Abstract
Aminoacyl-tRNA synthetases (aaRSs) are responsible for creating the pool of correctly charged aminoacyl-tRNAs that are necessary for the translation of genetic information (mRNA) by the ribosome. Each aaRS belongs to either one of only two classes with two different mechanisms of aminoacylation, making use of either the 2′OH (Class I) or the 3′OH (Class II) of the terminal A76 of the tRNA and approaching the tRNA either from the minor groove (2′OH) or the major groove (3′OH). Here, an asymmetric pattern typical of differentiation is uncovered in the partition of the codon repertoire, as defined by the mechanism of aminoacylation of each corresponding tRNA. This pattern can be reproduced in a unique cascade of successive binary decisions that progressively reduces codon ambiguity. The deduced order of differentiation is manifestly driven by the reduction of translation errors. A simple rule can be defined, decoding each codon sequence in its binary class, thereby providing both the code and the key to decode it. Assuming that the partition into two mechanisms of tRNA aminoacylation is a relic that dates back to the invention of the genetic code in the RNA World, a model for the assignment of amino acids in the codon table can be derived. The model implies that the stop codon was always there, as the codon whose tRNA cannot be charged with any amino acid, and makes the prediction of an ultimate differentiation step, which is found to correspond to the codon assignment of the 22nd amino acid pyrrolysine in archaebacteria.
Keywords: genetic code, evolution, aminoacylation mechanism, codon assignment, translation errors
INTRODUCTION
As the genetic code uses words (codons) of three letters, with four possible bases at each position of the codon, a table of 64 entries is needed to specify the correspondence of each codon with a given amino acid. One of the great achievements of experimental molecular biology in the 1960s was to decipher this code, which turns out to be almost universal. However, explaining how the genetic code evolved and why the codon assignment took its present form is one of the few theoretically challenging problems remaining in molecular biology (Crick 1968; Woese 2001). Since some of the key molecules needed to translate the genes (e.g., aminoacyl-tRNA synthetases, release factors, and ribosomal proteins) are proteins, i.e., the products of the translation process themselves, one is faced with a chicken-and-egg problem.
Obviously, the primordial translation apparatus need not be very accurate, and it seems reasonable to postulate an error-prone early genetic code with ambiguous codons that became gradually more accurate before finally reaching its final and current state (Woese 1965a). Moreover, because RNA can display all the required catalytic activities of a primary translation apparatus (Joyce 2002), it may be safely assumed that this apparatus was originally made entirely of RNA. However, such a primitive RNA translation apparatus was probably intrinsically limited in precision and, to increase in accuracy, had to evolve into a mixed protein–RNA apparatus. How exactly the transition from an RNA World to a protein–RNA World was made, resulting ultimately in the genetic code in its present-day form, is not yet understood. Furthermore, how the codon assignment process occurred in the first place remains mysterious; in particular, it is difficult to imagine that it was explored by evolution in a purely random way. Understanding how this process was explored in the RNA World might be useful for the design of RNA-based systems capable of solving difficult assignment tasks.
In this work, following others (Hornos and Hornos 1993; Bashford et al. 1998; Balakrishnan 2002), the problem is revisited by searching for symmetries in the table of codons that might indicate whether or not this evolution could have been driven by an underlying mechanism (rule), thus converging much more rapidly than by random trial and error. As the clues to such a hypothetical mechanism are very scarce, it is common practice to look for patterns in either one of the key constituents of the codon identity, namely, tRNAs (Eigen et al. 1989; Nicholas and McClain 1995) or aaRSs (Eriani et al. 1990; Ribas de Pouplana and Schimmel 2001a), and their respective evolution deduced from phylogenetic methods (Nagel and Doolittle 1995; Woese et al. 2000). In the case of aaRSs, the phylogeny is complicated by a number of horizontal transfers (Wolf et al. 1999).
All known aaRSs are built in a highly modular fashion (Delarue and Moras 1993) from several domains, but the aminoacylation domain always falls into one of either of two classes (Eriani et al. 1990; Cusack et al. 1991). The two classes are characterized by mutually exclusive sequence motifs, two very different molecular architectures, and a primary site of the aminoacylation reaction that can be either the 2′OH (Class I) or 3′OH (Class II) of the 3′ terminal base of the tRNA (Fig. 1). Recognition of tRNA occurs either from the minor groove (Class I) or the major groove (Class II) of the acceptor stem of the tRNA. The two classes of aaRSs therefore represent two “orthogonal” chemical solutions to the aminoacylation of the 3′ end of the tRNA in a sterically complementary way (Delarue and Moras 1992; Arnez and Moras 1997; Ribas de Pouplana and Schimmel 2001b). Since the original definition of the two classes of aaRSs, a number of structural studies have confirmed and further documented these concepts. Very recently, differences in the kinetic mechanisms of the two different classes were revealed (Zhang et al. 2006). Here I focus on these two different aminoacylation mechanisms, assuming that they must have appeared very early in its evolution, to find patterns in the genetic code.
FIGURE 1.
Main differences in the two aminoacylation mechanisms as seen in the present-day two classes of aaRSs (Eriani et al. 1990). The primary sites of aminoacylation for Class I aaRS (2′OH, green, left) and Class II aaRS (3′OH, red, right) are highlighted, corresponding to two different types of tRNA recognition through the minor groove (Class I) or the major groove (Class II) of the tRNA. The members of each class of aaRSs that stand out as exceptions in their own class are printed with a different color (PheRS and TyrRS) as discussed in the text.
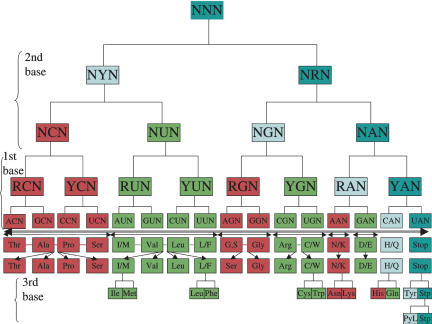
ASYMMETRIC PATTERN IN A COLORED CODON TABLE
In Figure 2 (left half), each codon in the classical genetic code is colored according to the aminoacylation mechanism of its tRNA with 2′OH codons colored in green and 3′OH codons colored in red (Fraser and Rich 1975). This is equivalent to coloring according to the Class I/Class II partition, except for PheRS, which is an outlier in its own class (Fig. 1). Indeed, the tRNA recognition mode of PheRS is clearly Class II (Goldgur et al. 1996), while it is aminoacylated on the 2′OH, as are all other Class I aaRSs. Also, extra care must be taken for those tRNAs whose aminoacylation has been shown to occur on both 2′OH and 3′OH. This concerns tRNATyr and tRNACys (Sprinzl and Cramer 1975; Hecht and Chinault 1976). In this respect, it is interesting to note that Tyr and Phe and Cys are considered to be late amino acids (Brooks and Fresco 2002). For tRNATyr, modeling (Bedouelle 1990) as well as crystallographic (Yaremchuk et al. 2002) structural studies of the TyrRS–tRNATyr complex clearly show that the former has a Class II tRNA recognition mode, even though TyrRS displays all the sequence motifs and the typical Rossmann fold of a normal Class I aaRS. Therefore Phe and Tyr codons are colored both red and green (hashed mode) in Figure 2. Actually, a recent compilation of tRNA sequences has identified the Phe–Tyr pair as the one paralogous pair displaying the highest sequence conservation, in an attempt to identify “alloacceptors” (Xue et al. 2003). This could be the mark of a recent assignment, or even a codon swapping phenomenon (Szathmary 1991). On the other hand, tRNACys, whose primary site of aminoacylation was also measured to be both 2′OH and 3′OH (Sprinzl and Cramer 1975; Hecht and Chinault 1976), has been shown to be recognized by CysRS (a Class I aaRS) in a purely Class I mode by crystallographic studies of the CysRS–tRNACys complex (Hauenstein et al. 2004). Also, more recent detailed kinetic studies showed that kcat (2′OH) ≫kcat (3′OH) by a factor of about 20 for CysRS (Shitivelband and Hou 2005). Therefore, Cys was assigned a green color (Fig. 2).
FIGURE 2.
The binary partition of the genetic code deduced from the aminoacylation mechanism of each corresponding tRNA. (Left half). The 2′OH mechanism (Class I) is in green, and the 3′OH (Class II) mechanism is in red. Ambiguous cases are both red and green, in the hash mode. The stop codon box is in white. Two variants of the mitochondrial codes have been adopted (printed in blue), so as to reduce the number of codons to 32 (only the pyrimidine/purine character of the third base matters), leaving only one stop codon. (Right half). Same as left half but with a different order for the bases of the three positions of the codon: U and C are permuted for the second base, and A, G, C, U order is adopted for the first base, instead of the usual U, C, A, G order. In addition, the rare AGR codons have been assigned to Gly/Ser as in most mitochondrial variants of the code (printed in white).
To simplify the discussion, the more symmetric code found in mitochondrial variants is adopted (Knight et al. 1999), with UGA as Trp instead of Ter and AUA as Met instead of Ile. Met and Ile are in the same class, and the case of the UGA stop codon will be briefly examined in the last part of the Discussion. This reduces the number of codons to 32, where only the purine/pyrimidine (R/Y) character of the third wobble base matters. In addition, a special case is made here for the AGR codons, which are the rarest ones in Escherichia coli, a possible mark for a codon reassignment process in progress (Sengupta and Higgs 2005). Six out of the 16 known mitochondrial variants of the code involve the AGR codons, where they are assigned to Ser or Gly instead of the canonical Arg (Knight et al. 1999). Therefore, it was decided to examine the consequences of assigning the AGR codons to Ser/Gly, i.e., a 3′OH mechanism (AGR-encoded amino acids are printed in white in Fig. 2 to highlight this fact).
Looking at the U, C, and G columns in Figure 2 (left half), a specific pattern begins to emerge with the alternance of green and red colors in finer and finer subdivisions. Any order of the bases can be chosen for the three positions of the codon, as already mentioned by several authors (e.g., Volkenstein 1966) to underline specific features of the genetic code (Trinquier and Sanejouand 1998). Furthermore, if a different permutation of the four bases for each position of the codon is allowed, then the pattern spotted in Figure 2 (left half) can be made almost perfect (see Fig. 2, right half): the four columns successively display 8 reds, 8 greens, 4 reds, 4 greens, 2 reds, 2 greens (but the Asp, i.e., GAY, codons are clearly an exception here), 1 red, 1 green, 1 red/green, and the stop codon, suggesting a binary mechanism for dividing (assigning) the codon table with a progressively finer and finer grid size. The fact that LysRS is a Class I aaRS (Ibba et al. 1997) in some archeabacteria can be taken as a sign of some flexibility in class assignment, which renders the Asp exception plausible. In the rest of this article, the implications of the pattern uncovered by coloring the genetic code according to Figure 2 (right half) are fully examined.
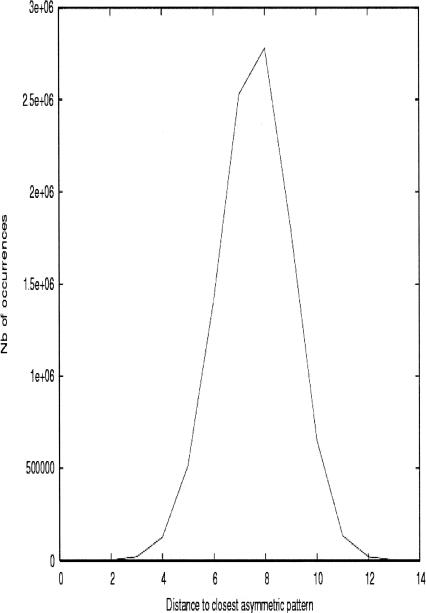
What is the significance of this asymmetric pattern and what is the probability of observing it by chance? One can calculate the number of such patterns to be 24 × 24 × 2 × 6, because they are generated (not counting the factor of 2 coming from switching all red positions into green and vice versa) by all the different permutations of the four bases at the first and second positions (24 × 24) and the Y/R choice (2) at the third position; the remaining factor (6) accounts for the different ways of choosing the order of differentiation in the three different positions of the codon (see below). Numerical simulations have been undertaken to quantify the probability of observing such asymmetric patterns, allowing for exceptions: A large number (107) of 32 + 32 binary partitions of the 64 codon table with Y and R degeneracy at the third position were randomly generated and, for each of them, their distance to the closest of the 6912 × 2 possible asymmetric tables was recorded (Fig. 3). It turns out that asymmetric codon tables with 1, 2, or 3 exceptions (Asp and maybe Phe and Ser/Gly in Fig. 2) occur with a probablility of 2 × 10−5, 2 × 10−4, and 2 × 10−3, respectively; these figures should be divided by 32 if the stop codon is known, because this fixes the last base of each permutation (see Fig. 4).
FIGURE 3.
Histogram of distances of random binary partitions of the codon table with the closest asymmetric distribution. Some 107 different binary partitions of the table of codons were generated randomly, with a probability of 1/2 for each color. For each one of them, the distance to the closest perfectly asymmetric table was recorded, scanning all 24 × 24 × 2 × 6 × 2 = 13,824 possible ones, and a histogram was built.
FIGURE 4.
A series of binary choices leading to the colored table of codons as in Figure 2 (left half). At each differentiation step, one daughter is a copy of the original phenotype (dark blue), while the other daughter (light blue) will differentiate at the next generation into two codons of different colors (green and red). From then on, red or green codons can never switch back. Only a few instances of the fifth differentiation are shown because of space limitations. There is a unique order of differentiation that reproduces the pattern of Figure 2. The two exceptions in comparing Figure 2 (left half) and this figure are Asp and possibly Phe codons, although the latter one can be considered both green (2′OH) and red (Class II sequence motifs and tRNA recognition mode). The underlying mechanism implies that the dark blue codon is the stop codon (i.e., the one that can be charged neither on the 2′OH nor on the 3′OH sites) while the light blue codon is an ambiguous one that can be charged on both 2′OH and 3′OH of A76 of tRNA.
A SERIES OF BINARY DIFFERENTIATION STEPS PROGRESSIVELY REDUCING CODON AMBIGUITY
There is a unique way to obtain the colored codon table of Figure 2 (left half), which is actually the result of a differentiation-like process that reduces progressively the ambiguity of all the codons (Fig. 4). At each differentiation step, the same following rule is applied: upon each division, one daughter (right) is the exact copy of the mother and maintains the ability of self-renewal, but the other one (left) gets “activated.” The activated one gains the ability to differentiate at the next generation and gives two daughters that are aminoacylated in a different way, using either the Class I (2′OH, minor groove approach) or Class II (3′OH, major groove approach) aminoacylation mechanism of A76 of the tRNA. From then on, the differentiated codon can never switch its color back. A cascade of five consecutive binary decisions following the same simple rule can account almost perfectly for the partition that is observed in the codon table (cf. Fig. 2, right half, and Fig. 4). The order of events leading to Figure 2 (right half) is (1) (Y/R) symmetry breaking at the second base; then (2) (C/U) and (G/A) specification on the same second base; then (3) (R/Y) symmetry breaking at the first base; followed by (4) (A/G) and (C/U) differentiation on the same base; and then finally (5) (Y/R) symmetry breaking on the third “wobble” base. The mechanism described in Figure 4 is clearly reminiscent of a “codon ambiguity reduction” scheme described earlier (Fitch and Upper 1987), but differs in many respects from it. What is clear, however, is that the driving force at work in the progressive reduction of codon ambiguity is the reduction of errors in translation (see below).
What are the unique features of this mechanism? One can clearly see that a modest but still significant number of binary partitions of the codon table can be systematically explored with this mechanism by choosing the permutation of the four bases on the first and second positions and the Y/R order on the third position, generating 6912 different tables. Among all the possible trees that follow this mechanism, the observed one is clearly special in that the deduced order of differentiation events is strictly correlated with the diminution of translation errors, as independently predicted by an argument developed by Woese (1965b), which states that the main problem in the evolution of the genetic code was to reduce reading errors. Indeed, experimental data on the translation of several homopolymers under suboptimal conditions, i.e., in the presence of Mn++ instead of Mg++, show that errors are made more frequently on the second, then the first, then the third base, in increasing order. Also, at each position, transitions between pyrimidines (C, U) or purines (A, G) occur more frequently than transversions (for review, see Woese 1965a). The physical basis for this may be found in the present-day decoding mechanism in the ribosome, which is based on the so-called A-minor RNA motifs (for review, see Lescoute and Westhof 2006); this motif probes the correctness of the codon–anticodon minihelix and makes more interactions with the second base than with the first or the third one.
Another feature concerns the fact that the colored codon table is already arranged in such a way that the rule “near codons encode near amino acids,” where “near” means “of the same color,” is indeed satisfied, as in the “adaptation idea” (Freeland and Hurst 1998). Indeed, the cumulated score of color changes upon mutation of each codon at either one of the three positions (27 mutations for each codon) of the colored codon table of Figure 2 is among the top 0.1% of 109 randomly generated different binary partitions of the codon table obtained by swapping the colors of randomly chosen pairs of codons (data not shown).
On a simple information-based level, the specificity of the mechanism underlying Figure 4 can be described as follows: a (0,0) mother codon gives two daughters: another (0,0) codon (copy of mother, dark blue codon) and a (−1,+1) codon (the activated one, light blue codon), which then divides into (+1,0) (green, Class I) and (−1,0) (red, Class II), in the asymmetric step. In the following, an interpretation of the nongreen and nonred codons is provided.
PREDICTIONS OF THE DIFFERENTIATION MODEL
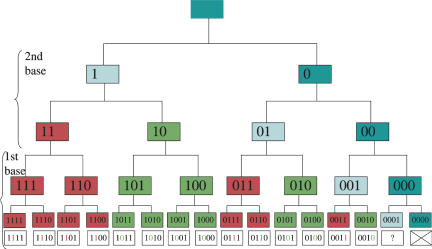
It is actually possible to find a rule allowing each codon to be “decoded” in its color as in Figure 2 (left half). Each of the 32 codons is represented by a five-bit long string (25 = 32). The order of the bits is chosen by going down the tree of Figure 4: The first bit is used to distinguish a purine from a pyrimidine at the second position of the codon, then the second bit is used to distinguish C from U and A from G at the same position, and so on (Fig. 5). There are other ways to represent the four bases with a string of bits, especially to account for hydrogen-bonding capabilities (see MacDonaill 2003, and his four-bit string representation of the bases), which would make it more straightforward to interpret in molecular terms. Here, a simple model with just two bits per position is presented, for illustration purposes. It turns out that there is a simple way to extract the color of each codon from this simple binary representation: the color bit is the one after the first nonzero bit, reading from left to right. This, in turn, not only provides the key to decode the codon table but also illuminates the role of the nonred and nongreen codons in Figure 3:
FIGURE 5.
Decoding each codon in a single (color) bit. Upon each differentiation step, the daughter on the left receives a “1,” the daughter on the right receives a “0.” The order of the bits is the one encountered when descending down the tree (two bits for second base, then two bits for first base). The color bit is the bit immediately after the first nonzero bit, reading from left to right: 1 is for red and 0 is for green. Only four divisions are shown here, but the same rule is valid if there are more divisions. The special roles of the stop codon and the ambiguous codon can also be deduced from the same rule.
(1) The rightmost codon (dark blue) cannot be decoded, as there is no nonzero bit in it, resulting in an always-uncharged tRNA. This is a termination codon because the translation machinery naturally has to stop upon meeting such an uncharged tRNA, leading eventually to the release of the newly synthesized polypeptide (today, this job is performed by polypeptides called release factors, RF1 and RF2). It is remarkable that the stop codon receives such a special interpretation, as nothing of the sort was imposed on it. The scheme in Figure 4 implies that the stop codon was present since the first step of the differentiation process. Retrospectively, this is quite reassuring because there is no guarantee that multiple translations of the same gene will give a reproducible form and function if its length is not fixed and somehow coded. This is, however, in contrast with a popular hypothesis that states that there was no stop codon at the beginning because any mutation to (or from) a stop codon would have been too deleterious (Sonneborn 1965). Nevertheless, as the number of stop codons quickly decreased (see below), the risk to mutate into it also decreased. Conversely, the possible loss of the stop codon by mutation may have been taken care of early on by simply placing several stop codons in a row at the end of each message.
(2) The penultimate (light blue) codon is ambiguous: the “reading head” (first nonzero bit) can be positioned but there is nothing to read, as it is located at the far end of the bit string. This results in the random addition of either a Class I (2′OH) or Class II (3′OH) amino acid. The relative importance of this ambiguous codon decreases as the number of differentiation steps increases. Retrospectively, the presence of an ambiguous codon (a reservoir of ambiguity) in the tree of Figure 2 (right half) may not be so surprising, as it has been already suggested that it can confer an evolutionary advantage to bacteria (Pezo et al. 2004). The selective advantage of codon ambiguity has also been discussed in yeast (Santos et al. 1999).
The model makes a definite prediction that there should be an “ultimate” stage of differentiation, namely, the one of the stop codon UAR (Fig. 3). Indeed the so-called 22nd amino acid, pyrrolysine, which is incorporated into a methyltransferase in some archaebacteria (Srinivasan et al. 2002), has its own aaRS and uses one of the stop codons (UAG), as predicted by the model. Although PylRS was later identified as a Class II aaRS (Polycarpo et al. 2004), it was also shown that Pyl-tRNA can be charged with lysine by a combination of both the Class I and Class II LysRS, namely, LysRS-I and LysRS-II. Apparently, the two of them are necessary, as neither LysRS-I nor LysRS-II works alone (Polycarpo et al. 2003). Strikingly, the corresponding light blue codon color (Fig. 3) predicts it should indeed be chargeable by both Class I (2′OH) and Class II aaRSs (3′OH). So, the situation of pyrrolysine in some archaebacteria may be a snapshot of an intermediate step, “caught in the act,” in the codon assignment mechanism described here (see above: the interpretation of the light blue codon).
Other binary patterns in the genetic code and translation apparatus have been previously noted by other authors. Indeed, the presence of an “operational code” on the acceptor stem of the tRNA has been hypothesized because this stem often contains determinants sufficient for specific aminoacylation (de Duve 1988; Schimmel et al. 1993). In addition a so-called (G/C) “operational code” has been described in the tRNA acceptor stem (Rodin and Ohno 1997), as well as a “steric code” for the recognition of aaRSs on each side of the tRNA acceptor stem (Ribas de Pouplana and Schimmel 2001b). More importantly, the possible presence of a remnant of the anticodon–codon pair in the acceptor stem of tRNAs (Rodin et al. 1996) fits very well with the model in Fig. 4. Indeed, if there is an “echo” of the codon identity in the acceptor stem of each tRNA, then a binary color code operating on the codon sequence, such as the one described in Figure 5, can very well dictate the physicochemical class and polarity of the amino acid to be charged by a proto-aaRS, assuming that the 2′OH (resp. 3′OH) mechanism is associated with large and hydrophobic (resp. small and polar) amino acid binding pockets (Woese 1965b; Crick 1968).
INVASION OF THE CODON TABLE: A TWO-STEP MODEL?
At least three different types of ideas are classically invoked (Knight et al. 1999; Di Giulio 2005) to explain the evolution of the genetic code—they are actually not mutually exclusive and may all be true but correspond to different periods during this evolution. In the “adaptation idea,” the codon assignment problem is described as a typical optimization problem with an error minimization scheme stating: “Near codons should encode similar amino acids.” This represents the simplest feedback loop of the optimization process at work. Upon one mutation of any one of the three bases of the codon, the encoded amino acid should be “similar” or synonymous to the one encoded by the unmutated codon, so as to minimize the structural and functional deleterious effects of this mutation on the resulting protein(s), including aaRSs. Indeed, as noted very early after the establishment of the genetic code (Woese 1965b), it is striking that chemically similar amino acids occupy close positions in the genetic code. A number of studies have assessed more quantitatively the importance of this criterion in understanding the genetic code (Freeland and Hurst 1998; Trinquier and Sanejouand 1998). In the resulting physicochemical and ambiguity reduction theory (Sonneborn 1965; Woese 1965a; Di Giulio 2005) the present-day genetic code is seen as mainly the result of physicochemical forces (and, of course, natural selection). However, Wong made the point that the relationships between amino acids encoded by near codons more faithfully reflect known biosynthetic pathways rather than physicochemical similarities (Wong 1975). This is the so-called “coevolution theory.” Finally, a stereochemical origin of the code (“the escaped triplet theory”) has been postulated, based on the existence of chemical and physical complementarity of amino acids and their codons. Some experimental evidence for this theory exists because RNA aptamers artificially evolved to bind some amino acids (arginine, isoleucine, etc.) display sequence similarity with either their codon or anticodon well above the level predicted by chance (Yarus et al. 2005).
How does the Figure 4 scheme compare with these theories? First of all, the underlying assumption for deriving a scenario for the evolution of the genetic code from Figure 4 is that the division into two different aminoacylation mechanisms predates the apparition of aaRSs. When they appeared, the two types of aaRSs merely copied a system that already existed in the RNA World. The fact that the extant aaRS mode of tRNA aminoacylation is used to extract clues to a mechanism that is postulated to have been done solely by RNA in the beginning is justified a posteriori by the fact that there are several situations where such a molecular mimicry phenomenon between proteins and tRNA, in e.g., RF1 and RF2 and EF-Tu, is known and documented (Liang and Landweber 2005).
However, it remains to be specified in Figure 4 what is meant by the color of a (degenerate) codon at intermediate steps of the differentiation process in terms of encoded amino acid(s), especially at the second and third steps of the tree. Is it a unique amino acid of a given class, beginning with the more “primordial” ones (Miller 1987), or a degenerate type of amino acid within a given class, e.g., the sum of its progeny? Several scenarios are possible.
The simplest scenario suggested by Figure 4 is a two-step mechanism that consists of invading as quickly and efficiently as possible the table of codons in a degenerate fashion, with codons assigned only to a class (color) of an aminoacylation mechanism in an essentially binary code, before fine tuning and going to a gradually more specific codon assignment. Both lattice and off-lattice simulations of binary heteropolymers (HP models) indeed suggest that stable conformations can be encoded by binary sequences (Chan and Dill 1996; Miller et al. 2002). The second stage would be driven by physicochemical forces, shaping differently the amino acid binding pocket for different amino acids of the same physicochemical class (Crick 1968). One might imagine then a gradual assignment of amino acid to (classes of) codons, descending the tree of Figure 4. This implies the temporary use of degenerate sequences in the encoded polypeptides. This is very similar to the transient existence of “statistical” ur-proteins postulated by Woese (1965a). In fact, one can argue that present-day known protein folds, which are especially resistant to mutations, may have been selected this way (Finkelstein et al. 1993). The novelty here is that there is a clear underlying mechanism supporting the early assignment of pools of similar amino acids to different sets of (related) codons. In this scenario, the time scale is probably very short for the first stage (color assignment) but still quite long for the second stage (assignment inside each class). It is interesting to compare this scenario with the theory of the coevolution of the code (Wong 1975), where only a small number of amino acids that are readily formed in the primordial soup are encoded in the first stage. In a subsequent stage, each precursor subdivides its codons among amino acids that are related by some biosynthetic pathway. Therefore the concession of codons should contain an echo of biosynthetic relationships between amino acids. In Figure 4, the families of amino acids are clearly related by physicochemical properties but not by some biosynthetic pathway. Therefore, the model of Figure 4 is at variance with the Wong coevolution theory but plainly compatible with the physicochemical theory.
However, in this scenario, the information content of the sequence after the first step is probably insufficient to produce efficient polypeptides able to play a role in the translation process itself. So when did the encoded-protein world emerge from the RNA World? It is tempting to attribute the top half of Figure 4 to a process performed solely in the RNA World, where partial and gradual diffentiation of the colored codons into amino acid codons can be postulated to be driven by stereochemical factors playing a major role in early tRNA–RNA–amino acid recognition—as in the stereochemical idea (Yarus et al. 2005) —while the lower part (see the separation arrow in Fig. 4) signals the entry into a world where RNA and encoded proteins coexisted and where approximate but functional aaRSs took over the role of decoding codons. Until the number of encoded (families of) amino acids was <5–7, the system was free to run, but the resulting polypeptides were of insufficient quality to be used for crucial tasks such as translation. Therefore, these tasks were still accomplished by a (then) more reliable RNA apparatus. Upon the assignment of amino acids to the families of codons generated after just four differentiation steps, one obtains six families of amino acids encoded plus one uncertain one, which is enough to code heteropolymers with a form and a function (Chan 1999; Wang and Wang 1999). The combinatorics of the remaining exact assignation of such a partition (4! × 4! × 2! × 2! × 1! × 1! = 2304) is small compared to the 14! = 8.7 × 1010 ways to assign 14 amino acids to 14 codons, even when one takes into account the initial block assignment (driven by the reduction of translation errors) and the selection of pools of amino acids for each block (governed by the volume and the polarity of amino acids).
A recent article (Vetsigian et al. 2006) mentions the possibility of horizontal and communal evolution of the code, through different subspecies using different codes and exchanging information. Clearly, different populations using the same codon colored table (where just the color counts) would indeed be able to exchange information even if the amino acid meaning of each codon were different for different subspecies. In other words, the colored codon table of Figure 4 provides a common “grammar” so that different subpopulations using different “vocabularies” could still understand each other and share inventions. It is interesting to note that other asymmetric partitions of the code have a large overlap with the one described in Figure 2 (right half). In particular, the C, U, A, G permutation in the second base would generate a stop codon at UGA while still maintaining 78% of the codon colors of the whole table.
CONCLUSION AND PERSPECTIVE
The scheme presented here allows one to envision how a quick preliminary binary assignment of the 16 X1X2N codons could be made using a series of binary decisions concerning the second and then the first base. The existence of two different mechanisms of aminoacylation of the tRNA is here postulated to be the first manifestation of a code. The partition of the codon table between the two classes always follows the same repetitive rule at each differentiation and ambiguity reduction step and is therefore entirely deterministic. However, this mechanism is not trivial as it ensures the presence of a stop codon since the beginning, as well as an ambiguous codon whose importance quickly decreased with time. This scheme also suggests that the takeover of some functions of the translation apparatus by proteins did not occur before the crude assignment of the 16 X1X2N codons to 6–7 families of amino acids was made. The rest of the assignment involves only evolution within classes, but may very well have been achieved by the competition between subpopulations sharing the same color scheme (grammar) in the codon tables. The crude assignment of amino acids to each of the codons is viewed essentially as a downhill process, where the translation error rate was optimized first and stereochemical factors drove an initial and crude assignment. Here, however, the role of chance in the first step is reduced to a minimum because the invasion of the codon repertoire by the two classes of aminoacylation mechanisms is entirely deterministic. Altogether, this is very similar to the scenario of Crick (1968) but answers one of its main weaknesses, which is “that the early step needed to get the system going requires a lot of chance effect” (Crick 1968).
A discussion of the possible molecular mechanisms at work for this preliminary assignment in only four (or five) “generations” is clearly outside the scope of this article, but it is of interest to note that all known mechanisms involved in the generation of asymmetric division involve a replication step (Klar 1987), thereby creating a curious link between gene replication and expression. Another link is the fact that structural homologs of both Class I and Class II aaRSs have been found as proteins associated with the replication (e.g., bacterial dnaG primase and the beta subunit of mitochondrial polymerase, respectively, see Carrodeguas et al. 1999). Also, it has been suggested (Rodin and Ohno 1995; Carter and Duax 2002) that the two ancestors of Class I and Class II aaRSs were in fact originally encoded by the same gene on the two complementary strands. If true, the mechanism for “switching class” may be understood by a simple recombination event, assuming the gene was inserted between two inverted repeats.
The codon assignment problem under the biological constraints invoked above (near codons encode near amino acids, metabolic pathways, etc.) is typically an example of NP-complete problems, a class of NP-hard problems (see, e.g., Berger and Leighton 1998) that are at the core of combinatorial optimization theory (Mezard et al. 2002). The asymmetric mechanism described here offers a way to explore efficiently a drastically (but intelligently) reduced search space, if the initial set of elements to be assigned can be subdivided into two chemically distinct subsets (classes). This may in turn prove useful in the emerging field of synthetic biology to design a biological system capable of quickly finding approximate solutions to various analoguous, but less vital, assignment problems under specific constraints, e.g., building a library of RNA sequences (stickers) in correspondance to ligands, such that errors in the sequence would have minimal effects in the retrieval of the associated ligand.
ACKNOWLEDGMENTS
Thanks are due to Benoit Arcangioli for many useful discussions and suggestions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.257607.
REFERENCES
- Arnez, J.G., Moras, D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- Balakrishnan, J. Symmetry scheme for amino acids codons. Phys. Rev. E. 2002;65:21912–21916. doi: 10.1103/PhysRevE.65.021912. [DOI] [PubMed] [Google Scholar]
- Bashford, J.D., Tsohantjis, I., Jarvis, P.D. A supersymmetric model for the evolution of the genetic code. Proc. Natl. Acad. Sci. 1998;95:987–992. doi: 10.1073/pnas.95.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedouelle, H. Recognition of tRNA-Tyr by tyrosyl-tRNA synthetase. Biochimie. 1990;72:589–598. doi: 10.1016/0300-9084(90)90122-w. [DOI] [PubMed] [Google Scholar]
- Berger, B., Leighton, T. Protein folding in the hydrophobic–hydrophilic HP model is NP-complete. J. Comput. Biol. 1998;5:27–40. doi: 10.1089/cmb.1998.5.27. [DOI] [PubMed] [Google Scholar]
- Brooks, D.J., Fresco, J.R. Increased frequency of Cysteine, Tyrosine and Phenylalanine residues since the last universal ancestor. Mol. Cell. Proteomics. 2002;12:125–131. doi: 10.1074/mcp.m100001-mcp200. [DOI] [PubMed] [Google Scholar]
- Carrodeguas, J.A., Kobayashi, R., Lim, S.E., Copeland, W.C., Bogenhagen, D.F. The accessory subunit of Xenopus laevis mitochondrial DNA polymerase γ increases processivity of the catalytic subunit of human DNA polymerase γ and is related to Class II aminoacyl-tRNA synthetases. Mol. Cell. Biol. 1999;19:4039–4046. doi: 10.1128/mcb.19.6.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, C.W., Jr., Duax, W.L. Did tRNA synthetase classes arise on opposite strands of the same gene? Mol. Cell. 2002;10:705–708. doi: 10.1016/s1097-2765(02)00688-3. [DOI] [PubMed] [Google Scholar]
- Chan, H.S. Folding alphabets. Nat. Struct. Biol. 1999;6:994–996. doi: 10.1038/14876. [DOI] [PubMed] [Google Scholar]
- Chan, H.S., Dill, K.A. Comparing folding codes for proteins and polymers. Proteins. 1996;24:335–344. doi: 10.1002/(SICI)1097-0134(199603)24:3<335::AID-PROT6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Cusack, S., Hartlein, M., Leberman, R. Sequence, structure and evolutionary relationships between Class 2 aminoacyl-tRNA synthetases. Nucleic Acid Res. 1991;19:3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve, C. tRNA: The second genetic code. Nature. 1988;333:117–118. doi: 10.1038/333117a0. [DOI] [PubMed] [Google Scholar]
- Delarue, M., Moras, D. Aminoacyl-tRNA synthetases: Partition into two classes. In: Eckstein F., Lilley D.M.J., editors. Nucleic acids and molecular biology. vol. 6. Springer; Berlin: 1992. pp. 203–224. [Google Scholar]
- Delarue, M., Moras, D. The aminoacyl-tRNA synthetase family: Modules at work. Bioessays. 1993;15:675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- Di Giulio, M. The origin of the genetic code: Theories and their relationships, a review. Biosystems. 2005;80:175–184. doi: 10.1016/j.biosystems.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Eigen, M., Lindemann, B., Tietze, M., Winkler-Oswatitsch, R., Dress, A., von Haesler, A. How old is the genetic code? Statistical geometry provides an answer. Science. 1989;244:673–678. doi: 10.1126/science.2497522. [DOI] [PubMed] [Google Scholar]
- Eriani, G., Delarue, M., Poch, O., Gangloff, J., Moras, D. Partition of aminoacyl-tRNA synthetases into two classes based on mutually exclusive sets of conserved motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Finkelstein, A.V., Gutin, A.M., Badretdinov, A.Ya. Why are the same protein folds used to perform different functions? FEBS Lett. 1993;325:23–28. doi: 10.1016/0014-5793(93)81407-q. [DOI] [PubMed] [Google Scholar]
- Fitch, W.M., Upper, K. The phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1987;52:759–767. doi: 10.1101/sqb.1987.052.01.085. [DOI] [PubMed] [Google Scholar]
- Fraser, T.H., Rich, A. Amino acids are not all initially attached to the same position on transfer RNA molecules. Proc. Natl. Acad. Sci. 1975;72:3044–3048. doi: 10.1073/pnas.72.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeland, S.J., Hurst, L.D. The genetic code is one in a million. J. Mol. Evol. 1998;47:238–248. doi: 10.1007/pl00006381. [DOI] [PubMed] [Google Scholar]
- Goldgur, Y., Mosyak, L., Reshetnikova, L., Ankilova, V., Lavrik, O., Khodyreva, S., Safro, M.G. Crystal structure of phenylalanyl-tRNA synthetase from Th. thermophilus complexed with cognate tRNAPhe. Structure. 1996;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- Hauenstein, S., Zhang, C.M., Hou, Y.M., Perona, J.J. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2004;11:1134–1141. doi: 10.1038/nsmb849. [DOI] [PubMed] [Google Scholar]
- Hecht, S.M., Chinault, A.C. Position of aminoacylation of individual Escherichia coli and yeast tRNAs. Proc. Natl. Acad. Sci. 1976;73:405–409. doi: 10.1073/pnas.73.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornos, J.E.M., Hornos, Y.M.M. Algebraic model for the evolution of the genetic code. Phys. Rev. Lett. 1993;71:4401–4404. doi: 10.1103/PhysRevLett.71.4401. [DOI] [PubMed] [Google Scholar]
- Ibba, M., Morgan, S., Curnow, A.W., Pridmore, D.R., Vothknecht, U.C., Gardner, W., Lin, W., Woese, C.R., Söll, D. An euryarchaeal lysyl-tRNA synthetase: Resemblance to Class I synthetase. Science. 1997;278:1119–1122. doi: 10.1126/science.278.5340.1119. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- Klar, A. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987;326:466–470. doi: 10.1038/326466a0. [DOI] [PubMed] [Google Scholar]
- Knight, R.D., Freeland, S.J., Landweber, L.F. Selection, history and chemistry: The three faces of the genetic code. Trends Biochem. Sci. 1999;24:241–247. doi: 10.1016/s0968-0004(99)01392-4. [DOI] [PubMed] [Google Scholar]
- Lescoute, A., Westhof, E. The A-minor motifs in the decoding recognition process. Biochimie. 2006;88:993–999. doi: 10.1016/j.biochi.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Liang, H., Landweber, L.F. Molecular mimicry: Quantitative methods to study similarity between tRNA and proteins. RNA. 2005;11:1167–1172. doi: 10.1261/rna.7207205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonaill, D.A. Why nature chose A, C, G, T/U: An error-coding perspective of nucleotide alphabet composition. Orig. Life Evol. Biosph. 2003;33:433–455. doi: 10.1023/a:1025715209867. [DOI] [PubMed] [Google Scholar]
- Mezard, M., Parisi, G., Zecchina, R. Analytic and algorithmic solution of random satisfiability problems. Science. 2002;297:812–815. doi: 10.1126/science.1073287. [DOI] [PubMed] [Google Scholar]
- Miller, S.L. Which organic compounds could have occurred on the prebiotic earth? Cold Spring Harb. Symp. Quant. Biol. 1987;52:17–27. doi: 10.1101/sqb.1987.052.01.005. [DOI] [PubMed] [Google Scholar]
- Miller, J., Zeng, C., Wingreen, N.S., Tang, C. Emergence of highly designable protein-backbone conformations in an off-lattice model. Proteins. 2002;47:506–512. doi: 10.1002/prot.10107. [DOI] [PubMed] [Google Scholar]
- Nagel, G.M., Doolittle, R.F. Phylogenetic analysis of aminoacyl-tRNA synthetases. J. Mol. Evol. 1995;40:487–498. doi: 10.1007/BF00166617. [DOI] [PubMed] [Google Scholar]
- Nicholas, H.B., McClain, W.H. Searching tRNA sequences for relatedness to aminoacyl-tRNA synthetase families. J. Mol. Evol. 1995;40:482–486. doi: 10.1007/BF00166616. [DOI] [PubMed] [Google Scholar]
- Pezo, V., Metzgar, D., Hendrickson, T.L., Waas, W.F., Hazebrouck, S., Doring, V., Marlière, P., Schimmel, P., de Crécy-Lagard, V. Artificially ambiguous genetic code confers growth yield advantage. Proc. Natl. Acad. Sci. 2004;101:7593–7597. doi: 10.1073/pnas.0402893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpo, C., Ambrogelly, A., Ruan, B., Tumbula-Hansen, D., Ataide, S.F., Ishitani, R., Yokoyama, S., Nureki, O., Ibba, M., Söll, D. Activation of Pyrrolysine suppressor tRNA requires formation of a ternary complex with Class I and Class II Lysyl-tRNA synthetases. Mol. Cell. 2003;12:287–294. doi: 10.1016/s1097-2765(03)00280-6. [DOI] [PubMed] [Google Scholar]
- Polycarpo, C., Ambrogelly, A., Berube, A., Winbush, S.M., McCloskey, J.A., Crain, P.F., Wood, J.L., Söll, D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas de Pouplana, L., Schimmel, P. Aminoacyl-tRNA synthetases: Potential markers of genetic code development. Trends Biochem. Sci. 2001a;26:591–596. doi: 10.1016/s0968-0004(01)01932-6. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana, L., Schimmel, P. Two classes of tRNA synthetases suggested by sterically compatible docking on tRNA acceptor stem. Cell. 2001b;104:191–193. doi: 10.1016/s0092-8674(01)00204-5. [DOI] [PubMed] [Google Scholar]
- Rodin, S., Ohno, S. Two types of aminoacyl-tRNA synthetases could be originally encoded by complementary strands on the same nucleic acid. Orig. Life Evol. Biosph. 1995;25:565–589. doi: 10.1007/BF01582025. [DOI] [PubMed] [Google Scholar]
- Rodin, S., Ohno, S. Four primordial modes of tRNA-synthetase recognition, determined by the (G, C) operational code. Proc. Natl. Acad. Sci. 1997;94:5183–5188. doi: 10.1073/pnas.94.10.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin, S., Rodin, A., Ohno, S. The presence of codon–anticodon pairs in the acceptor stem of tRNAs. Proc. Natl. Acad. Sci. 1996;93:4537–4542. doi: 10.1073/pnas.93.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, M.A.S., Cheesman, C., Costa, V., Morades-Feirrara, P., Tuite, M.F. Selective advantage created by codon ambiguity allowed for the evolution of an alternative code in Candida spp. Mol. Microbiol. 1999;31:937–947. doi: 10.1046/j.1365-2958.1999.01233.x. [DOI] [PubMed] [Google Scholar]
- Schimmel, P., Giégé, R., Moras, D., Yokoyama, S. An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl. Acad. Sci. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, S., Higgs, P.G. A unified model of codon reassignment in alternative genetic codes. Genetics. 2005;170:831–840. doi: 10.1534/genetics.104.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitivelband, S., Hou, Y.M. Breaking the stereo barrier of amino acid attachment to tRNA by a single nucleotide. J. Mol. Biol. 2005;348:513–521. doi: 10.1016/j.jmb.2005.02.023. [DOI] [PubMed] [Google Scholar]
- Sonneborn, T. Degeneracy of the genetic code: Extent, nature and genetic implications. In: Bryson V., Vogel H.J., editors. Evolving genes and proteins. Academic; New York: 1965. pp. 377–397. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Cramer, F. Site of aminoacylation of tRNAs from Escherichia coli with respect to the 2′- or 3′-hydroxyl group of the terminal adenosine. Proc. Natl. Acad. Sci. 1975;72:3049–3053. doi: 10.1073/pnas.72.8.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, G., James, C.M., Krzycki, J.A. Pyrrolysine encoded by UAG in Archea: Charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- Szathmary, E. Codon swapping as a possible evolutionary mechanism. J. Mol. Evol. 1991;32:178–182. [Google Scholar]
- Trinquier, G., Sanejouand, Y.H. Which effective property of amino acids is best preserved by the genetic code? Protein Eng. 1998;11:153–169. doi: 10.1093/protein/11.3.153. [DOI] [PubMed] [Google Scholar]
- Vetsigian, K., Woese, C.R., Goldenfeld, N. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. 2006;103:10696–10701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkenstein, M.V. The genetic coding of protein structures. Biochim. Biophys. Acta. 1966;119:421–424. doi: 10.1016/0005-2787(66)90204-8. [DOI] [PubMed] [Google Scholar]
- Wang, J., Wang, W. A computational approach to simplifying the protein folding alphabet. Nat. Struct. Biol. 1999;6:1033–1038. doi: 10.1038/14918. [DOI] [PubMed] [Google Scholar]
- Woese, C.R. On the evolution of the genetic code. Proc. Natl. Acad. Sci. 1965a;54:1546–1552. doi: 10.1073/pnas.54.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R. Order in the genetic code. Proc. Natl. Acad. Sci. 1965b;54:71–75. doi: 10.1073/pnas.54.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R. Translation: In retrospect and prospect. RNA. 2001;7:1055–1067. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R., Olsen, G.J., Ibba, M., Söll, D. Aminoacyl-tRNA synthetases, the genetic code and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, Y.I., Aravind, L., Grishin, N.V., Koonin, E.V. Evolution of aminoacyl-tRNA synthetases: Analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfers. Genet. Res. 1999;9:689–710. [PubMed] [Google Scholar]
- Wong, J.T.F. A co-evolution theory of the genetic code. Proc. Natl. Acad. Sci. 1975;72:1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, H., Tong, K.L., Marck, C., Grosjean, H., Wong, J.T. Transfer RNA paralogs: Evidence for genetic code–amino acid biosynthesis coevolution and an archeal tree of life. Gene. 2003;310:59–66. doi: 10.1016/s0378-1119(03)00552-3. [DOI] [PubMed] [Google Scholar]
- Yaremchuk, A., Kriklivyi, I., Tukhalo, M., Cusack, S. Class I tyrosyl-tRNA synthetase has a Class II mode of tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus, M., Caporaso, J.G., Knight, R. Origins of the genetic code: The escaped-triplet theory. Annu. Rev. Biochem. 2005;74:179–198. doi: 10.1146/annurev.biochem.74.082803.133119. [DOI] [PubMed] [Google Scholar]
- Zhang, C.M., Perona, J.J., Ryu, K., Francklyn, C., Hou, Y.M. Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 2006;361:300–311. doi: 10.1016/j.jmb.2006.06.015. [DOI] [PubMed] [Google Scholar]