingsley D.M., Blackman B.K., Shapiro M.D., Schluter D., Kingsley D.M., Shapiro M.D., Schluter D., Kingsley D.M., Schluter D., Kingsley D.M., Kingsley D.M. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2004;2:e109. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo P.F., Hosemann K.E., Balabhadra S., Villarreal G., Jr., Dickson M., Grimwood J., Schmutz J., Myers R.M., Schluter D., Kingsley D.M., Hosemann K.E., Balabhadra S., Villarreal G., Jr., Dickson M., Grimwood J., Schmutz J., Myers R.M., Schluter D., Kingsley D.M., Balabhadra S., Villarreal G., Jr., Dickson M., Grimwood J., Schmutz J., Myers R.M., Schluter D., Kingsley D.M., Villarreal G., Jr., Dickson M., Grimwood J., Schmutz J., Myers R.M., Schluter D., Kingsley D.M., Dickson M., Grimwood J., Schmutz J., Myers R.M., Schluter D., Kingsley D.M., Grimwood J., Schmutz J., Myers R.M., Schluter D., Kingsley D.M., Schmutz J., Myers R.M., Schluter D., Kingsley D.M., Myers R.M., Schluter D., Kingsley D.M., Schluter D., Kingsley D.M., Kingsley D.M. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Cresko W.A., Amores A., Wilson C., Murphy J., Currey M., Phillips P., Bell M.A., Kimmel C.B., Postlethwait J.H., Amores A., Wilson C., Murphy J., Currey M., Phillips P., Bell M.A., Kimmel C.B., Postlethwait J.H., Wilson C., Murphy J., Currey M., Phillips P., Bell M.A., Kimmel C.B., Postlethwait J.H., Murphy J., Currey M., Phillips P., Bell M.A., Kimmel C.B., Postlethwait J.H., Currey M., Phillips P., Bell M.A., Kimmel C.B., Postlethwait J.H., Phillips P., Bell M.A., Kimmel C.B., Postlethwait J.H., Bell M.A., Kimmel C.B., Postlethwait J.H., Kimmel C.B., Postlethwait J.H., Postlethwait J.H. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko W.A., McGuigan K.L., Phillips P.C., Postlethwait J.H., McGuigan K.L., Phillips P.C., Postlethwait J.H., Phillips P.C., Postlethwait J.H., Postlethwait J.H. Studies of threespine stickleback developmental evolution: Progress and promise. Genetica. 2006 doi: 10.1007/s10709-006-0036-z. doi: 10.1007/s10709-006-0036-z. [DOI] [PubMed] [Google Scholar]
- Davidson E.H. Genomic regulatory systems: Development and evolution. Academic Press; San Diego, CA: 2001. [Google Scholar]
- Doerge R.W. Mapping and analysis of quantitative trait loci in experimental populations. Nat. Rev. Genet. 2002;3:43–52. doi: 10.1038/nrg703. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. The genetical theory of natural selection. Clarendon Press; Oxford: 1930. [Google Scholar]
- Hollich V., Johnson E., Furlong E.E., Beckmann B., Carlson J., Celniker S.E., Hoheisel J.D., Johnson E., Furlong E.E., Beckmann B., Carlson J., Celniker S.E., Hoheisel J.D., Furlong E.E., Beckmann B., Carlson J., Celniker S.E., Hoheisel J.D., Beckmann B., Carlson J., Celniker S.E., Hoheisel J.D., Carlson J., Celniker S.E., Hoheisel J.D., Celniker S.E., Hoheisel J.D., Hoheisel J.D. Creation of a minimal tiling path of genomic clones for Drosophila: Provision of a common resource. Biotechniques. 2004;37:282–284. doi: 10.2144/04372MT01. [DOI] [PubMed] [Google Scholar]
- Jaccoud D., Peng K., Feinstein D., Kilian A., Peng K., Feinstein D., Kilian A., Feinstein D., Kilian A., Kilian A. Diversity arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001;29:e25. doi: 10.1093/nar/29.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley D.M., Zhu B.L., Osoegawa K., De Jong P.J., Schein J., Marra M., Peichel C., Amamiya C., Schluter D., Balabhadra S., Zhu B.L., Osoegawa K., De Jong P.J., Schein J., Marra M., Peichel C., Amamiya C., Schluter D., Balabhadra S., Osoegawa K., De Jong P.J., Schein J., Marra M., Peichel C., Amamiya C., Schluter D., Balabhadra S., De Jong P.J., Schein J., Marra M., Peichel C., Amamiya C., Schluter D., Balabhadra S., Schein J., Marra M., Peichel C., Amamiya C., Schluter D., Balabhadra S., Marra M., Peichel C., Amamiya C., Schluter D., Balabhadra S., Peichel C., Amamiya C., Schluter D., Balabhadra S., Amamiya C., Schluter D., Balabhadra S., Schluter D., Balabhadra S., Balabhadra S., et al. New genomic tools for molecular studies of evolutionary change in threespine sticklebacks. Behaviour. 2004;141:1331–1344. [Google Scholar]
- Luikart G., England P.R., Tallmon D., Jordan S., Taberlet P., England P.R., Tallmon D., Jordan S., Taberlet P., Tallmon D., Jordan S., Taberlet P., Jordan S., Taberlet P., Taberlet P. The power and promise of population genomics: From genotyping to genome typing. Nat. Rev. Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Lynch M., Walsh B., Walsh B. Genetics and analysis of quantitative traits. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- Mackay T.F. The genetic architecture of quantitative traits. Annu. Rev. Genet. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H., Dong S., Loi H., Di X., Liu G., Hubbell E., Law J., Berntsen T., Chadha M., Hui H., Dong S., Loi H., Di X., Liu G., Hubbell E., Law J., Berntsen T., Chadha M., Hui H., Loi H., Di X., Liu G., Hubbell E., Law J., Berntsen T., Chadha M., Hui H., Di X., Liu G., Hubbell E., Law J., Berntsen T., Chadha M., Hui H., Liu G., Hubbell E., Law J., Berntsen T., Chadha M., Hui H., Hubbell E., Law J., Berntsen T., Chadha M., Hui H., Law J., Berntsen T., Chadha M., Hui H., Berntsen T., Chadha M., Hui H., Chadha M., Hui H., Hui H., et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- Orr H.A., Presgraves D.C., Presgraves D.C. Speciation by postzygotic isolation: Forces, genes and molecules. Bioessays. 2000;22:1085–1094. doi: 10.1002/1521-1878(200012)22:12<1085::AID-BIES6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Peichel C.L. Fishing for the secrets of vertebrate evolution in threespine sticklebacks. Dev. Dyn. 2005;234:815–823. doi: 10.1002/dvdy.20564. [DOI] [PubMed] [Google Scholar]
- Peichel C.L., Nereng K.S., Ohgi K.A., Cole B.L., Colosimo P.F., Buerkle C.A., Schluter D., Kingsley D.M., Nereng K.S., Ohgi K.A., Cole B.L., Colosimo P.F., Buerkle C.A., Schluter D., Kingsley D.M., Ohgi K.A., Cole B.L., Colosimo P.F., Buerkle C.A., Schluter D., Kingsley D.M., Cole B.L., Colosimo P.F., Buerkle C.A., Schluter D., Kingsley D.M., Colosimo P.F., Buerkle C.A., Schluter D., Kingsley D.M., Buerkle C.A., Schluter D., Kingsley D.M., Schluter D., Kingsley D.M., Kingsley D.M. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Raghavachari N., Bao Y.P., Li G., Xie X., Muller U.R., Bao Y.P., Li G., Xie X., Muller U.R., Li G., Xie X., Muller U.R., Xie X., Muller U.R., Muller U.R. Reduction of autofluorescence on DNA microarrays and slide surfaces by treatment with sodium borohydride. Anal. Biochem. 2003;312:101–105. doi: 10.1016/s0003-2697(02)00440-2. [DOI] [PubMed] [Google Scholar]
- Sandmann T., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E., Eichenlaub M.P., Bork P., Furlong E.E., Bork P., Furlong E.E., Furlong E.E. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Sive H.L., St John T., St John T. A simple subtractive hybridization technique employing photoactivatable biotin and phenol extraction. Nucleic Acids Res. 1988;16:10937. doi: 10.1093/nar/16.22.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J.F. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., de van Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Hogers R., Bleeker M., Reijans M., de van Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Bleeker M., Reijans M., de van Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Reijans M., de van Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., de van Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Frijters A., Pot J., Peleman J., Kuiper M., Pot J., Peleman J., Kuiper M., Peleman J., Kuiper M., Kuiper M., et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.G., Fan J.B., Siao C.J., Berno A., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., Fan J.B., Siao C.J., Berno A., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., Siao C.J., Berno A., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., Berno A., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J., Ghandour G., Perkins N., Winchester E., Spencer J., Perkins N., Winchester E., Spencer J., Winchester E., Spencer J., Spencer J., et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Wenzl P., Carling J., Kudrna D., Jaccoud D., Huttner E., Kleinhofs A., Kilian A., Carling J., Kudrna D., Jaccoud D., Huttner E., Kleinhofs A., Kilian A., Kudrna D., Jaccoud D., Huttner E., Kleinhofs A., Kilian A., Jaccoud D., Huttner E., Kleinhofs A., Kilian A., Huttner E., Kleinhofs A., Kilian A., Kleinhofs A., Kilian A., Kilian A. Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. 2004;101:9915–9920. doi: 10.1073/pnas.0401076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A., Richards D.R., Conway A.R., Goldstein A.L., Kalman S., McCullough M.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., Richards D.R., Conway A.R., Goldstein A.L., Kalman S., McCullough M.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., Conway A.R., Goldstein A.L., Kalman S., McCullough M.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., Goldstein A.L., Kalman S., McCullough M.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., Kalman S., McCullough M.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., McCullough M.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., McCusker J.H., Stevens D.A., Wodicka L., Lockhart D.J., Stevens D.A., Wodicka L., Lockhart D.J., Wodicka L., Lockhart D.J., Lockhart D.J., et al. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- Wootton R.J. The biology of the sticklebacks. Academic Press; London, New York: 1976. [Google Scholar]