Abstract
The role of 2′-hydroxyl groups in stabilizing the tightly kinked geometry of the kink-turn (K-turn) has been investigated. Individual 2′-OH groups have been removed by chemical synthesis, and the kinking of the RNA has been studied by gel electrophoresis and fluorescence resonance energy transfer. The results have been analyzed by reference to a database of 11 different crystallographic structures of K-turns. The potential hydrogen bonds fall into several classes. The most important are those in the core of the turn and ribose–phosphate interactions around the bulge. Of these the single most important hydrogen bond is one donated from the 2′-OH of the 5′ nucleotide of the bulge to the N1 of the adenine of the kink-proximal A•G pair. This is present in all known K-turn structures, and removal of the 2′-OH completely prevents metal ion-induced folding. Hydrogen bonds formed in the minor grooves of the helical stems are less important, and removal of the participating 2′-OH groups leads to reduced impairment of folding. These interactions are generally more polymorphic, and hydrogen bonds probably form where possible, as permitted by the global structure.
Keywords: RNA structure, RNA folding, hydrogen bonds
INTRODUCTION
The kink-turn (K-turn) motif (Klein et al. 2001) introduces a very tight kink into the axis of helical RNA. It clearly plays an important structural role in RNA and is important in many aspects of RNA function, including translation, modification, and splicing, as well as genetic regulation. The K-turn was first identified as a novel motif occurring multiple times in the ribosome. Further examples have been found in mRNA (Mao et al. 1999; Winkler et al. 2001; Allmang et al. 2002; Montange and Batey 2006), guide RNAs (Kuhn et al. 2002; Watkins et al. 2002; Bortolin et al. 2003; Marmier-Gourrier et al. 2003; Rozhdestvensky et al. 2003), and spliceosomal RNA (Vidovic et al. 2000; Watkins et al. 2000).
Most of the known K-turns serve as the binding sites for one or more proteins. The most common K-turn binding protein is the ribosomal L7Ae and related proteins. These form a family of RNA-binding proteins including the eukaryotic and archaeal proteins L7Ae, L30e, and S12e (Koonin et al. 1994), the yeast Nhp2 and Snu13p proteins, and the human 15.5-kDa protein (Nottrott et al. 1999). K-turns are found in the box C/D and H/ACA guide snoRNA species that direct site-specific methylation and pseudouridylation, respectively, of RNA in archaea and eukaryotes (Kiss-Laszlo et al. 1996; Ganot et al. 1997). These modifications require the formation of a nucleoprotein complex, the assembly of which requires initial binding of an L7Ae-related protein to the K-turn motif. The 15.5-kDa protein also binds a K-turn within the U4 stem–loop within the U4-U6.U5 tri-snRNP (Nottrott et al. 1999).
The canonical K-turn comprises a 3-nucleotide (nt) bulge flanked on its 3′ side by two consecutive trans-Hoogsteen-sugar edge A•G pairs (the N-C stem), and on its 5′ side by a section of regular base pairing (the C stem) (Klein et al. 2001; Fig. 1). Crystal structures reveal a pronounced kink in the RNA, with an included angle between the axes of ∼60°. The minor grooves of the two helices are juxtaposed, and the conformation appears to be stabilized by interactions between the stacked adenosines of the A•G pairs and the C stem and by stacking of the 5′ and central bases of the bulge on the ends of the C and N-C stems, respectively. The structural principles of K-turns have been analyzed in detail using isostericity matrices by Westhof and colleagues (Lescoute et al. 2005).
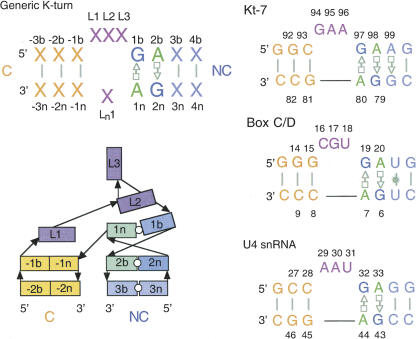
FIGURE 1.
A nomenclature for the K-turn. The sequence of a generic K-turn with the numbering system is shown, together with the sequences of Kt-7, box C/D, and U4 snRNA K-turns as found in the crystal structures analyzed. The schematic presents a view of the structure of a K-turn in its strongly kinked conformation.
The structure of free K-turn RNA is very dynamic and dependent on the binding of metal ions (Goody et al. 2003) or L7Ae-related protein (Turner et al. 2005; Wozniak et al. 2005). In the absence of protein binding, K-turn RNA exists in an equilibrium between the tightly kinked and a more extended structure, and the position of equilibrium is determined by metal ion concentration. The kinked conformation confers strongly retarded electrophoretic mobility to the RNA, whereas the extended structure migrates with a mobility typical of a simple 3-nt bulge (Goody et al. 2003; Matsumura et al. 2003). Fluorescence resonance energy transfer (FRET) measurements (Goody et al. 2003) show that the RNA undergoes an ion-induced two-state transition between the two structures, consistent with molecular dynamics studies that have predicted structural bistability in K-turn RNA (Cojocaru et al. 2005; Razga et al. 2005, 2006). Single-molecule FRET data indicate that transitions between the kinked and extended forms are fast (B. Turner, J.C. Penedo, and D.M.J. Lilley, unpubl.).
The K-turn provides an excellent opportunity to explore the role of individual hydrogen bonds in the folding of an RNA motif. The two-state folding process is readily followed by electrophoretic or FRET methods (Goody et al. 2003), and there are a number of crystal structures of K-turns in their folded conformation that provide structural insight. In this study we have examined the importance of selected 2′-hydroxyl groups in the stabilization of the kinked structure by systematic chemical substitution experiments. Examination of crystal structures of the eight K-turns found in the ribosome (Ban et al. 2000; Wimberly et al. 2000; Brodersen et al. 2002) and those of the box C/D snoRNA (Moore et al. 2004), U4 snRNA (Vidovic et al. 2000), and the SAM riboswitch (Montange and Batey 2006), reveals a number of common direct hydrogen bonding interactions involving 2′-OH groups (potential solvent-mediated contacts have not been considered here). These are tabulated for three representative K-turns, those of Kt-7 of the 50S ribosomal subunit (the sequence of which is closest to the consensus), box C/D, and U4 in Table 1. The hydrogen bonds can be divided into three groups. There are interactions occurring in the center of the K-turn that involve the bases of the A•G pairs, those around the backbone of the tight turn made by the nucleotides of the bulge, and those in the juxtaposed minor grooves of the C and NC helices. In our experiments we have examined the folding of the Kt-7 sequence with selected single 2′-deoxyribose substitutions. We find that ion-induced folding is impaired to various degrees. The most debilitating substitutions are those that affect interactions within the K-turn core, while the least significant modifications disrupt potential hydrogen bonding between the helical stems.
TABLE 1.
Hydrogen bonds observed in the crystal structures of three K-turn elements (Ban et al. 2000; Vidovic et al. 2000; Moore et al. 2004)a
RESULTS AND DISCUSSION
A general nomenclature for the K-turn
In the course of these studies we have referred to crystallographic data from a number of different K-turn structures. Since all use different nucleotide numbering systems, this can become confusing, and we therefore devised a universal nomenclature shown in Figure 1. The unpaired bulge nucleotides are labeled Lj, where the index j is numbered sequentially from the 5′ side. Unpaired nucleotides on the opposite strand (found in a small fraction of K-turns, exemplified by Kt-58 of the 50S ribosomal subunit) are designated Ln j, with j increasing 5′ to 3′. For all the remaining nucleotides, the two strands are differentiated by the suffix of b for the bulge-containing strand or n for the nonbulged strand. The nucleotides of the noncanonical stem are positively numbered from the first G•A pair in the 5′ to 3′ direction relative to the bulged strand, while those of the canonical stem are negatively numbered in the 3′ to 5′ direction. The numbering system can accommodate loops of different sizes.
Analysis of folding by gel electrophoresis
We have previously shown that RNA duplexes containing a central K-turn sequence exhibit strongly retarded electrophoretic mobility in polyacrylamide in the presence of Mg2+ ions (Goody et al. 2003), consistent with the adoption of a highly kinked conformation. Sequence changes that disrupt the canonical K-turn sequence lead to normal electrophoretic mobility for an RNA duplex of the same length interrupted by a 3-nt bulge. In previous studies we have prepared the RNA by in vitro transcription, but in this study we needed to introduce 2′-deoxynucleotide substitutions at selected positions, and therefore made the RNA by chemical synthesis. We constructed duplexes of 65 base pairs (bp) (including three A•G pairs), of which both outer segments of 20 bp comprised DNA and the inner section was RNA. The K-turn sequence lay at the center of the RNA section; this was the Kt-7 sequence that we have used previously in our studies, as it is very close to the consensus K-turn sequence. Radioactively labeled K-turn-containing duplexes were electrophoresed in 15% polyacrylamide gels in 90 mM Tris.borate (pH 8.3) with metal salts added as required.
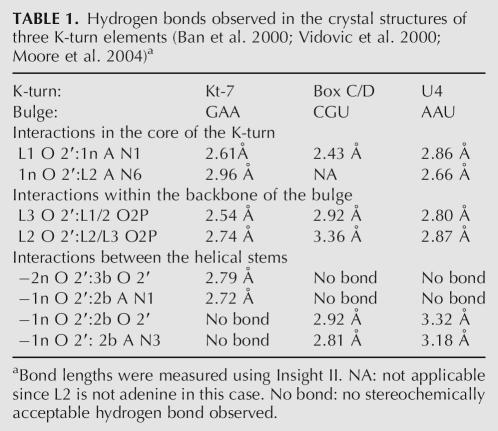
A series of K-turn species was made with single 2′-deoxyribose substitutions at points of interest (discussed below). The results are shown in Figure 2. In the presence of EDTA used to chelate metal ions, most of the species exhibited unretarded electrophoretic mobility (data not shown). Kt-7 does not adopt its tightly kinked conformation under these conditions (Goody et al. 2003), and therefore the substitutions cannot destabilize the conformation further. Upon addition of Mg2+ ions, pronounced differences in electrophoretic mobility become apparent. Each modified species exhibits a characteristic mobility intermediate between those of the unmodified K-turn and a corresponding duplex. However, the widths of the bands do not change for any species. This indicates that there is a rapid equilibrium between folded and unfolded species, the position of which is affected by the 2′-deoxy substitutions to differing extents. The relative mobilities do not change greatly between 500 μM and 2 mM Mg2+ ions (Fig. 2A,B), except for those of the L1- and 2b-deoxyribose species discussed below. A very similar pattern of mobilities was also obtained in the presence of 10 μM Co(III) hexammine (Fig. 2C).
FIGURE 2.
Electrophoretic analysis of K-turn folding. Radioactively 5′-32P-labeled 65-bp DNA/RNA/DNA duplex species with a central Kt-7 sequence were electrophoresed in 15% polyacrylamide gels in 90 mM Tris.borate (pH 8.3) plus 500 μM (A) or 2 mM (B) Mg2+, or 10 μM Co(III) hexammine (C) salts. Phosphorimages of the dried gels are shown. The simple duplex species was loaded at the left, followed by the unmodified Kt-7 sequence. These two species define the range of electrophoretic mobilities. Single 2′-deoxyribose variants were electrophoresed, indicated by the position of the modification in the K-turn nomenclature (written above tracks) or in the Kt-7 numbering system (below tracks). The original gels included three tracks not related to the experiments discussed here between the tracks containing L3 and 2b variants; these have been removed for clarity and the position indicated by the bold line. The gray lines have been added to aid identification of different tracks. Two versions of the L1 modification were used, with the natural guanine (dG94) and variant adenine (dA94).
Analysis of folding by steady-state fluorescence resonance energy transfer
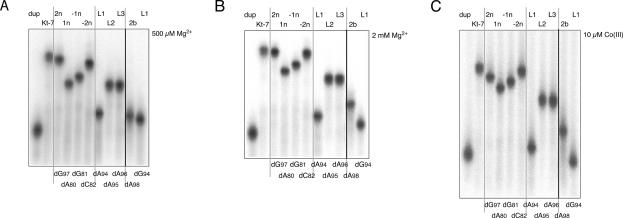
FRET provides an alternative method to examine the folding of K-turn-containing RNA. In this approach we have used 25-bp RNA duplexes containing a central Kt-7 sequence (identical to that used in the electrophoretic experiments), with fluorescent donor (fluorescein) and acceptor (Cy3) groups covalently attached at the 5′ termini of the bulged and nonbulged strands, respectively. When the K-turn sequence adopts the strongly kinked conformation, the fluorophores are brought closer together, resulting in a marked increase in energy transfer efficiency (Fig. 3). The ion-induced folding can be followed as a function of metal ion concentration, and the resulting data fitted to a two-state folding process from which the cooperativity and midpoint ionic concentration may be calculated. As observed previously (Goody et al. 2003), the unmodified Kt-7 sequence exhibits a large increase in FRET efficiency (ΔE FRET ∼ 0.33 for this construct) upon addition of Mg2+ or Na+ ions, with [Mg2+]1/2 = 78 μM and [Na+]1/2 = 30 mM under present conditions. The variant K-turns with single 2′-deoxyribose substitutions exhibited two differences from the natural sequence (Table 2). In general the half-ionic concentration was found to increase, consistent with the position of the equilibrium being shifted to the unfolded form due to the 2′-deoxyribose modifications, and therefore requiring greater reduction in electrostatic repulsion to achieve the same degree of folding of the population. The more disabling substitutions also resulted in a reduced change in FRET efficiency, consistent with a lower extent of folding achievable at saturating ionic concentration. The largest effect resulted from the removal of the 2′-oxygen atom at the L1 position (dA94), where no folding was detectable with either Mg2+ or Na+ ions.
FIGURE 3.
Analysis of metal ion-induced folding of Kt-7 and single 2′-deoxyribose variants by fluorescence resonance energy transfer in the steady state. Each species comprises an RNA duplex terminally labeled with fluorescein and Cy3 as shown in the schematic. Upon folding into the kinked structure the fluorophores become closer in space, resulting in an increase in the efficiency of energy transfer (E FRET). E FRET was measured for each species over a range of Mg2+ or Na+ concentrations, and plotted in A and B, respectively. Each curve (except for L1-2′-deoxyribose) has been fitted to a two-state model from which the extent of change in FRET efficiency and the half-ionic concentration were calculated. The L2-2′-deoxyribose variant of Kt-7 has a significantly higher E FRET at low salt concentrations, and is therefore plotted separately (insert) for clarity. The following variants are shown (indicated by the position of the 2′-deoxyribose): Kt-7 unmodified (black, filled circles); −2n (purple, open squares); −1n (cyan, closed triangles); 1n (green, open circles); L3 (orange, filled squares); 2b (blue, open triangles); L1 (red, filled circles). (Insert) L2 (black, filled circles). Note that the L1 2′-deoxyribose variant exhibits no increase in FRET at any ionic concentration, indicating that it is totally unable to fold into the strongly kinked conformation.
TABLE 2.
Changes in FRET efficiency (ΔE FRET) and half-ionic concentrations for Kt-7 and 2′-deoxyribose derivatives obtained from two-state fitting FRET titrations in Mg2+ and Na+ ions
Hydrogen bonding between C and NC stems
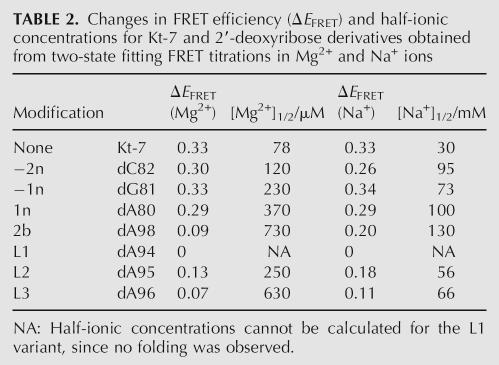
There is significant variability in the pattern of potential hydrogen bonding between the C and NC stems in the different crystal structures of K-turns. The structure of Kt-7 found in the Haloarcula marismortui 50S ribosomal subunit has an evident hydrogen bond (O–O distance of 2.79 Å) between the 2′-hydroxyl groups of 3b and −2n (A99 of the third non-Watson–Crick pair of the NC stem and C82 in the C stem) (Fig. 4A). In addition, there is a hydrogen bond donated from the 2′-OH of −1n (G81) to N1 of adenine 2b (A98, the conserved adenine of the second G•A pair). Removal of the 2′-hydroxyl from −2n of Kt-7 resulted in a minimal effect on the electrophoretic mobility of the RNA under all ionic conditions and a very small difference in the Mg2+- or Na+-induced folding observed by FRET. This is not consistent with a requirement for a hydrogen bond involving this oxygen atom, and strongly suggests that this interaction is not important for the stabilization of free Kt-7 in solution.
FIGURE 4.
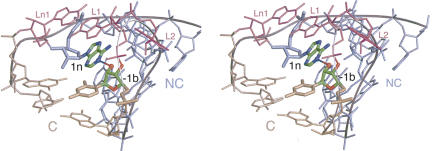
Interactions between the C and NC helices found in crystal structures of Kt-7 (Klein et al. 2001), box C/D (Moore et al. 2004), and U4 (Vidovic et al. 2000) K-turns. (A,B) Parallel-eye stereo view of Kt-7 (A) and box C/D (B) K-turns looking down into the minor groove interface between the C and NC helices, with the bulge shown at the top. The C helix is colored gold, NC colored blue, and bulge nucleotides are colored purple; this scheme has been used in most graphics images in this article. The nucleotides participating in hydrogen bonding interactions are highlighted, and the bond drawn in. In Kt-7 (A) hydrogen bonds are seen between the 2′-OH groups of nucleotides −2n and 3b, and between the 2′-OH of −1n and N1 of adenine 2b. In box C/D (B) there is no interaction between nucleotides −2n and 3b. However, there are hydrogen bonds between the 2′-OH of −1n and N3 and 2′-O of adenine 2b. (C) Close-up view of the hydrogen bonding between the 2′-OH of −1n and N3 and 2′-O of adenine 2b in the U4 K-turn. (D) The chemical structure of the hydrogen bonding pattern between the 2′-OH of −1n and N3 and 2′-O of adenine 2b in box C/D and U4 K-turns.
The pattern of interactions in the box C/D (Fig. 4B) or U4 RNA K-turns is different from that in Kt-7. In the box C/D and U4 K-turns the potential ribose–ribose interaction is effectively translocated 1 nt toward the loop of the K-turn (Fig. 4B), between the 2′-hydroxyl groups of 2b and −1n (A20: C8, and A33: G45 in box C/D and U4, respectively). The 2′-O of −1n could still hydrogen bond with the adenine nucleobase of 2b, but the acceptor is now N3. Thus the 2′-O of −1n acts as both hydrogen bond donor and acceptor (Fig. 4C). Removal of the 2′-O of −1n (G81) from Kt-7 resulted in a greater reduction in electrophoretic retardation compared to the corresponding substitution at −2n (Fig. 2), and a twofold increase in the [Mg2+]1/2 concentration in FRET experiments (Fig. 3). This effect is in better agreement with the structure observed for the box C/D and U4 RNAs than that of Kt-7 in the crystal structure of the 50S ribosomal subunit.
Removal of the 2′-OH from the putative partner in the interaction, 2b (A98), had an even greater effect on electrophoretic mobility under all conditions (Fig. 2). Mg2+- or Na+-induced folding observed by FRET was also seriously impaired, requiring a 10-fold higher Mg2+ ion concentration to get maximal folding compared to the unmodified sequence (Fig. 3A). Interestingly, a higher extent of folding was possible using a saturating Na+ concentration compared to Mg2+. The greater impairment of folding resulting from removal of the 2′-OH at position 2b compared to −1n suggests that the former results from more than the loss of the potential hydrogen bond with the −1n ribose; it is possible that a change of sugar pucker at 2b might reposition the adenine nucleobase that is involved in one of the critical A•G pairs.
These results suggest that the Kt-7 sequence in free solution adopts a conformation that is different from that observed in the crystal, and probably is more similar to those of the box C/D and U4 K-turns.
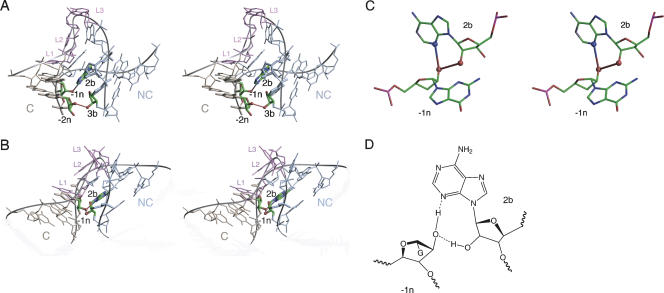
A very important ribose–base interaction between L1 and 1n
A potential hydrogen bond is observed between L1 2′-OH and the N1 of the adenine nucleobase in the 1n position (i.e., the first G•A pair) (Fig. 5). This distance is optimal in U4, but slightly shorter in Kt-7 and box C/D. This interaction is the best conserved of all the potential hydrogen bonds (discussed below). Removal of the 2′-OH from position L1 (with L1 as either A or G) in Kt-7 leads to a marked loss of electrophoretic retardation (Fig. 2). In the presence of 2 mM Mg2+ or 10 μM Co(III) hexammine ions, the L1-modified species were less retarded than any other variant K-turn, with a mobility closely similar to a simple duplex. The inability of the L1 2′-deoxyribo K-turn to adopt the kinked geometry is strongly supported by the FRET data, where no increase in FRET efficiency was found at any concentration of Mg2+ or Na+ ions (Fig. 3). All the data demonstrate that this species is totally unable to fold into the kinked structure, consistent with an important role for the L1:1n interaction in stabilizing the folded form of the K-turn in Kt-7. This indicates that the hydrogen bond is a significant part of the interactions stabilizing the folded conformation, consistent with the high conservation of this element.
FIGURE 5.
An important ribose–nucleobase interaction between L1 and 1n in the box C/D K-turn. (A) Parallel-eye stereoscopic view of the box C/D K-turn showing the L1:1n interaction. The K-turn is oriented in this view to look down the axis of the NC helix, with the C helix running horizontally to the left. The bulge is at the lower right of this view. The nucleotides participating in the interaction are highlighted. The L1 2′-OH proton is donated to N1 of adenine 1n. (B) The chemical structure of the hydrogen bonding interaction.
Backbone hydrogen bonding stabilizing the turn of the bulged strand
Two potential hydrogen bonds appear to stabilize the phosphodiester backbone of the bulge as it makes a very tight turn at the apex of the K-turn (Fig. 6), and are well conserved. An interaction between the 2′-O of L3 and the proS nonbridging O of the phosphate between L1 and L2 bridges the neck of the turn and is observed in most K-turn crystal structures, although it is short in Kt-7. Removal of the 2′O from the L3 ribose in Kt-7 (A96) led to a significant loss of electrophoretic retardation (Fig. 2). A marked impairment of ion-induced folding was also observed by FRET, with efficiency increases reduced to approximately one-third that of the natural Kt-7, and substantially increased [Mg2+]1/2 and [Na+]1/2 concentrations (Fig. 3).
FIGURE 6.
Ribose–phosphate interactions in the phosphodiester backbone of the bulge of the box C/D K-turn. Parallel-eye stereoscopic view of the isolated bulge region showing two hydrogen bonds. That from the 2′-OH of L3 to the proS nonbridging O atom of the L1/L2 phosphate bridges across the tight turn of the bulge. The other is from the 2′-OH of L2 to the proS nonbridging O atom of the L2/L3 phosphate.
The second interaction is between the 2′-O of L2 and the proS nonbridging O of the phosphate between L2 and L3, located on the outer edge of the bulge. While the length is optimal for hydrogen bonding in Kt-7 and U4, it is long in box C/D. Removal of the 2′O from the L2 ribose in Kt-7 (A95) led to a significant loss of electrophoretic retardation (Fig. 2), similar to that resulting from removal of the L3 2′OH under all ionic conditions. Removal of the L3 2′-OH resulted in a significantly increased FRET efficiency in the absence of added metal ions (Fig. 3), suggesting that the helix is more kinked than for the unmodified sequence. To a lesser degree, this was also true for the L3 modification, and both forms exhibited a degree of electrophoretic retardation in the presence of EDTA (data not shown). This complicates the analysis of the FRET data, but nevertheless it is apparent that the folding of the L2-2′-deoxy form is impaired, though to a lesser degree than the L3-2′-deoxy species.
These results demonstrate impaired folding of the K-turn upon removal of the 2′-O from either the L2 or L3 ribose positions, consistent with an important role for these ribose–phosphate interactions in stabilizing the tightly kinked backbone around the bulge of the K-turn. However, the FRET data indicate that the longer-range hydrogen bond donated by the 2′-O of L3 to the proS nonbridging O of the phosphate between L1 and L2 is more important than that involving the 2′-O of L2.
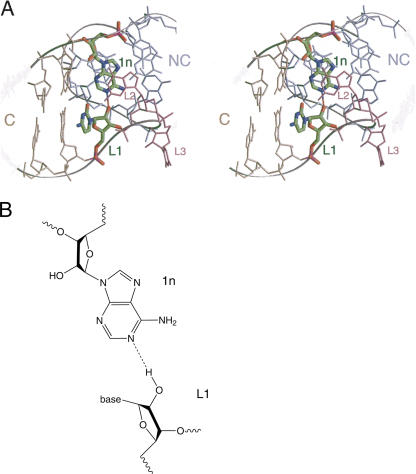
A sugar–base hydrogen bond involving 1n in Kt-7
An additional ribose–base interaction is found in Kt-7, between the 2′-OH of ribose 1n (A80) and the N6 of the adenine at L2 (the exocyclic amine of A95) (Fig. 7). Adenine is not conserved at the L2 position, being present in slightly less than half of the K-turns of the ribosome, for example, but it is adenine in the U4 K-turn where an equivalent interaction is found. However, in box C/D, the L2 nucleotide is guanine, and its O6 is not positioned to make a strong hydrogen bond with the 2′-OH at 1n (A7). Removal of the 2′-O from the 1n ribose of Kt-7 resulted in a marked impairment of K-turn folding, generating a major reduction in electrophoretic retardation (Fig. 2). The extent of the change in FRET efficiency of this variant was similar to the natural K-turn in both Mg2+ and Na+ ions, but a fivefold increase in [Mg2+]1/2 = 370 μM and threefold increase in [Na+]1/2 = 100 mM was observed (Fig. 3).
FIGURE 7.
A ribose–nucleobase interaction between nucleotides 1n and L2 in Kt-7. This interaction is only possible where L2 is adenine. (A) Parallel-eye stereoscopic view looking down onto the major groove at the apex of the turn, with the C helix extending forward. The nucleotides participating in the interaction are highlighted. (B) The chemical structure of the hydrogen bonding interaction.
Structural comparisons with other K-turn elements
Altogether we have examined the crystal structures of 11 different well-defined K-turns, allowing us to examine our results on the requirements of 2′-OH groups for the folding into the kinked geometry in the context of a wide structural base. The interactions are summarized schematically in Figure 8.
FIGURE 8.
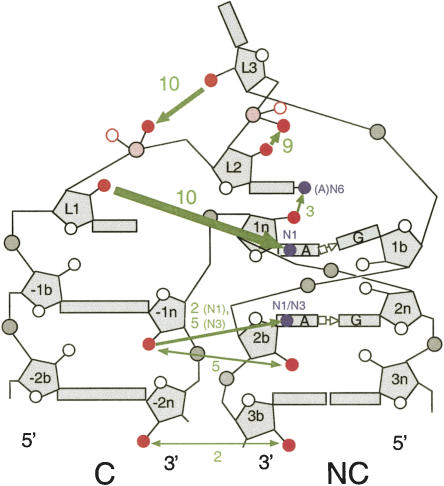
Cartoon summary of hydrogen bonding interactions involving 2′-OH groups. The green arrows depict hydrogen bonds; the thickness reflects the degree of impairment of folding upon removal of the participating 2′-OH group. The numbers written in green are the number of times each hydrogen bond is identifiable in the 11 K-turn structures analyzed. Participating oxygen and nitrogen atoms are shown in red and blue, respectively, in this scheme.
L1 2′-OH to 1n adenine N1
The hydrogen bond donated from L1 2′-OH to 1n adenine N1 is the only universal interaction of the K-turn structure. It is observed in every case, including the K-turn of the SAM riboswitch (see below) and a slight variation in Kt-58. The mean length is 2.70 ± 0.18 Å over the complete set. The most interesting test of the generality of this interaction is provided by Kt-58, where the loop is shortened to 2 nt and opposed by another formally unpaired nucleotide (Ln1). In this K-turn there is no loop nucleobase capping the C helix, and the L1 nucleobase caps the NC helix and is itself stacked by the Ln1 nucleobase (Fig. 9). In the absence of the L1 nucleotide stacked on the end of the C helix, −1b (C1602) assumes the normal role of L1 and donates its 2′-OH proton to adenine N1 at the 1n position to give a hydrogen bond 2.78 Å in length. Kt-58 is thus very much the exception that proves the rule. The near-invariant nature of this interaction is in excellent agreement with our experimental observation that folding is totally prevented by removal of the L1 2′-OH in Kt-7. Interestingly, removal of the L1 2′-OH has been found to be the most deleterious change for the binding of the human 15.5-kDa protein to box C/D U25 RNA in NAIM experiments (Szewczak et al. 2005).
FIGURE 9.
Interaction between the ribose 2′-OH of −1b and the N1 of adenine 1n in Kt-58. The axis of the C helix is approximately vertical on the left side, and the view presents the major groove at the apex of the turn. The bulge of Kt-58 is 2 nt in length, with a single opposed nucleotide on the opposite strand (Ln1). Exceptionally, there is no nucleotide stacked onto the end of the C helix in this K-turn. In this situation, the 2′-OH of the −1b ribose donates a hydrogen bond to N1 of adenine, thus performing the role of L1 in all the other K-turn structures analyzed (see Fig. 5).
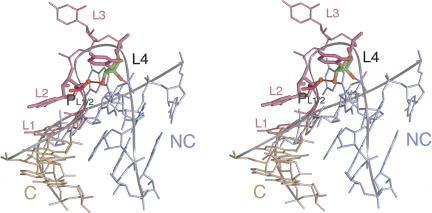
L3 2′-OH to the proS O of L1/L2 phosphate
This interaction is observed in the majority of K-turn structures, with a mean length of 2.83 ± 0.36 Å. An interesting test of this interaction is provided by Kt-38, where the loop is 4 nt in length. The nucleobase of L1 caps the C helix as normal, while it is the ribose of L2 that is stacked onto the end of the NC helix, with its nucleobase stacked onto L1 (Fig. 10). Both L3 and L4 are extended away from the RNA in the ribosome, and in this K-turn it is the ribose of L4 (U1030) that donates its 2′-OH proton to the proS nonbridging O of the L1/L2 phosphate, with a length of 2.63 Å. In almost every K-turn this interaction appears to stabilize the tight bend adopted by the bulge in the folded conformation, and the near-universal nature of this interaction is consistent with the great impairment to the folding of Kt-7 caused by removal of the 2′-OH of L3. The stereoselectivity of the hydrogen bonding to the phosphate groups has been further examined in preliminary experiments involving chemical substitution of racemic methyl phosphonate groups at the L2/L3 position, whereupon two species of different electrophoretic mobility were obtained (data not shown). However, detailed interpretation will require configurational assignment, analogous to our recent study of DNA junctions (Liu et al. 2005).
FIGURE 10.
Ribose–phosphate interactions in the backbone of the bulge of Kt-38. This K-turn has a 4-nt loop. A hydrogen bond is donated from the 2′-OH of L4 to the proS nonbridging O atom of the L1/L2 phosphate (PL1/2), bridging across the tight turn of the bulge. This is equivalent to the L3 2′-OH:L1/L2 phosphate interaction observed in 3-nt bulge K-turns (see Fig. 6).
L2 2′-OH to the proS O of L2/L3 phosphate
This interaction is observed in nine of 11 K-turn structures, with a mean length of 2.80 ± 0.36 Å. An equivalent interaction is not possible in Kt-38, and the bond length is too long in Kt-15.
Interactions between the minor grooves of the C and NC helices
These interactions are much less well conserved across the range of K-turn structures, but mainly fall into one of two classes. In the first class there are hydrogen bonds between the 2′-OH groups of −2n and 3b, plus one donated from the 2′-OH of −1n to N1 of adenine 2b. This is found in the crystal structures of Kt-7 and Kt-23 of the 30S ribosomal subunit. A possible interaction is also found in Kt-11, but the −2n:3b bond is long at 3.8 Å. The second class is larger, exemplified by box C/D, U4, Kt-42, Kt-46, and the SAM riboswitch. In these K-turns there is a hydrogen bond between the 2′-OH groups of −1n and 2b. There is also a hydrogen bond donated from the 2′-OH of −1n to the nucleobase of the adenine at 2b, but the acceptor is N3 in this case. The mean length over the five examples is 2.88 ± 0.16 Å. A few K-turns, such as Kt-58 and Kt-15, have no equivalent interactions. The very small effect on the folding of Kt-7 resulting from the removal of the 2′-OH from −2n suggests that the solution structure might put this K-turn into the second class above. However, the relatively small effect of the removal of the 2′-OH from −1n position coupled with the variation in hydrogen bonding patterns seen over the range of K-turns indicates that the interactions between the minor grooves of the C and NC helices beyond the region of the kinking are of minor importance.
1n 2′-OH to L2 adenine N6
This interaction is only possible when L2 is adenine, but this is the case for five of the 11 K-turns for which structural data are available. Of these, three clearly make this interaction (Kt-7, Kt-15, and U4), with a mean distance of 2.80 ± 0.12 Å. In Kt-11 the geometry appears to be consistent with hydrogen bonding, but the distance is long at 3.48 Å. Nevertheless, the significant effect upon the folding of Kt-7 when the 2′-OH was removed from 1n is consistent with stabilization by this interaction, when the sequence permits it.
CONCLUSIONS
The experimental data presented here demonstrate the importance of 2′-OH groups in the stabilization of the kinked geometry of the K-turn. However, there is a wide variation in the level of impairment of folding resulting from the removal of different individual 2′-hydroxyl groups from the Kt-7 sequence. Some (e.g., 2n or −2n) have little effect, while others result in very impaired folding (e.g., L3). The single largest effect arises when the 2′-OH of the L1 ribose is removed. This is consistent with the key role of a hydrogen bond donated to the N1 of adenine 1n, which is conserved in all available K-turn structures. The complete absence of ion-induced folding of the L1-deoxyribose variant (the only case where the impairment is total) suggests that this interaction may be required before other interactions can form productively. It may contribute to the absence of detectable folding in K-turn variants lacking adenine at the 1n position (Goody et al. 2003). The analysis presented here reveals the importance of other 2′-OH interactions in Kt-7, although none is as important as that between L1 and the base of 1n. Of these, the most important interaction appears to be the potential bonding between the L3 2′-OH and the proS nonbridging oxygen of the L1/L2 phosphate. This is well conserved in most of the structures, probably helping to draw the two interacting minor grooves together. Removal of the L3 2′-OH leads to a major impairment of folding.
While these interactions are conserved in the K-turn structures, there is some variability elsewhere in the structure. We have noted that hydrogen bonding between the minor grooves of the C and NC stems can take different forms, and indeed our data suggest that the structure of the Kt-7 sequence in free solution may differ from that found in the crystals of 50S ribosomal subunits. It is possible that longer-range tertiary interactions in the context of the ribosome may manipulate the stems so that alternative interactions form between their minor grooves. The relatively weak effects arising from removal of the 2′-OH of −1n suggest that these interactions are not critical to the formation of the kinked geometry, but form adventitiously. Moreover, molecular dynamics simulations have suggested a degree of hinge-like flexibility in the K-turn (Cojocaru et al. 2005; Razga et al. 2004, 2005, 2006), and it is possible that these interactions may be continually forming and rearranging.
The structural variability between K-turns is further illustrated by the recently presented structure of the SAM riboswitch (Montange and Batey 2006). This contains a K-turn that conforms well to the consensus sequence and includes an A•G pair at the 3b:3n position like Kt-7. The global shape is closely similar to the other K-turns, but the 2′-O interactions are largely different. None of the previously observed stem–stem interactions is found in the structure as presented. There is a significant change of geometry around the loop, so that the backbone is oriented quite differently around the L3, 1b region. This prevents the formation of the hydrogen bond donated from the L3 2′-OH to the L1/L2 proS nonbridging oxygen atom. However, the key interaction in which the L1 2′-OH proton is donated to the N1 of the adenine at 1n is still present, confirming the key importance of this interaction. Interestingly, the different backbone geometry makes further hydrogen bonds possible with the 2′-OH of 1b, forming a potential network that stabilizes the core of the K-turn.
In summary, the single most important interaction involving a 2′-OH group is the L1 2′-OH:1n base interaction. This is found even where the other potential hydrogen bonds are not. Without this interaction it appears that the core of the K-turn cannot be stabilized in the kinked geometry. If this is present, then the base interactions and the packing of the minor groove faces allow folding to occur, and the remaining hydrogen bonds form where they can. However, it is likely that the exact pattern of hydrogen bonding will depend upon the global geometry of the K-turn, and this may be subject to manipulation by longer-range tertiary interactions and protein binding.
MATERIALS AND METHODS
RNA synthesis and construction of K-turn species
The majority of oligonucleotides were synthesized using t-BDMS phosphoramidite chemistry (Beaucage and Caruthers 1981), as described by Wilson et al. (2001), with 2′-deoxyribose substitution as required. Fluorescein and Cy3-conjugated oligonucleotides were attached at 5′ termini as phosphoramidites during synthesis as required. A few oligonucleotides used for fluorescence were purchased from Dharmacon, made using 2′-ACE chemistry. Oligoribonucleotides were deprotected in 25% ethanol/ammonia solution at 55°C for 4 h (2 h for oligonucleotides with attached fluorophores), and evaporated to dryness. Oligoribonucleotides were redissolved in 300 μL 1 M tetrabutylammonium fluoride (Aldrich) in tetrahydrofuran to remove t-BDMS protecting groups and agitated at 20°C for 16 h prior to desalting by G25 Sephadex (NAP columns, Pharmacia) and ethanol precipitation. All oligonucleotides were purified by gel electrophoresis in polyacrylamide and recovered from gel fragments by electroelution or diffusion in buffer followed by ethanol precipitation. Fluorescently labeled RNA was subjected to further purification by reversed-phase HPLC on a C18 column (Waters μ-Bondapak) using an acetonitrile gradient with an aqueous phase of 100 mM triethylammonium acetate (pH 7.0). Oligonucleotides required for electrophoretic analysis were radioactively 5′-32P-labeled using T4 polynucleotide kinase and [γ-32P] ATP (Amersham).
Duplex species were prepared by mixing equimolar quantities of the appropriate oligoribonucleotides and annealing them in 50 mM Tris.HCl (pH 7.5), 25 mM NaCl by slow cooling from 80°C to 4°C. They were purified by electrophoresis in 8% or 12% polyacrylamide under nondenaturing conditions and recovered by either electroelution or diffusion into buffer, followed by ethanol precipitation.
Study of folding by gel electrophoresis
Radioactively 5′-32P-labeled RNA species were electrophoresed in 15% polyacrylamide gels (29:1, acrylamide:bis) in 90 mM Tris.borate (pH 8.3) plus added salts or 2 mM EDTA as required. Electrophoresis was performed at 120 V at room temperature for at least 72 h, with recirculation of the buffer at >1 L h−1. Gels were dried onto Whatman 3MM paper, exposed to storage phosphor plates, and imaged using a Fuji BAS-1500 PhosphorImager.
The sequences used for the electrophoretic experiments were (written 5′ to 3′):
Kt-7 upper strand: CGCAAGCGACAGGAACCTCGCCAGUCAGUGGCGAAGAACCAUGUCAGGGGACTGTCAAGTTGAACAGG;
Kt-7 lower strand: CCTGTTCAACTTGACAGTCCCCUGACAUGGGGAGCCACUGACUGGCGAGGTTCCTGTCGCTTGCG.
The DNA sections of these oligonucleotides are shown underlined. Single 2′-deoxyribose substitutions were introduced into the RNA sections as required.
Study of folding by fluorescence resonance energy transfer
Absorption spectra were measured in 90 mM Tris.borate (pH 8.3) at 4°C in 150 μL volumes using a Cary 1E spectrophotometer. Spectra were deconvoluted using a corresponding RNA species labeled only with Cy3, and fluorophore absorption ratios were calculated using a MATLAB program. Fluorescence spectra were recorded in 90 mM Tris.borate (pH 8.3) at 4°C using an SLM-Aminco 8100 fluorimeter. Spectra were corrected for lamp fluctuations and instrumental variations, and polarization artifacts were avoided by setting excitation and emission polarizers crossed at 54.7°. Values of FRET efficiency (E FRET) were measured using the acceptor normalization method (Clegg 1992) implemented in MATLAB. FRET efficiency (E FRET) as a function of metal ion concentration was analyzed on the basis of a model in which the fraction of folded molecules corresponds to a simple two-state model for ion-induced folding, i.e.,
where E 0 is the FRET efficiency of the RNA in the absence of added metal ions, ΔE FRET is the increase in FRET efficiency at saturating metal ion concentration, [M] is the prevailing metal ion concentration, KA is the apparent association constant for metal ion binding, and n is a Hill coefficient. Data were fitted to this equation by nonlinear regression. The metal ion concentration at which the transition is half complete ([M]1/2) is given by (1/KA)1/n.
The sequences used in the FRET analyses were (written 5′ to 3′):
Kt-7 upper strand: Fluorescein-CCAGUCAGUGGCGAAGAACCAUGUCAGG
Kt-7 lower strand: Cy3-CCUGACAUGGGGAGCCACUGACUGG
Single 2′-deoxyribose substitutions were introduced as required.
Analysis of K-turn structures
Analysis of K-turn structures was performed using Insight II (Accelrys Software Inc.) and PyMOL (DeLano Scientific LLC) using the following coordinate files. 50S ribosomal subunit of H. marismortui at 2.4 Å resolution, file 1FFK.pdb (Ban et al. 2000); 30S ribosomal subunit of Thermus thermophilus at 3.05 Å resolution, file 1J5E.pdb (Wimberly et al. 2000; Brodersen et al. 2002); box C/D (bound to protein L7Ae) of Archeoglobus fulgidus at 2.7 Å resolution, file 1RLG.pdb (Moore et al. 2004); human U4 snRNA (bound to 15.5-kDa protein) at 2.9 Å resolution, file 1E7K.pdb (Vidovic et al. 2000); SAM-binding riboswitch of Thermoanaerobacter tengcongensis at 2.9 Å resolution, file 2GIS.pdb (Montange and Batey 2006). Molecular graphics images were generated using Insight II and annotated in Photoshop.
ACKNOWLEDGMENTS
We thank Dr. David Norman for valuable discussions and Cancer Research-UK for financial support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.285707.
REFERENCES
- Allmang, C., Carbon, P., Krol, A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: Identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Beaucage, S.L., Caruthers, M.H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- Bortolin, M.L., Bachellerie, J.P., Clouet-d'Orval, B. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp) Nucleic Acids Res. 2003;31:6524–6535. doi: 10.1093/nar/gkg860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, D.E., Clemons W.M., Jr, Carter, A.P., Wimberly, B.T., Ramakrishnan, V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Clegg, R.M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- Cojocaru, V., Klement, R., Jovin, T.M. Loss of G-A base pairs is insufficient for achieving a large opening of U4 snRNA K-turn motif. Nucleic Acids Res. 2005;33:3435–3446. doi: 10.1093/nar/gki664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot, P., Bortolin, M.L., Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- Goody, T.A., Melcher, S.E., Norman, D.G., Lilley, D.M.J. The kink-turn motif in RNA is dimorphic, and metal ion dependent. RNA. 2003;10:254–264. doi: 10.1261/rna.5176604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., Steitz, T.A. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V., Bork, P., Sander, C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, J.F., Tran, E.J., Maxwell, E.S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute, A., Leontis, N.B., Massire, C., Westhof, E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Déclais, A.-C., McKinney, S.A., Ha, T., Norman, D.G., Lilley, D.M.J. Stereospecific effects determine the structure of a four-way DNA junction. Chem. Biol. 2005;12:217–228. doi: 10.1016/j.chembiol.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Mao, H., White, S.A., Williamson, J.R. A novel loop–loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 1999;6:1139–1147. doi: 10.1038/70081. [DOI] [PubMed] [Google Scholar]
- Marmier-Gourrier, N., Clery, A., Senty-Segault, V., Charpentier, B., Schlotter, F., Leclerc, F., Fournier, R., Branlant, C. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003;9:821–838. doi: 10.1261/rna.2130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura, S., Ikawa, Y., Inoue, T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–5551. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montange, R.K., Batey, R.T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Moore, T., Zhang, Y., Fenley, M.O., Li, H. Molecular basis of box C/D RNA-protein interactions; Cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Nottrott, S., Hartmuth, K., Fabrizio, P., Urlaub, H., Vidovic, I., Ficner, R., Lührmann, R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razga, F., Spackova, N., Reblova, K., Koca, J., Leontis, N.B., Sponer, J. Ribosomal RNA kink-turn motif—A flexible molecular hinge. J. Biomol. Struct. Dyn. 2004;22:183–194. doi: 10.1080/07391102.2004.10506994. [DOI] [PubMed] [Google Scholar]
- Razga, F., Koca, J., Sponer, J., Leontis, N.B. Hinge-like motions in RNA Kink-turns: The role of the second A-minor motif and nominally unpaired bases. Biophys. J. 2005;5:3466–3485. doi: 10.1529/biophysj.104.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razga, F., Zacharias, M., Reblova, K., Koca, J., Sponer, J. RNA kink-turns as molecular elbows: Hydration, cation binding, and large-scale dynamics. Structure. 2006;14:825–835. doi: 10.1016/j.str.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky, T.S., Tang, T.H., Tchirkova, I.V., Brosius, J., Bachellerie, J.-P., Hüttenhofer, A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczak, L.B., Gabrielsen, J.S., Degregorio, S.J., Strobel, S.A., Steitz, J.A. Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein. RNA. 2005;11:1407–1419. doi: 10.1261/rna.2830905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, B., Melcher, S.E., Wilson, T.J., Norman, D.G., Lilley, D.M.J. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–1200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic, I., Nottrott, S., Hartmuth, K., Lührmann, R., Ficner, R. Crystal structure of the spliceosomal 15.5 kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Segault, V., Charpentier, B., Nottrott, S., Fabrizio, P., Bachi, A., Wilm, M., Rosbash, M., Branlant, C., Lührmann, R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., Dickmanns, A., Lührmann, R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.J., Zhao, Z.-Y., Maxwell, K., Kontogiannis, L., Lilley, D.M.J. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons W.M., Jr, Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Grundy, F.J., Murphy, B.A., Henkin, T.M. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, A.K., Nottrott, S., Kuhn-Holsken, E., Schroder, G.F., Grubmuller, H., Lührmann, R., Seidel, C.A., Oesterhelt, F. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA. 2005;11:1545–1554. doi: 10.1261/rna.2950605. [DOI] [PMC free article] [PubMed] [Google Scholar]