Abstract
Cytoplasmic polyadenylation regulates mRNA stability and translation and is required for early development and synaptic plasticity. The GLD-2 poly(A) polymerase catalyzes cytoplasmic polyadenylation in the germline of metazoa. Among vertebrates, the enzyme is encoded by two isoforms of mRNA that differ only in the length of their 3′-UTRs. Here we focus on regulation of vertebrate GLD-2 mRNA. We show that the 3′-UTR of GLD-2 mRNA elicits its own polyadenylation and translational activation during frog oocyte maturation. We identify the sequence elements responsible for repression and activation, and demonstrate that CPEB and PUF proteins likely mediate repression in the resting oocyte. Regulated polyadenylation of GLD-2 mRNA is conserved, as are the key regulatory elements. Poly(A) tails of GLD-2 mRNA increase in length in the brain in response to neuronal stimulation, suggesting that a comparable system exists in that tissue. We propose a positive feedback circuit in which translation of GLD-2 mRNA is stimulated by its polyadenylation, thereby reinforcing the switch to polyadenylate and activate batteries of mRNAs.
Keywords: CPEB, GLD-2, PUF, cytoplasmic polyadenylation, poly(A) polymerase
INTRODUCTION
The 3′-untranslated region (3′-UTR) of mRNAs plays a key role in controlling nuclear export, localization, translation, and stability of mRNAs (for review, see Sonenberg 1994; Hentze 1995; Wickens et al. 2002; Kuersten and Goodwin 2003). Cytoplasmic lengthening of the poly(A) tail increases translation and mRNA stability, while its removal can cause translational repression and mRNA decay. These regulated changes in poly(A) length are critical in early development, and participate in pattern formation and cell cycle control (for review, see Richter 2000; Wickens et al. 2000). In the nervous system, repeated stimulation of synapses activates polyadenylation and local translation (Wu et al. 1998; Huang et al. 2002; Si et al. 2003a; Theis et al. 2003), events that are critical in long-term potentiation (LTP) and learning (Alarcon et al. 2004). The widespread anatomical distribution of polyadenylation factors suggests that the same system may operate in many other biological contexts as well.
Meiotic maturation of Xenopus oocytes and mitotic timing in embryos require cytoplasmic polyadenylation of cyclin and c-mos mRNAs (for review, see Mendez and Richter 2001; Wickens et al. 2002). The polyadenylation of these mRNAs requires elements in their 3′-UTRs, including a uridine-rich cytoplasmic polyadenylation element (CPE) and the hexanucleotide sequence AAUAAA (Fox et al. 1989; McGrew and Richter 1990). CPE-binding protein (CPEB) binds target mRNAs and elicits their polyadenylation and activation (Hake and Richter 1994). A cytoplasmic form of cleavage and polyadenylation specificity factor (CPSF) complex, a multiprotein complex, binds AAUAAA (Bilger et al. 1994; for review, see Mendez and Richter 2001). Neuronal CPEB is required for polyadenylation of CPE-containing mRNAs (Wu et al. 1998; Si et al. 2003a) and is important for formation of hippocampal-dependent memories (Alarcon et al. 2004; Berger-Sweeney et al. 2006).
CPEB is both an activator and repressor. The inhibitory function is linked to several other translational repressor proteins, including maskin, PUF, and Nanos proteins. Members of each family, along with CPEB, are bound to specific mRNAs in Xenopus oocytes, and cause their repression prior to meiotic maturation (Stebbins-Boaz et al. 1999; Nakahata et al. 2001; Cao and Richter 2002; Nakahata et al. 2003; for review, see Richter 2000). These different factors may repress through redundant mechanisms. Maskin directly interacts with CPEB and with eIF-4E, preventing initiation (Stebbins-Boaz et al. 1999; Cao and Richter 2002). PUF proteins can recruit the Ccr4 complex, containing both deadenylases and the inhibitory Dhh1p RNA helicase (Goldstrohm et al. 2006). CPEB and PUF proteins are required for learning (Dubnau et al. 2003) and localize to dendrites (Wu et al. 1998; Ye et al. 2004; Vessey et al. 2006).
GLD-2, a divergent cytoplasmic poly(A) polymerase (PAP), was first identified in Caenorhabditis elegans (Wang et al. 2002). The enzyme is related to the Cid1 and Cid13 PAPs of Schizosaccharomyces pombe (Read et al. 2002; Saitoh et al. 2002). GLD-2 enzyme activity is stimulated by interaction with an RNA-binding protein, GLD-3 (Wang et al. 2002). Together, GLD-2 and GLD-3 are thought to form a novel heterodimeric PAP, in which the RNA-binding component GLD-3 recruits the catalytic subunit GLD-2 to specific mRNAs (Wang et al. 2002). GLD-2 homologs in Saccharomyces cerevisiae, Trf4p and Trf5p, localize to the nucleus. Like GLD-2/GLD-3 in C. elegans, Trf4p's polymerase activity is stimulated by RNA-binding proteins Air1p or Air2p. Trf4p, Air2p, and Mtr4p comprise the TRAMP complex (Vanacova et al. 2005). In contrast to GLD-2, however, polyadenylation by this complex leads to RNA degradation by the nuclear exosome, rather than to activation (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005).
GLD-2 poly(A) polymerases were recently identified in vertebrates, including frogs, mice, and humans (Barnard et al. 2004; Kwak et al. 2004; Rouhana et al. 2005). The mouse (mGLD-2) and Xenopus GLD-2 (XlGLD-2) proteins are both nuclear and cytoplasmic in oocytes (Rouhana et al. 2005; Nakanishi et al. 2006). Cytoplasmic XlGLD-2 protein is physically associated with CPEB and CPSF (Barnard et al. 2004; Rouhana et al. 2005), but not with CPEB-protein-bound repressors Pumilio and maskin (Rouhana et al. 2005). Vertebrate GLD-2 proteins are important for meiotic maturation of oocytes, and likely for activation of many mRNAs throughout early development (Barnard et al. 2004; Rouhana et al. 2005; Nakanishi et al. 2006). In the mouse brain, GLD-2 mRNA is expressed in anatomical regions necessary for long-term cognitive and emotional learning (Rouhana et al. 2005). In the hippocampus, mGLD-2 mRNA co-localizes with CPEB1 and Pumilio1 mRNAs, both of which are involved in synaptic plasticity. In the cerebellum, mGLD-2 mRNA is present within the major neuronal cell populations, coincident with the distribution of other CPEB isoforms.
In this study, we focus on the post-transcriptional regulation of GLD-2 mRNA. We show that two previously identified GLD-2 mRNA isoforms [XlGLD-2A(L) and XlGLD-2A(S)] differ in their regulation during oocyte maturation. In particular, the mRNA with a long 3′-UTR causes repression before maturation, then elicits polyadenylation-dependent activation once maturation begins. This polyadenylation-dependent activation of a poly(A) polymerase's mRNA may comprise a positive-feedback circuit that reinforces a regulatory switch that supports activation of many mRNAs.
RESULTS
XlGLD-2A(L) 3′-UTR causes translational stimulation during oocyte maturation
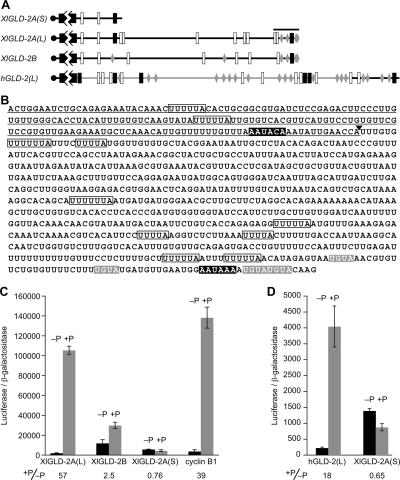
The XlGLD-2A gene encodes two mRNAs, termed GLD-2A(S) and GLD-2A(L). These mRNAs differ solely in the length of their 3′-UTRs, due to the use of alternative 3′-end cleavage in the nucleus. The long 3′-UTR extends an additional 747 nucleotides (nt) beyond that of the short form (Fig. 1A,B). The structures and polyadenylation sites of the two mRNAs were confirmed by isolation of multiple ESTs, directed RT-PCR, and Northern blotting (data not shown; Rouhana et al. 2005). The 3′-termini of both mRNA forms are preceded by polyadenylation hexanucleotides, either AAUACA or AAUAAA. A second Xenopus GLD-2 gene, XlGLD-2B, produces a single mRNA with a long 3′-UTR that is 83% identical to that of XlGLD-2A(L). Human (hGLD-2) and mouse GLD-2 genes produce two mRNA isoforms analogous to XlGLD-2A(L) and (S); these, too, encode identical proteins. The human and frog GLD-2 3′-UTRs diverge substantially. Despite only 3% identity throughout the 3′-UTRs, the human 3′-UTR contains islands of sequence conserved in the two frog mRNAs. For example, the last 32 nt of human and frog mRNAs are 87% identical.
FIGURE 1.
GLD-2 mRNA architecture and activation during oocyte maturation. (A) GLD-2 3′-UTRs. Diagram of vertebrate GLD-2 mRNA 3′-UTRs and the position of putative cytoplasmic polyadenylation elements (CPEs; white boxes), PUF binding elements (PBEs; gray diamonds), and cleavage and polyadenylation specificity sequences (black boxes) are shown. The last 90 nt of the XlGLD-2A(L) 3′-UTR, analyzed in later figures, is indicated by the black bar over XlGLD-2A(L) mRNA. Regions of high homology with XlGLD-2A(L) sequence are indicated by the black backbone. (B) XlGLD-2A 3′-UTR sequence. The 932-nt sequence of the XlGLD-2A(L) 3′-UTR is presented. The XlGLD-2A(S) 3′-UTR sequence is underlined; an arrowhead indicates the poly(A) addition site of that mRNA. The elements depicted in A are shown here using the same color key. (C) The XlGLD-2A(L) 3′-UTR increases translation of an mRNA during oocyte maturation. Luciferase reporter mRNAs, bearing different 3′-UTRs, were injected into Xenopus oocytes and incubated overnight in the absence or presence of progesterone. LacZ mRNA was coinjected as an internal control. Luciferase activity was measured and normalized to β-galactosidase activity and is shown for progesterone-treated and untreated oocytes. The ratio of luciferase activity in treated over untreated oocytes is shown under the X-axis. (D) Elements required for translational stimulation by the XlGLD-2A(L) 3′-UTR are conserved in the hGLD-2(L) 3′-UTR. A luciferase reporter mRNA bearing the hGLD-2(L) 3′-UTR was injected and analyzed as in C.
The conserved pattern of isoforms and sequence islands suggested that the 3′-UTRs might have a regulatory role. To test this hypothesis, we prepared mRNAs in vitro bearing the luciferase open reading frame (ORF) followed by either the long or short form of the XlGLD-2A 3′-UTR (Fig. 1C). The mRNAs were injected into the cytoplasm of Xenopus laevis oocytes. Luciferase activity was determined after incubation in the absence or presence of progesterone, which induces meiotic maturation. A β-galactosidase mRNA carrying 15 adenosines was used as an internal control. Translation of the mRNA bearing the XlGLD-2A(L) 3′-UTR was stimulated >50-fold during oocyte maturation (Fig. 1C). The XlGLD-2B 3′-UTR stimulated translation only two- to threefold, while the XlGLD-2A(S) had no significant effect (Fig. 1C). An mRNA bearing the cyclin B1 3′-UTR, used as a positive control, stimulated translation ∼40-fold.
The human GLD-2 gene produces two mRNAs with different 3′-UTRs, analogous in lengths to those of the Xenopus GLD-2A gene. As mentioned above, the 3′-UTR sequence of the human GLD-2(L) 3′-UTR is only 3% identical to that of the frog GLD-2A(L) 3′-UTR overall; however, both 3′-UTRs possess putative CPEs and PUF protein-binding sites near their 3′-termini. To test whether the 3′-UTR of hGLD-2(L) conferred translational regulation, we injected mRNAs bearing that 3′-UTR into oocytes. The human 3′-UTR enhanced luciferase activity 18-fold after progesterone treatment (Fig. 1D). An mRNA bearing the XlGLD-2A(S) 3′-UTR, used as a control, did not increase activity. We conclude that despite the great sequence divergence between the human and frog 3′-UTRs, both specifically stimulate translation during oocyte maturation.
XlGLD-2A(L) is polyadenylated during oocyte maturation
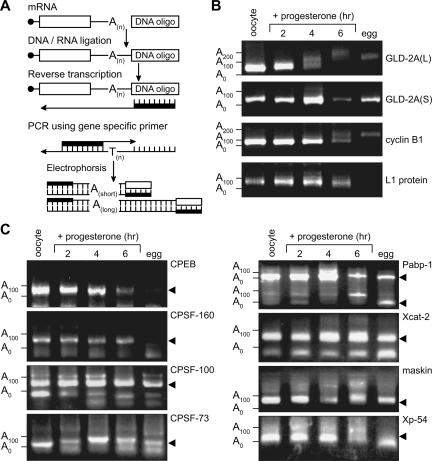
To determine whether endogenous GLD-2A mRNA was polyadenylated during oocyte maturation, we determined the length of poly(A) on endogenous XlGLD-2A mRNA using a PCR-based assay (Fig. 2A; Charlesworth et al. 2004). Total RNA was extracted from X. laevis oocytes at various times after addition of progesterone. A DNA oligonucleotide was ligated to the 3′-end of the RNA, and a complementary primer was used to initiate cDNA synthesis by reverse transcriptase. The primer possessed four T residues at its 3′ terminus, and thus would detect only polyadenylated mRNAs. Gene-specific primers in the 3′-UTR of the mRNA of interest were used to prime second-strand synthesis. After PCR, the lengths of poly(A) tails were deduced from the length of DNA fragments.
FIGURE 2.
Polyadenylation of endogenous XlGLD-2A(L) mRNA during oocyte maturation. (A) Measurements of poly(A) length. Total RNA from Xenopus oocytes, maturing oocytes, and eggs was ligated to the 5′-end of a modified DNA oligo. Reverse transcriptase was used to synthesize DNA complementary to the mRNA, using a primer anti-sense to the ligated DNA oligonucleotides; this newly synthesized DNA contained the poly(A) tail region. A gene-specific primer, along with a primer anti-sense to the ligated DNA oligo, was used to amplify the 3′-end of the gene of interest by PCR. The lengths of PCR products from gene-specific primers were determined using 2.5% agarose gels. (B) XlGLD-2A(L) is polyadenylated during oocyte maturation. mRNA was prepared at the indicated times after addition of progesterone or from unfertilized eggs. To determine poly(A) lengths of each endogenous mRNA, reactions were performed using primers specific for each of the four indicated mRNAs. The lengths of products obtained in resting oocytes were consistent with the known ends of the mRNA. The PCR products of XlGLD-2A(L) and cyclin B1 show slower electrophoretic mobility during maturation, indicating that those mRNAs are polyadenylated during maturation. The PCR products obtained with XlGLD-2A(S) and ribosomal protein L1 primers do not increase in length, indicating that these mRNAs are not polyadenylated. (C) Analysis of mRNAs encoding factors involved in translational regulation during oocyte maturation. The poly(A) tail of mRNAs for CPEB, CPSF-160, CPSF-100, CPSF-73, PABP-1, Xcat-2, and maskin were analyzed during oocyte maturation, using the same protocol as in A and B. The lengths of the specific 3′-end fragments, as determined by EST analysis, are indicated with arrowheads.
The poly(A) tails of endogenous XlGLD-2A(L) mRNA became progressively longer at successive times during oocyte maturation (Fig. 2B); after 6 h, the tails had been extended by 150–200 adenosines. In contrast, the poly(A) tails of XlGLD-2A(S) mRNA did not appear to change in length during maturation (Fig. 2B), consistent with the lack of translational stimulation by that mRNA's 3′-UTR (Fig. 1C). As controls, the poly(A) tails of cyclin B1 mRNA were extended by ∼150 residues, while polyadenylated forms of ribosomal protein-L1 mRNA apparently disappeared, as expected (Fig. 2B). We conclude that polyadenylation of the endogenous mRNAs mirrors the translational properties of the 3′-UTR reporters, and that endogenous XlGLD-2A(L) mRNA is polyadenylated during maturation.
Polyadenylation of XlGLD-2A(L) mRNA is specific, as it was not observed with other mRNAs, including ones involved in translational control during maturation (Fig. 2C). The poly(A) tails of CPEB, CPSF-160, CPSF-100, PABP-1, Xcat-2, maskin, and Xp54 mRNAs were not dramatically different before and after maturation. Xp54, maskin, and CPSF-73 mRNAs appeared to undergo transient polyadenylation, during or after germinal vesicle breakdown, but were later deadenylated (Fig. 2C). The multiple bands observed with PABP-1 mRNA reflect different 3′-UTR lengths, consistent with cDNA and EST databases.
GLD-2 control is mediated by CPE- and AAUAAA-dependent cytoplasmic polyadenylation
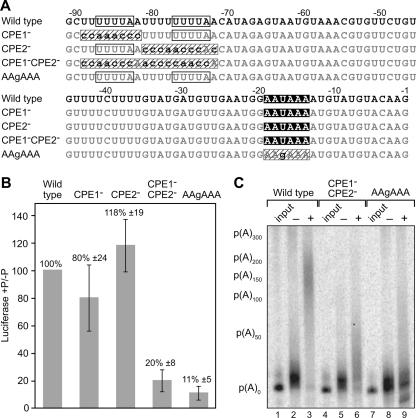
To identify the sequence elements in XlGLD-2A(L) mRNA that promote its translational activation, we first analyzed the roles of CPEs and the polyadenylation signal AAUAAA. Both of these elements are conserved in the long isoforms of GLD-2 mRNA in vertebrates (Fig. 1A). We focused on the last 90 nt of the XlGLD-2A(L) 3′-UTR because it stimulated translation during maturation as efficiently as the full-length XlGLD-2A(L) 3′-UTR and is highly conserved between Xenopus and humans. The conservation includes tandem CPEs and AAUAAA (Fig. 3A), and the spacings between them (data not shown).
FIGURE 3.
Translational stimulation by the XlGLD-2A(L) 3′-UTR is due to CPE-dependent polyadenylation. (A) Mutational analysis of the last 90 nt of the XlGLD-2A(L) 3′-UTR. The RNA sequence of the last 90 nt of the XlGLD-2A(L) 3′-UTR and of mutant 3′-UTRs. Two CPEs (UUUUA; white boxes) in this region were mutated into CA-rich sequences, either individually (CPE1− and CPE2−) or together (CPE1−CPE2−). The cleavage and polyadenylation specificity hexanucleotide sequence AAUAAA (black box) was mutated to AAgAAA. Mutated elements are highlighted with dashed boxes. (B) The last 90 nt of the XlGLD-2A(L) 3′-UTR increases translation during oocyte maturation, in a CPE- and AAUAAA-dependent manner. Luciferase reporter mRNAs carrying the last 90-nt sequence of XlGLD-2A(L), and the mutations indicated in A, were injected into Xenopus oocytes and incubated overnight in the absence or presence of progesterone. Luciferase activity was measured and normalized to the co-injected β-galactosidase mRNA, as described in Figure 1C. Translational stimulation of luciferase by the wild-type sequence was set as 100%. (C) XlGLD-2A(L) undergoes CPE- and AAUAAA-mediated cytoplasmic polyadenylation. 32P-labeled mRNAs, containing a mini-ORF and the indicated 3′-UTRs, were injected into Xenopus oocytes and incubated overnight in the absence (−) or presence (+) of progesterone. “Input” lanes are the RNA immediately after injection. mRNAs were extracted and analyzed for changes in electrophoretic mobility in a 6% polyacrylamide gel. (Lanes 1–3) Wild-type XlGLD-2A(L) sequence was polyadenylated in response to progesterone treatment (average An = 150). mRNAs with mutated (lanes 4–6) CPEs (CPE1−CPE2−) or (lanes 7–9) AAUAAA element (AAgAAA) showed reduced polyadenylation.
Mutant forms of the 3′-UTR linked to the luciferase ORF were injected into oocytes, which were then induced to mature using progesterone (Fig. 3B). mRNAs with mutations in either CPE alone did not perturb translational stimulation; however, simultaneous mutations in both CPEs decreased stimulation substantially, as did a point mutation in AAUAAA (Fig. 3B). We conclude that the two CPEs act redundantly, and in concert with AAUAAA, to promote translational activation.
To assess polyadenylation of these RNA sequence variants directly, we injected short, radiolabeled RNAs encoding a 21-amino acid-long micro-ORF linked to each terminal 3′-UTR segment. The 32P-labeled mRNAs, extracted from oocytes after overnight treatment with or without progesterone, were analyzed by electrophoresis (Fig. 3C). RNAs containing the wild-type sequence of the XlGLD-2A(L) 3′-UTR increased by ∼150 nt in length after oocyte maturation (Fig. 3C, lanes 1–3), consistent with the length of poly(A) added to the endogenous mRNA (Fig. 2B). mRNAs that lacked both CPE1 and CPE2 were much less active as substrates (Fig. 3C, lanes 4–6). Mutation of AAUAAA essentially abolished polyadenylation (Fig. 3C, lanes 7–9). Thus the sequence specificities of translational activation and polyadenylation are similar.
GLD-2 protein is bound to XlGLD-2A(L) mRNA in Xenopus oocytes
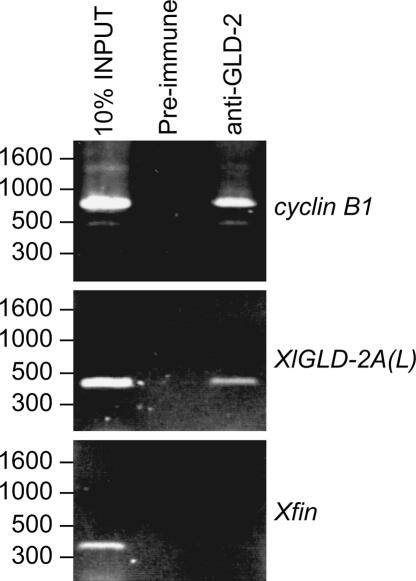
The GLD-2 enzyme catalyzes polyadenylation of certain mRNAs during oocyte maturation and acts in collusion with CPEB and CPSF (Barnard et al. 2004; Rouhana et al. 2005; Nakanishi et al. 2006). We reasoned that GLD-2 protein might polyadenylate GLD-2 mRNA. To test whether XlGLD-2A(L) mRNA was bound to GLD-2 protein, we incubated oocyte lysates with anti-GLD-2 antibodies, extracted immunoprecipitated RNAs, and analyzed them by semiquantitative RT-PCR (Fig. 4). XlGLD-2A(L) mRNA was readily detected in immunoprecipitates obtained with α-GLD-2, but not with control, pre-immune antibodies (Fig. 4). cyclin B1 mRNA, used as a positive control, was also enriched in α-GLD-2 immunoprecipitates (Fig. 4; Rouhana et al. 2005). Xfin, an mRNA that undergoes deadenylation during oocyte maturation (Ruiz i Altaba et al. 1987; Fox and Wickens 1990), was not specifically immunoprecipitated by α-GLD-2, as it was absent in both the α-GLD-2 and pre-immune immunoprecipitates (Fig. 4). We conclude that XlGLD-2A(L) mRNA is bound by GLD-2 protein in Xenopus oocytes.
FIGURE 4.
GLD-2 protein is bound to XlGLD-2A(L) mRNA. Oocyte extracts were incubated with α-GLD-2 or pre-immune guinea pig serum and bound to Protein A–Sepharose. RNA was extracted and used as a template for reverse transcription. mRNAs were detected by semiquantitative PCR on cDNA with gene-specific primers. XlGLD-2A(L) mRNA is detected in RNAs immunoprecipitated by α-GLD-2, but not by pre-immune serum. The lengths of products are consistent with the known 3′-termini of the mRNAs.
CPEs and PUF-binding elements are required for repression of GLD-2 mRNA
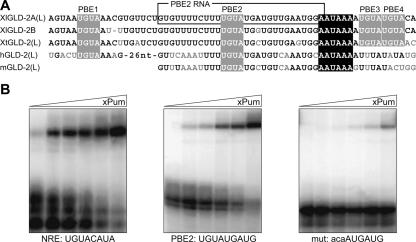
The last 90 nt of XlGLD-2A(L) contains four putative PUF-protein-binding elements (PBEs), as does GLD-2 mRNA in Xenopus tropicalis (Fig. 5A). Mouse and human GLD-2(L) mRNAs possess a single PBE in the same location as PBE2 (Fig. 5A). To determine whether PBE2 bound Xenopus Pumilio, we performed gel retardation assays using purified recombinant GST-Xl Pumilio (Fig. 5B). The protein bound to both PBE2 and the NRE (derived from hunchback mRNA). Binding was specific, since it was reduced by at least an order of magnitude by mutation of the UGU to ACA, as expected for a PUF interaction (Fig. 5B; e.g., Bernstein et al. 2005).
FIGURE 5.
Xenopus Pumilio binds to PBEs in the GLD-2(L) 3′-UTR. (A) A highly conserved region of the GLD-2(L) 3′-UTR contains multiple PBEs. Putative PBEs (PBE1-4 for Xenopus GLD-2; gray boxes) and cleavage and polyadenylation hexanucleotides (black boxes) in GLD-2(L) mRNAs. The chemically synthesized RNA used for xPum/RNA-binding assays is indicated by the black rectangle on the XlGLD-2A(L) sequence. (B) Binding to purified PUF protein. Various concentrations of purified, recombinant GST-xPum were incubated with labeled RNAs in vitro and analyzed by electrophoretic mobility shift assays. The RNAs correspond to the NRE of hunchback mRNA, PBE2, and a mutant form of PBE2 in which the UGU has been changed to ACA. The protein concentrations were 10 nM, 25 nM, 50 nM, 100 nM, 270 nM, and 810 nM.
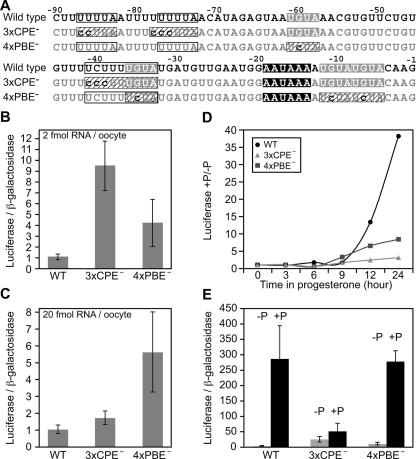
Both CPEB and PUF proteins can inhibit translation of mRNAs to which they bind. To test whether the CPEs or PBEs of XlGLD-2A(L) mRNA repressed its translation, we prepared luciferase mRNAs linked to mutant and wild-type 3′-UTRs (Fig. 6A). For repression studies, we prepared mutant mRNAs containing mutations in either all four putative PUF-binding elements (PBE1–4) or both CPEs and a third degenerate CPE. The degenerate CPE was included because it partially overlaps PBE2 and because residual polyadenylation was detected with a 3′-UTR carrying mutations in the first two CPEs (Fig. 3C, lanes 4–6). These mRNAs were injected into resting oocytes, without progesterone treatment. The mRNA that carried CPE mutations was translated ninefold more efficiently than a wild-type mRNA, suggesting that these CPEs repress translation of GLD-2 mRNA prior to maturation. Repression by the CPEs was dose-dependent: it was observed at low (2 fmol per oocyte) but not high (20 fmol per oocyte) concentrations, consistent with earlier reports that CPE-mediated repression is saturable (Fig. 6B,C; Barkoff et al. 2000; Cao and Richter 2002). An mRNA bearing mutations in all four PBEs also was de-repressed in the resting oocyte, yielding four- to sixfold more luciferase than the wild-type mRNA. This effect was not dose dependent (Fig. 6B,C).
FIGURE 6.
CPE- and PBE-mediated repression of the XlGLD-2A(L) 3′-UTR. (A) Mutational analysis of the last 90 nt of the XlGLD-2A(L) 3′-UTR. Wild-type sequence of the last 90 nt of the XlGLD-2A(L) 3′-UTR is shown on the top line. CPE1, CPE2, and a downstream degenerate CPE (white boxes) were mutated to generate 3xCPE−. Four putative PBEs (gray boxes) in this region were mutated to generate 4xPBE−. The second PBE and the degenerate CPE sequences overlap; the mutations were designed to abolish one or the other element in isolation. Mutated elements are highlighted with dashed boxes. (B) PBEs and CPEs of the XlGLD-2A(L) 3′-UTR repress translation in the oocyte. Luciferase reporter mRNAs carrying the 3′-UTRs shown in A were injected at 2 fmol of RNA per oocyte and incubated overnight at room temperature. Luciferase activity was measured and normalized to β-galactosidase activity, as in Figure 1C. The luciferase/β-galactosidase ratio of the reporter carrying the wild-type 3′-UTR was set to 1 for normalization. (C) Repression by CPEs, but not PBEs, is abolished at high mRNA concentrations. mRNAs described in A were injected at 10-fold higher concentration than in Figure 6B (20 fmol per oocyte), and tested for their effect on luciferase translation. (D) Translational stimulation during oocyte maturation requires CPEs, but not PBEs. The luciferase reporter mRNAs indicated above were injected at 2 fmol per oocyte and incubated at room temperature in the presence or absence of progesterone. Luciferase and β-galactosidase measurements were taken at the time points indicated. The ratios of luciferase/β-galactosidase activities in progesterone treated versus untreated oocytes are plotted on the Y-axis. (E) Translational stimulation during oocyte maturation requires CPEs, but not PBEs. The luciferase reporter mRNAs indicated in A were injected at 2–3 fmol per oocyte and incubated overnight in the presence or absence of progesterone. Luciferase and β-galactosidase activities were measured and normalized as indicated for Figure 1C. The activities of wild-type and 4xPBE− mRNA increased with progesterone treatment, and both mRNAs were translated at similar levels in mature oocytes (+P). The activity of the 3xCPE− mRNA was higher in untreated oocytes, but significantly lower than wild-type and 4xPBE− mRNAs in mature oocytes.
The CPEs in the XlGLD-2A(L) 3′-UTR also are required for translational activation once progesterone is added. mRNAs that lacked CPEs (injected at 2–3 fmol per oocyte) did not increase in activity significantly during maturation (Fig. 6D,E). In contrast, PBE mutant mRNAs did increase in activity in the same period (Fig. 6D,E). The magnitude of stimulation (the ratio of activities with and without progesterone) was less with the PBE mutant than with the wild type; this difference arises because the PBE mutations increased the “basal” level of translation prior to maturation. We conclude that both CPE- and PBE-dependent mechanisms repress XlGLD-2A(L) mRNA prior to maturation. The CPEs are required to stimulate expression during maturation, while the PBEs are not.
Polyadenylation in resting oocytes
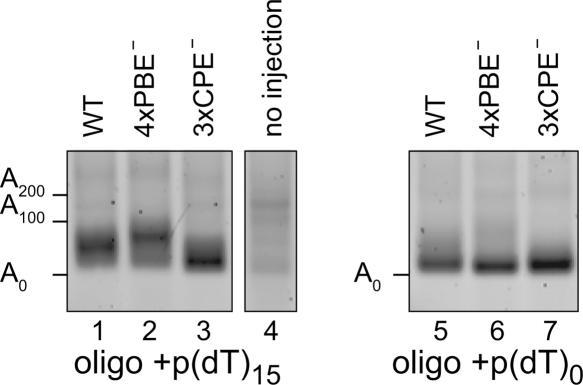
Mutations in either the CPEs or the PBEs increase translation in oocytes prior to maturation (Fig. 6). To assay polyadenylation of these RNAs sensitively, we extracted total RNA from oocytes 4 h after injection and performed PCR-based poly(A) length assays as in Figure 2. An oligonucleotide was first ligated to the total RNA. To specifically detect polyadenylated mRNAs, we used a primer complementary to the ligated oligonucleotide plus 15 3′-terminal thymidine residues. Surprisingly, in resting oocytes, both wild-type and PBE mutant mRNAs were polyadenylated. Roughly 40 adenosines were added to the wild-type mRNA and 50 to the PBE mutant mRNA (Fig. 7, lanes 1,2). The length of poly(A) on endogenous XlGLD-2A(L) mRNA is comparable (see Fig. 2B). This polyadenylation reaction was largely CPE dependent (Fig. 7, lane 3). As a control, we used a primer that was complementary to the ligated primer, but without an oligo(dT) segment. With this primer, the products obtained correspond in majority to nonadenylated mRNAs: wild-type and mutant mRNAs yielded identical distributions of PCR products, as expected. Together, these observations suggest that XlGLD-2A(L) mRNA, prior to maturation, receives a poly(A) tail of limited length. This polyadenylation event is detected on a minority of molecules. We conclude that CPE-mediated polyadenylation occurs in oocytes prior to maturation. This “resting polyadenylation” reaction is counteracted in part by PBEs, presumably acting through the Xenopus PUF protein.
FIGURE 7.
CPE-dependent polyadenylation in resting oocytes. Wild-type, 4xPBE−, and 3xCPE− mutant mRNAs were injected into resting oocytes. mRNA from oocytes injected with the reporters shown in Figure 6A was analyzed using the PCR-based poly(A) length assay (Fig. 2A). Reverse transcription and PCR were performed with a primer selective for poly(A) tails [oligo+p(dT)15; lanes 1–3] revealed longer tails for the 4xPBE− mRNA than for wild-type mRNA (lanes 1,2). The poly(A) tail length of 3xCPE− mRNA is minimal (lane 3). The background signal from oocytes not injected with reporter mRNA is shown (lane 4). The polyadenylation difference is not observed when using an oligo that is not selective for mRNAs with a poly(A) tail [oligo+p(dT)0; lanes 5–7].
GLD-2 expression and polyadenylation in the brain
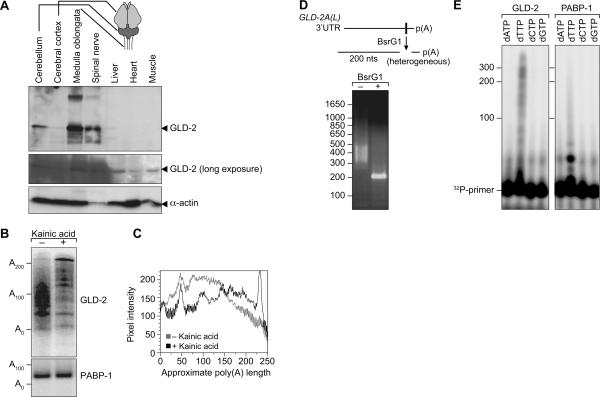
In the nervous system, translation of dendritically localized mRNAs is enhanced upon synaptic stimulation and is thought to require cytoplasmic polyadenylation (Si et al. 2003a). We reasoned that GLD-2 mRNA itself might be regulated by polyadenylation in the brain. The components of the regulatory machinery—GLD-2, CPEB, and PUF proteins—co-localize at the mRNA level in the hippocampus, cerebellum, and olfactory bulb of the mouse brain (Rouhana et al. 2005). To detect the tissue distribution of GLD-2 protein in Xenopus, we extracted total protein from various tissues, including dissected subregions of the brain, and detected GLD-2 by Western blotting with affinity-purified α-GLD-2 antibodies. GLD-2 was most abundant in medulla, spinal nerve, cerebellum, and cerebrum (Fig. 8A). It was present at lower levels in other somatic tissues, including liver, heart, and muscle. α-Actin antibodies served as a loading control.
FIGURE 8.
Poly(A) tail length of neuronal GLD-2(L) mRNA increases upon stimulation. (A) Western blot analysis of GLD-2 protein in tissues of frog brain, spinal nerve, liver, heart, and muscle. GLD-2 protein was enriched in tissues of the nervous system (top), but was also seen expressed in other tissues after longer exposures (middle). Actin was also analyzed as a loading control (bottom). (B) The XlGLD-2A(L) poly(A) tail is increased in response to kainic acid in frog brain. PCR-based poly(A)-length assay (performed as in Fig. 2) on cerebellar RNA from kainic acid treated and untreated frogs. High resolution was obtained by 32P-end-labeling of DNA products and electrophoresis in a 6% polyacrylamide gel. (C) Poly(A)-length distribution for XlGLD-2A(L) was created from PhosphorImager scans for the kainic acid treated and untreated PCR-based poly(A)-length measuring assay. (D) Heterogeneous XlGLD-2A(L) PCR products from poly(A) length analysis due to different lengths at 3′-ends. XlGLD-2A(L) DNA products from the poly(A)-length assay were digested with BsrG1, whose substrate sequence is present downstream from the AAUAAA hexanucleotide on XlGLD-2A(L). The BsrG1 digest creates a homogeneous 200-nt product, as is expected from the XlGLD-2A(L) UTR sequence. (E) Primer extension analysis confirms heterogeneity of XlGLD-2A(L) poly(A) lengths. A 32P-labeled DNA oligonucleotide anti-sense to the oligonucleotide ligated to the end of the mRNA was used for primer extension analysis of XlGLD-2A(L) and PABP-1 PCR products. Primer extensions were performed in the presence of one of dATP, dTTP, dCTP, or dGTP. Primers were extended only in the presence of dTTP, indicative of poly(A) at the end of both XlGLD-2A(L) and PABP-1 substrates. The XlGLD-2A(L) products were extended by 50–300 residues in the presence of dTTP, while PABP-1 products were extended by 20–30 residues. These lengths are consistent with the measurements in B.
To determine whether brain XlGLD-2A(L) mRNA was polyadenylated after neuronal stimulation, we examined its polyadenylation state after treating intact animals with kainic acid. Kainic acid, a seizure-inducing agent, dramatically stimulates neuronal activity (Watkins and Evans 1981). Male frogs were injected with kainic acid, which induced tremors, as expected; injections of saline were used as controls. After 45 min, the cerebellum was dissected. The lengths of poly(A) on GLD-2 mRNA were determined using the PCR-based assays described previously. As expected, the lengths of poly(A) on XlGLD-2A(L) mRNA were heterogeneous both with and without kainic acid, ranging from ∼50 to 200 nt (Fig. 8B,C). However, the average poly(A) tail length for XlGLD-2A(L) mRNA increased in response to kainic acid and included a new discrete species of ∼220 residues (Fig. 8B,C). The poly(A) tails of PABP-1 mRNA, used as a control, were homogeneous (∼30 adenosines) and were unaffected by kainic acid treatment (Fig. 8B).
To ensure that the heterogeneous PCR products were derived from XlGLD-2A(L) and contained poly(A), we exploited a BsrG1 restriction enzyme site located at the 3′-most end of the XlGLD-2A(L) 3′-UTR (Fig. 8D, diagram). Treatment of the GLD-2 PCR products with BsrG1 transformed the heterogeneous population into a homogeneous-sized DNA of the predicted length, showing that the PCR products were, indeed, derived from the XlGLD-2A(L) 3′-UTR (Fig. 8D). To determine whether the heterogeneity was due to heterogeneity in poly(A) tails, we performed primer extension analysis of the GLD-2 PCR products in the presence of dATP, dTTP, dCTP, or dGTP (Fig. 8E). The primer, which was complementary to the ligated oligonucleotide, was extended only in the presence of dTTP, corresponding to poly(A) tails in the mRNA (Fig. 8E). We conclude that the poly(A) tails of XlGLD-2A(L) mRNA in the brain increase in length after kainic acid treatment. These data suggest that GLD-2 mRNA is selectively polyadenylated after neuronal stimulation, as in oocytes; however, it remains possible that the increase in poly(A) length after stimulation reflects new transcription of mRNAs with longer tails, or inhibition of deadenylation.
DISCUSSION
XlGLD-2A(L) mRNA is repressed in the oocyte, then polyadenylated and activated during maturation. This process is conserved, since the human GLD-2(L) 3′-UTR behaves comparably (Fig. 1) and mouse GLD-2 mRNA is polyadenylated during oocyte maturation (Nakanishi et al. 2006). CPEs and AAUAAA are required for polyadenylation of GLD-2 mRNA and for the increase in translational activity during maturation; this implies that GLD-2 polyadenylates its own mRNA, presumably via protein–protein interactions with CPSF and CPEB (Barnard et al. 2004; Rouhana et al. 2005). Indeed, GLD-2 protein is bound to its own mRNA in the resting oocyte (Fig. 4). A similar circuit may exist in the brain, since the poly(A) tails of XlGLD-2A(L) mRNA increase in length in response to the brain in response to neuronal stimuli.
We propose that a positive feedback circuit activates and sustains GLD-2 protein, both during oocyte maturation and in response to synaptic stimulation (Fig. 9). This circuit would enhance translation not only of XlGLD-2A(L), but also of its other, CPE-containing mRNA targets on which GLD-2 acts. In the nervous system, GLD-2 autoactivation may complement the CPEB prion switch (Si et al. 2003b).
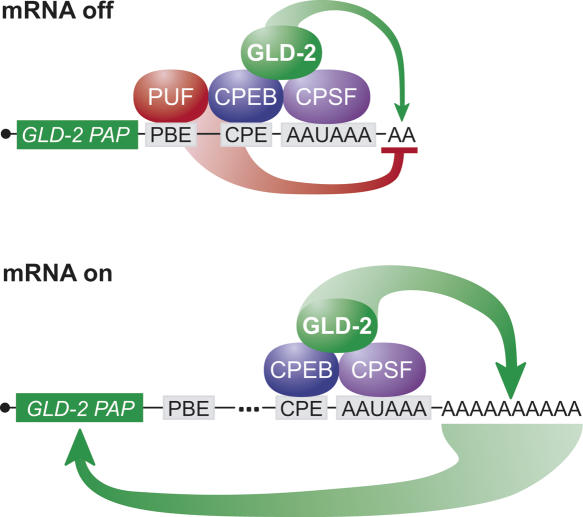
FIGURE 9.
Working model: a positive feedback loop activates GLD-2 mRNA. In the “off” state, translation of GLD-2 mRNA is repressed via PUF and CPEB, bound to specific sites in the long 3′-UTR isoform of GLD-2A mRNA. The bound PUF protein promotes removal of the tail, as suggested by the measurements of poly(A) length on PBE mutant mRNAs. GLD-2 protein also is bound to the mRNA and modestly promotes poly(A) lengthening prior to maturation. The net result is that the poly(A) tail of the mRNA is short. The PUF and CPEB proteins may also repress by deadenylation-independent mechanisms not depicted in the figure. After stimulation, GLD-2 mRNA is de-repressed and becomes a substrate for CPE-mediated polyadenylation by GLD-2 cytoplasmic PAP. The ability of GLD-2 to polyadenylate its own mRNA creates a positive feedback mechanism for the polyadenylation of this (and other) CPE-encoding mRNAs.
The GLD-2(L) mRNA isoform is particularly abundant in mouse and human brains (Rouhana et al. 2005). Other tissues, such as pituitary gland, heart, placenta, ovary, testis, liver, spleen, and kidney; possess abundant GLD-2(S) as well as GLD-2(L) mRNA (Rouhana et al. 2005). The preponderance of the long form in the brain may reflect the requirement for regulation in that tissue.
Alternative 3′-UTRs in certain other mRNAs also mediate developmental, physiological, and pathological controls (for review, see Hughes 2006). For example, an alternative 3′ exon in the Drosophila elav gene generates a longer 3′-UTR that is autoregulated by ELAV protein (Borgeson and Samson 2005). Alternative 3′-end formation is often conserved and tissue-specific (for review, see Edwalds-Gilbert et al. 1997; Tian et al. 2005; Zhang et al. 2005). Longer brain-specific 3′-UTRs have been reported for the Huntington disease gene, beta-adducin, and mCPEB3 (Lin et al. 1993; Theis et al. 2003; Costessi et al. 2006). Recently, a genome-wide study suggested differential repression of transcripts with alternative 3′-UTRs by microRNAs (Legendre et al. 2006). The differential control imposed by alternative 3′-UTRs, as seen with GLD-2 mRNA, may be common.
The addition of poly(A) in resting oocytes, observed here for the first time, may be catalyzed by previously reported complexes containing CPEB, CPSF, and GLD-2 (G complexes) (Rouhana et al. 2005). Indeed, this polyadenylation process is CPE dependent and partially counteracted by PBEs. Only a short tail is added prior to maturation, suggesting that either the addition process is blocked or that a deadenylase compensates and balances the tail length, as suggested during oocyte maturation (Fox et al. 1989; Varnum and Wormington 1990).
The presence of two comparable isoforms of GLD-2 mRNA in frog, mice, and humans suggests a conserved difference in the function of short and long GLD-2 mRNAs. Here we have shown that in Xenopus, the long isoform of XlGLD-2A is polyadenylated during oocyte maturation; the short isoform is not. CPEs, presumably acting through CPEB, inhibit translation of XlGLD-2A(L) (Cao and Richter 2002); PBEs may effect poly(A) tail length through a PUF–deadenylase interaction (Goldstrohm et al. 2006). Both mechanisms would prevent translation of XlGLD-2A(L) mRNA until stimulation occurs. Once polyadenylation did occur, GLD-2 protein would then polyadenylate its own GLD-2(L) mRNA, creating a positive feedback cascade (Fig. 9). The short isoform of mRNA may contribute a low level of latent poly(A) polymerase, whose activation initiates the cascade.
The level of total GLD-2 protein detected by α-GLD-2 antibodies does not change significantly during maturation (data not shown). This might be due to the fact that GLD-2A(L) mRNA is repressed in the resting oocyte, while GLD-2A(S) and GLD-2B mRNAs are readily translatable (Fig. 1C). It is possible that the protein accumulated from GLD-2A(S) and GLD-2B mRNAs during oogenesis obscures the newly synthesized protein produced from activation of GLD-2A(L) mRNA during oocyte maturation, as analyzed by Western blotting. Direct measurements of the relative contributions of GLD-2A and GLD-2B proteins to the total GLD-2 in an oocyte have not been possible. The GLD-2 polyadenylation circuit may be conserved in part because of its role in other tissues. The initial switch in the circuit may involve activation of latent GLD-2 protein, perhaps produced from the GLD-2A(S) or GLD-2B isoforms. It is unclear why the 3′-UTR of XlGLD-2B is largely inactive, despite its high sequence similarity to that of XlGLD-2A(L).
The positive feedback circuit may be reinforced at multiple levels to achieve a sustained and robust switch in polyadenylation activity. Many mRNAs that undergo cytoplasmic polyadenylation during oocyte maturation encode factors involved in protein phosphorylation cascades (e.g., protein kinases Cdk-2, c-mos, Wee1, FGFR1, and Eg2; and cyclins A1, B1, B2, and B4) (Sheets et al. 1994; for review, see Wormington 1994; Stebbins-Boaz et al. 1996; Charlesworth et al. 2004). Polyadenylation of GLD-2 mRNA presumably serves as the upstream activator of the cascade, turning on the translation of these mRNAs, as well as its own. Interestingly, GLD-2 protein activity is regulated in part by the targets it controls, as Eg2, whose mRNA is regulated by polyadenylation (Charlesworth et al. 2004), phosphorylates CPEB and thereby enhances polyadenylation (Mendez et al. 2000a,b). At synapses, CaMKII mRNA undergoes CPEB-protein-mediated polyadenylation (Wu et al. 1998) and CaMKII protein phosphorylates CPEB (Atkins et al. 2004). The robustness of the switch may be critical both in development and in the nervous system. In oocytes, once polyadenylation is activated, the system maintains that state and leads to polyadenylation of batteries of mRNAs. GLD-2, CPEB, and PUF proteins act in concert to establish that circuit.
MATERIALS AND METHODS
DNA constructs
pLG-CB1
pLG-MS2 (Gray et al. 2000) was digested with SpeI and BglII, in order to remove the MS2 sequence, blunt ended with Klenow DNA polymerase (Promega), and re-ligated. This construct is used for transcription of the luciferase-cyclinB1 3′-UTR.
pLuci-XlGLD-2A(L)
The XlGLD-2A(L) 3′-UTR was amplified from IMAGE:6638921 using primers CAGCACTAGTACTAGTACTGGAATCTGCAGAGAAATAC and ACAGGATCCGGATCCGTACATACATTTTATTCCATTCAACATCAT, digested with SpeI and BamHI, and ligated into SpeI- and BamHI-digested pLG-MS2. This construct is used for transcription of the luciferase-XlGLD-2A(L) 3′-UTR.
pLuci-XlGLD-2A(S)
The XlGLD-2A(S) 3′-UTR was amplified from IMAGE:6638921 using primers CAGCACTAGTACTAGTACTGGAATCTGCAGAGAAATAC and ACAGGATCCGGATCCGGTTCAATATTTGTATTTAAAC, digested with SpeI and BamHI, and ligated into SpeI- and BamHI-digested pLG-MS2. This construct is used for transcription of the luciferase-XlGLD-2A(S) 3′-UTR.
pLuci-XlGLD-2B
The XlGLD-2B 3′-UTR was amplified from IMAGE:6643643 using primers CAGCACTAGTACTAGTACTGGAATCTGCAGAGAAATAC and ACAGGATCCGGATCCGGTACATACATTTTATTCCATTCAACACC, digested with SpeI and BamHI, and ligated into SpeI- and BamHI-digested pLG-MS2. This construct is used for transcription of the luciferase-XlGLD-2B 3′-UTR.
pLuci-hGLD-2(L)
The hGLD-2(L) 3′-UTR was amplified from IMAGE:4824607 using primers CAGCACTAGTACTCTGGCCTCTATTTCTTAATAAATTCTTCC and CGGATCCGGATCCGTCCATATATAAATTTTATTGCATTTGACATC, digested with SpeI and BamHI, and ligated into SpeI- and BamHI-digested pLG-MS2. This construct is used for transcription of the luciferase-hGLD-2(L) 3′-UTR.
pLR9 series
The wild-type 3′-most 90-nt sequence from XlGLD-2A(L) (as well as mutations describe in Figs. 3A, 6A), were oligo-cloned into SpeI- and BamHI-digested pLG-MS2. This series of constructs is used for transcription of the luciferase last 90 nt of the XlGLD-2A(L) 3′-UTR (and mutations thereof).
pT7-LR9 series
The pT7-LR9 series is a series of clones based on the pLR9 series, but with removal of most of the luciferase ORF, created by blunt-end ligation and cloning. This series of constructs is used for transcription of a mini-ORF with the last 90 nt of the XlGLD-2A(L) 3′-UTR (and mutations thereof; see Fig. 3A).
pJK350 (β-galactosidase)
This construct was previously described (Gray et al. 2000).
In vitro transcription
The pLG-CB1, pLuci-XlGLD-2A(L), pLuci-XlGLD-2A(S), pLuci-XlGLD-2B, pLuci-hGLD-2(L), pLR9 series, and pT7-LR9 series constructs were cleaved with BamHI and transcribed with T7 RNA polymerase. Transcriptions were performed in the presence of 6 mM m7GpppG. In some cases, [α-32P]UTP was incorporated during in vitro transcription as described (Dickson et al. 2001).
mRNA injection and manipulation of Xenopus oocytes
Oocyte removal, injection, and the induction of meiotic maturation were performed as previously described (Ballantyne et al. 1997). mRNAs were injected at either 2–3 fmol/50 nL, or 20–30 fmol/50 nL for the luciferase reporter mRNAs, along with 12 fmol/50 nL for the β-gal reporter mRNA.
Extraction and analysis of mRNAs from oocytes
Total RNA was extracted from frog tissues using 10 oocytes, or eggs, per 1 mL of TRI reagent (Sigma) following the manufacturer's protocol. After ethanol precipitation, the RNA was solubilized in 10 μL of DEPC-treated water, and either used for the PCR-based poly(A)-length assay, or loaded to a 6% polyacrylamide gel (short 32P-labeled RNAs) and analyzed by autoradiography.
Luciferase and β-galactosidase measuring assay
These assays were performed as described by Gray et al. (2000).
PCR-based poly(A)-length measuring assay
This technique was performed as described by Charlesworth et al. (2004). Briefly, 4 μg of oocyte (or 2 μg of brain) total RNA was ligated to 0.4 μg of a 3′-amino-modified DNA anchor primer (Operon). The ligated products were then reverse-transcribed using a primer anti-sense to the anchor primer plus four thymidine residues on its 3′-end, using Superscript II (Invitrogen). Two microliters of this reaction were used as a template for gene-specific PCR, using the primers listed in Table 1, and the primer anti-sense to the anchor primer, used in the reverse-transcriptase reaction (unless specified differently). The annealing temperature was 56°C for all genes except for CPSF-73, for which the annealing temperature was 48°C. DNA products from the brain XlGLD-2A(L) PCR-based poly(A)-length measuring assay, were end labeled with T4 Polynucleotide Kinase (Promega) and [γ-32P]ATP.
TABLE 1.
Oligonucleotides used in PCR-based poly(A)-length assay
RNA coimmunoprecipitation
Coimmunoprecipitation and extraction of RNA with α-GLD-2 antibodies were performed as described previously (Rouhana et al. 2005). cDNA was made from total and coimmunoprecipitated oocyte RNA by oligo-coupled reverse transcription with Superscript II (Invitrogen). One-twentieth of the reverse-transcription reaction was used as a template for gene-specific PCR. Oligos GCAGGTTTGCGCTTGAGAAAATGTCAC and GGAGGAAGCAGCTGCTATTTGGGAAGGCG were used as primers for cyclin B1; GCTGCCTATTTAATACTTATCCATGAG and CAAAGACACCAGATTGTGCCTTAATTG for XlGLD-2A(L); and CATTGACCGCTCATCAGTTGTC and CAGAGCGACTTCAGAAATCCAACC for Xfin.
Electrophoretic mobility shift assays
GST-fused Xenopus Pumilio (amino acids 824–1185) was purified as described for GST-FBF-2 (Bernstein et al. 2005) and combined with 100 fMol of 32P-end-labeled RNA oligoribonucleotides (Integrated DNA Technologies). The shift conditions and binding constants were calculated as described by Hook et al. (2005).
Kainic acid treatment and brain RNA extraction
Kainic acid was administered to males (15 mg/kg) via subcutaneous injection in the lymph sac. Sterile saline (pH 7.4) was administered as a negative control. Frogs were monitored for 45 min, made lethargic through hypothermia, and decapitated. Brain tissue was then dissected for cerebellum RNA extraction and analysis by PCR-based poly(A)-length assay.
ACKNOWLEDGMENTS
We thank Craig Schiltz and Charles Landry for help with the experiment in Figure 9. We appreciate the suggestions of Judith Kimble and Jerry Yin and their comments on the manuscript. We also thank members of the Kimble and Wickens laboratories for their advice and assistance. We are grateful to The Media Center at the Biochemistry Department, University of Wisconsin–Madison, particularly Laura Vanderploeg and her plow, for assistance with figures. This work was supported by NIH grants (GM50942 and GM31892) to M.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.333507.
REFERENCES
- Alarcon, J.M., Hodgman, R., Theis, M., Huang, Y.S., Kandel, E.R., Richter, J.D. Selective modulation of some forms of Schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn. Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins, C.M., Nozaki, N., Shigeri, Y., Soderling, T.R. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J. Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne, S., Daniel, D.L., Jr., Wickens, M. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell. 1997;8:1633–1648. doi: 10.1091/mbc.8.8.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoff, A.F., Dickson, K.S., Gray, N.K., Wickens, M. Translational control of cyclin B1 mRNA during meiotic maturation: Coordinated repression and cytoplasmic polyadenylation. Dev. Biol. 2000;220:97–109. doi: 10.1006/dbio.2000.9613. [DOI] [PubMed] [Google Scholar]
- Barnard, D.C., Ryan, K., Manley, J.L., Richter, J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Berger-Sweeney, J., Zearfoss, N.R., Richter, J.D. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn. Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- Bernstein, D., Hook, B., Hajarnavis, A., Opperman, L., Wickens, M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilger, A., Fox, C.A., Wahle, E., Wickens, M. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes & Dev. 1994;8:1106–1116. doi: 10.1101/gad.8.9.1106. [DOI] [PubMed] [Google Scholar]
- Borgeson, C.D., Samson, M.L. Shared RNA-binding sites for interacting members of the Drosophila ELAV family of neuronal proteins. Nucleic Acids Res. 2005;33:6372–6383. doi: 10.1093/nar/gki942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Q., Richter, J.D. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, A., Cox, L.L., MacNicol, A.M. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costessi, L., Devescovi, G., Baralle, F.E., Muro, A.F. Brain-specific promoter and polyadenylation sites of the β-adducin pre-mRNA generate an unusually long 3′-UTR. Nucleic Acids Res. 2006;34:243–253. doi: 10.1093/nar/gkj425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, K.S., Thompson, S.R., Gray, N.K., Wickens, M. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem. 2001;276:41810–41816. doi: 10.1074/jbc.M103030200. [DOI] [PubMed] [Google Scholar]
- Dubnau, J., Chiang, A.S., Grady, L., Barditch, J., Gossweiler, S., McNeil, J., Smith, P., Buldoc, F., Scott, R., Certa, U., et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert, G., Veraldi, K.L., Milcarek, C. Alternative poly(A) site selection in complex transcription units: Means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, C.A., Wickens, M. Poly(A) removal during oocyte maturation: A default reaction selectively prevented by specific sequences in the 3′-UTR of certain maternal mRNAs. Genes & Dev. 1990;4:2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- Fox, C.A., Sheets, M.D., Wickens, M.P. Poly(A) addition during maturation of frog oocytes: Distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes & Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- Goldstrohm, A.C., Hook, B.A., Seay, D.J., Wickens, M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Gray, N.K., Coller, J.M., Dickson, K.S., Wickens, M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake, L.E., Richter, J.D. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Hentze, M.W. Translational regulation: Versatile mechanisms for metabolic and developmental control. Curr. Opin. Cell Biol. 1995;7:393–398. doi: 10.1016/0955-0674(95)80095-6. [DOI] [PubMed] [Google Scholar]
- Hook, B., Bernstein, D., Zhang, B., Wickens, M. RNA–protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA. 2005;11:227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.S., Jung, M.Y., Sarkissian, M., Richter, J.D. N-Methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and α CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.A. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., Goodwin, E.B. The power of the 3′-UTR: Translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Kwak, J.E., Wang, L., Ballantyne, S., Kimble, J., Wickens, M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Legendre, M., Ritchie, W., Lopez, F., Gautheret, D. Differential repression of alternative transcripts: A screen for miRNA targets. PLoS Comput. Biol. 2006;2:e43. doi: 10.1371/journal.pcbi.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B., Rommens, J.M., Graham, R.K., Kalchman, M., MacDonald, H., Nasir, J., Delaney, A., Goldberg, Y.P., Hayden, M.R. Differential 3′ polyadenylation of the Huntington disease gene results in two mRNA species with variable tissue expression. Hum. Mol. Genet. 1993;2:1541–1545. doi: 10.1093/hmg/2.10.1541. [DOI] [PubMed] [Google Scholar]
- McGrew, L.L., Richter, J.D. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: Characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990;9:3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, R., Richter, J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mendez, R., Hake, L.E., Andresson, T., Littlepage, L.E., Ruderman, J.V., Richter, J.D. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000a;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Mendez, R., Murthy, K.G., Ryan, K., Manley, J.L., Richter, J.D. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell. 2000b;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Nakahata, S., Katsu, Y., Mita, K., Inoue, K., Nagahama, Y., Yamashita, M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 2001;276:20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- Nakahata, S., Kotani, T., Mita, K., Kawasaki, T., Katsu, Y., Nagahama, Y., Yamashita, M. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 2003;120:865–880. doi: 10.1016/s0925-4773(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Nakanishi, T., Kubota, H., Ishibashi, N., Kumagai, S., Watanabe, H., Yamashita, M., Kashiwabara, S., Miyado, K., Baba, T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Read, R.L., Martinho, R.G., Wang, S.W., Carr, A.M., Norbury, C.J. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, J.D. The influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 785–805. [Google Scholar]
- Rouhana, L., Wang, L., Buter, N., Kwak, J.E., Schiltz, C.A., Gonzalez, T., Kelley, A.E., Landry, C.F., Wickens, M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba, A., Perry-O'Keefe, H., Melton, D.A. Xfin: An embryonic gene encoding a multifingered protein in Xenopus . EMBO J. 1987;6:3065–3070. doi: 10.1002/j.1460-2075.1987.tb02613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W.H., Thelander, L., Yates, J.R., Russell, P. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- Sheets, M.D., Fox, C.A., Hunt, T., Vande Woude, G., Wickens, M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes & Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- Si, K., Giustetto, M., Etkin, A., Hsu, R., Janisiewicz, A.M., Miniaci, M.C., Kim, J.H., Zhu, H., Kandel, E.R. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003a;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Si, K., Lindquist, S., Kandel, E.R. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003b;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N. mRNA translation: Influence of the 5′ and 3′ untranslated regions. Curr. Opin. Genet. Dev. 1994;4:310–315. doi: 10.1016/s0959-437x(05)80059-0. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz, B., Hake, L.E., Richter, J.D. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus . EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- Stebbins-Boaz, B., Cao, Q., de Moor, C.H., Mendez, R., Richter, J.D. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Theis, M., Si, K., Kandel, E.R. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc. Natl. Acad. Sci. 2003;100:9602–9607. doi: 10.1073/pnas.1133424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, B., Hu, J., Zhang, H., Lutz, C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum, S.M., Wormington, W.M. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: A default mechanism for translational control. Genes & Dev. 1990;4:2278–2286. doi: 10.1101/gad.4.12b.2278. [DOI] [PubMed] [Google Scholar]
- Vessey, J.P., Vaccani, A., Xie, Y., Dahm, R., Karra, D., Kiebler, M.A., Macchi, P. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J. Neurosci. 2006;26:6496–6508. doi: 10.1523/JNEUROSCI.0649-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Eckmann, C.R., Kadyk, L.C., Wickens, M., Kimble, J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans . Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Watkins, J.C., Evans, R.H. Excitatory amino acid transmitters. Annu. Rev. Pharmacol. Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- Wickens, M., Goodwin, E.B., Kimble, J., Strickland, S., Hentze, M. Translational control in developmental decisions. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 295–370. [Google Scholar]
- Wickens, M., Bernstein, D.S., Kimble, J., Parker, R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wormington, M. Unmasking the role of the 3′-UTR in the cytoplasmic polyadenylation and translational regulation of maternal mRNAs. Bioessays. 1994;16:533–535. doi: 10.1002/bies.950160804. [DOI] [PubMed] [Google Scholar]
- Wu, L., Wells, D., Tay, J., Mendis, D., Abbott, M.A., Barnitt, A., Quinlan, E., Heynen, A., Fallon, J.R., Richter, J.D. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Wyers, F., Rougemaille, M., Badis, G., Rousselle, J.C., Dufour, M.E., Boulay, J., Regnault, B., Devaux, F., Namane, A., Seraphin, B., et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Ye, B., Petritsch, C., Clark, I.E., Gavis, E.R., Jan, L.Y., Jan, Y.N. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Lee, J.Y., Tian, B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]