Abstract
Nucleotides are specifically and efficiently targeted for modification from C to U within transcripts of chloroplasts in higher plants. Although the enzymatic apparatus responsible for altering C to U has not been identified, the sequences surrounding editing sites are known to contain information essential for efficient editing. We set out to determine the nucleotides that are critical for editing of a particular C, NTpsbE C214, in chloroplast transcripts in tobacco. Assay of editing of substrates with different lengths of 5′ and 3′ sequence around the target C was carried out to delimit the region of sequence critical for editing in vitro. Mutated substrates were then constructed with an altered nucleotide at each position within the previously defined region around NTpsbE C214. In individual nucleotides, both 5′ and 3′ of the edited nucleotide were found to be important for editing. The sequence GCCGUU, which occurs 5′ of the editing site, was discovered to be critical for editing. Editing substrates mutated to alter the distance between the GCCGUU sequence and NTpsbE C214 resulted in the generation of a new editing target, the 3′ adjacent nucleotide. These data are consistent with a model in which the selection of the C target for editing is determined by its distance from a crucial 5′ sequence.
Keywords: psbE mRNA, chloroplast, RNA editing, in vitro, tobacco, poisoned primer extension
INTRODUCTION
C-to-U RNA editing is a vital component of chloroplast gene expression in higher plant species. First observed in maize chloroplasts (Hoch et al. 1991), chloroplast RNA editing is now known to occur also in other seed plants as well as in hornworts, bryophytes, true ferns, and fern allies (Freyer et al. 1997). Although C-to-U RNA editing of chloroplast transcripts occurs in all vascular plants that have been examined, the particular C targets of editing are not conserved across different plant species. Some species carry T at the homologous location where other plants encode a C that is modified to U at the transcript level. A typical number of editing targets in the chloroplast transcripts of an angiosperm species is 30–40, while many more Cs are targeted in mitochondria (Maier et al. 1995; Wakasugi et al. 1996; Corneille et al. 2000; Tsudzuki et al. 2001; Schmitz-Linneweber et al. 2002; Inada et al. 2004; Tillich et al. 2005; Sasaki et al. 2006; Kahlau et al. 2006).
When the sequences around all known chloroplast editing sites are aligned, no common consensus sequence can be detected. The most extensive information on the effect of cis-acting sequences on RNA editing has been obtained in tobacco, where chloroplast transformation and active chloroplast editing extracts have allowed analysis of mutated substrates both in vivo and in vitro (Chaudhuri and Maliga 1996; Bock et al. 1997; Hirose and Sugiura 2001; Hayes et al. 2006). Of primary importance for editing efficiency is the sequence 5′ of the C target of editing. Previous analyses of editing in vitro by Miyamoto et al. (2002) revealed the presence of important editing cis elements in the sequence −15/−1 region 5′ to the single target of editing in tobacco psbE transcripts. The −15/−5 sequence affects the specific UV cross-linking of a 56-kDa protein to psbE transcripts (Miyamoto et al. 2004). We set out to further define the sequence requirements within the critical 5′ and 3′ regions of the tobacco psbE editing site.
Not only has the identity of particular nucleotides around a C target been shown to affect editing efficiency, but also the particular C that is targeted for editing can be altered from one to another by insertion or deletion of mutations in a substrate. Hermann and Bock (1999) found that mutated ndhB transcripts expressed in vivo were edited at novel sites if the distance between an unidentified 5′ cis element and the normal C target of editing was altered. In ndhB transcripts, a “molecular ruler” might be critical to specify the selection of the C to be edited. To gain more information concerning the mechanism of selection of the C target in chloroplast transcripts, we have assayed the effect of nucleotide (nt) insertions and deletions on editing of tobacco psbE transcripts in vitro.
RESULTS
The sequence 13 nt upstream and 10 nt downstream of the tobacco psbE C214 is critical for editing
The 5′ sequence requirements of Arabidopsis psbE had been examined using Arabidopsis extracts and six substrates we constructed to carry different lengths of 5′ sequence (Hegeman et al. 2005). Tobacco and Arabidopsis psbE sequences are very similar, with only 7 nt differences between tobacco and Arabidopsis psbE in the −150/+15 region around the editing site. Tobacco extracts are known to efficiently edit Arabidopsis psbE editing substrates (Hegeman et al. 2005). Because we already had produced six Arabidopsis psbE substrates, we assayed their editing efficiencies in tobacco chloroplast extracts to obtain an initial indication of the 5′ sequence essential for editing (Fig. 1).
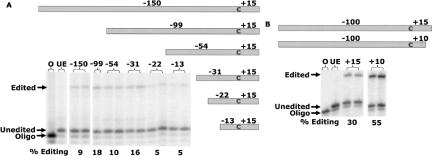
FIGURE 1.
Editing in RNA substrates with various lengths of 5′ (A) and 3′ (B) sequence. Gray boxes represent the substrates containing psbE sequence from Arabidopsis (A) and tobacco (B) with the number of nucleotides 5′ and 3′ indicated. Electrophoretograms of the poisoned primer extension (PPE) reactions used to assay editing efficiencies are shown below the substrate diagrams, with the % editing calculated as described previously (Peeters and Hanson 2002). (Lane O) PPE without template, indicating the size of the labeled oligo; (lane UE) PPE of substrate incubated without competent editing extract. All other lanes are labeled according to the length of sequence 5′ (A)and 3′ (B).
All six substrates exhibited some level of editing, including templates with only 13 nt 5′ of the edited nucleotide (Fig. 1A). The substrates containing only 23 or 13 nt 5′ of the edited nucleotide were significantly less edited than the longer substrates. Because the −99/+15 substrate was edited most efficiently, two additional substrates were produced, each carrying 100 nt of tobacco psbE sequence 5′ to the editing site (Fig. 1B). The shorter substrate (−100/+10) was edited more highly than the longer one (−100/+15), although both substrates are edited. Ten nucleotides 3′ of NTpsbE C214 are evidently sufficient for efficient editing in vitro.
Individual nucleotides 5′ and 3′ of the edited nucleotide are important for editing
Our initial experiments with substrates of different lengths stimulated us to focus more closely on the importance of specific nucleotides in the sequence from −20 to +10 surrounding the C214 editing target. Thirty substrates were constructed, each with 1 nt substituted with the complementary nucleotide in the −20/+10 region. In order to easily observe the effect of mutations, we analyzed substrates containing the −100/+10 region around the edited nucleotide, due to the high editing efficiency of wild-type substrates carrying the −100/+10 sequence (Fig. 1).
Substrates with changes in either 5′ or 3′ nucleotides were reduced in editing compared with substrates with wild-type sequences (Fig. 2). Alterations in the sequence from −11 to −2, +2 to +4, and +8 to +9 around C214 were all harmful to editing. However, changes within the −11/−7 region were the most inimical to editing in vitro. Alteration of the 5′ adjacent nucleotide to the C target from a T to an A did not significantly reduce the ability for the substrate to be edited, in agreement with prior observations using a different RNA substrate in vitro (Miyamoto et al. 2004). However, Miyamoto et al. (2004) found that RNA substrates with a mutation at the 5′ adjacent nucleotide from T to G or T to C exhibited greatly decreased editing efficiency in vitro. Therefore, we can conclude that the sequence from −11 to −1 to NTpsbE C214 as well as sequences from +2 to +4 and +8 to +9 are critical for editing at this site.
FIGURE 2.
Effect of mutations in nucleotides surrounding NTpsbE C214 on editing efficiency. The gray bars represent percentage of editing relative to the wild-type substrate of various substrates that contain 1 nt altered from wild-type tobacco sequence. The wild-type sequence at each position around the edited nucleotide is listed below each bar, and the alteration contained in the mutant substrate is listed below the wild-type nucleotide. Letters representing the nucleotides are colored from light to dark gray, with darker color indicating those nucleotides that exhibit more detrimental effects on editing than nucleotides shown in lighter color.
Sequences in common between NTpsbE C214 and editing sites in rpoB and ndhB are not critical for editing of NTpsbE C214
The sequence 5′ to NTpsbE C214 exhibits some identity to sequences 5′ to NTndhB C467 and NTrpoB C338 (Fig. 3A; Hegeman et al. 2005). Editing of sites exhibiting some 5′ sequence identity has previously been found to be affected when one of the “cluster” of sites is overexpressed in vivo. For example, overexpressing NTrpoB C473 in vivo results in decreased editing of NTpsbL C2 and NTrps14 C80, presumably due to competition for a common factor required for editing of these sites (Chateigner-Boutin and Hanson 2002). Because no chloroplast transgenic plant overexpressing NTpsbE C214 is available, we did not know whether the sequence similarity between the psbE site and NTndhB C467 and NTrpoB C338 was merely fortuitous or actually is significant to editing.
FIGURE 3.
Similarity of 5′ regions of clusters of tobacco editing sites. (A) Alignment of editing sites with common sequences with NTpsbE C214. (B) Sequences common in 5′ region of three editing sites known to be affected when RpoB C473 is overexpressed (Chateigner-Boutin and Hanson 2002). Bold characters indicate common sequences, and the edited nucleotides are represented in a larger font.
Inspection of editing of the mutated substrates shown in Figure 2 reveals that some of the 5′ nucleotides in common between the three sites are not important for editing of NTpsbE C214, namely, the TAATAAC sequence. To investigate further whether these three editing sites might share factors that interact with the common nucleotides, we decided to carry out competition experiments in vitro to test whether excess amounts of transcripts carrying either NTrpoB C338 or NTndhB C467 would be inimical to NTpsbE C214 editing, implying limiting amounts of a shared factor.
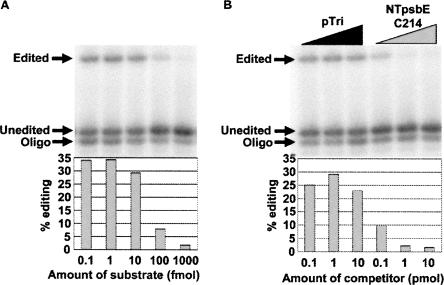
In order to carry out competition experiments that mimic the parameters of editing in vitro, we investigated how much NTpsbE C214 substrate we could add to the reaction in vitro before the percent of edited molecules decreased (Fig. 4). At 10 fmol of RNA editing substrate, the editing percentage in vitro was equivalent to that observed at 1 fmol and 0.1 fmol. However, the amount of editing was greatly reduced at 100 fmol and undetectable at 1 pmol.
FIGURE 4.
Increasing amounts of substrate (A) or self-competitor RNA (B) reduces the percentage of substrate edited in vitro. Competitor RNAs were added at 0.1, 1, and 10 pmol amounts to reactions in standard in vitro conditions with 10 fmol of editing substrate. Competitor RNA amounts correspond to 10×, 100×, and 1000× of the editing substrate, respectively. pTri RNA is a nonspecific competitor transcribed from the control plasmid from the T7 MegaShortscript kit (Ambion). NTpsbE C214 self-competitor contains the same −100/+10 region as the substrate, but SK and KS flanking sequences have been swapped to prevent amplification by RT-PCR.
We then determined the amount of additional psbE RNA needed to reduce editing activity when the initial concentration of editing substrate is 10 fmol (Fig. 4). When either 100 fmol or 1 pmol of psbE RNA is added, the editing in vitro of the 10 fmol of input RNA is reduced. In other words, limitations on editing efficiency become evident when 10× more substrate is present than can be handled by the editing apparatus present in the chloroplast extracts. Thus, if another editing site can compete for the same factors needed for psbE editing, we would expect that adding 10× or 100× more of the competitor RNA should result in a decrease in psbE editing.
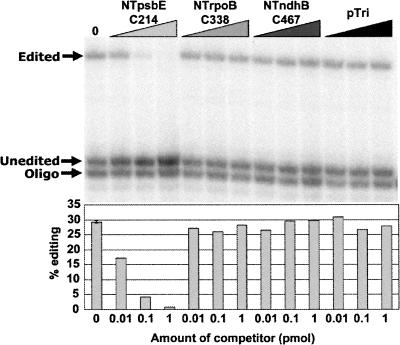
We created competitor RNAs containing −100/+10 regions of sequence around the editing sites NTrpoB C338 and NTndhB C467. Addition of these RNAs in amounts up to 1 pmol had no effect on editing of 10 fmol of NTpsbE C214 substrate (Fig. 5). The lack of competitor effect suggests that the common region of sequence is not a critical editing cis element recognized by a limiting editing factor in vitro.
FIGURE 5.
Effect of the presence of transcripts carrying NTndhB C467 and NTrpoB C338 on editing of NTpsbE C214 in vitro. Competitor RNAs were added to reactions in standard in vitro conditions at amounts of 0.01, 0.1, and 1 pmol, corresponding to 1×, 10×, and 100× of editing substrate, respectively.
Nucleotide preference for editing at key positions around the editing site
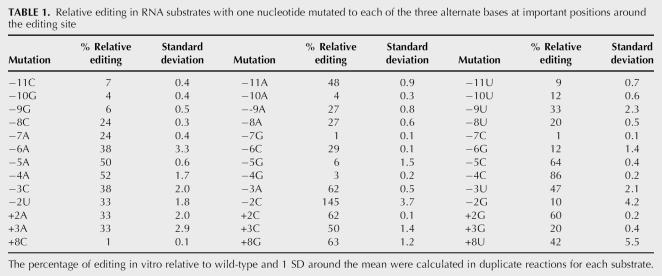
Both the competitor experiments (Fig. 5) and the mutated substrate experiments (Fig. 2) suggested that the 5′ and 3′ nucleotides of most importance around NTpsbE C214 are the 13 nt located at −11/−2, +2, +3, and +8. We therefore more intensively examined these nucleotide positions by making substrates in which each of the nucleotide positions was altered to carry each of the three other possible nucleotides. Editing efficiency in vitro of these 39 different substrates was assayed and then calculated relative to a wild-type substrate (Fig. 6; Table 1). The editing apparatus at nucleotide positions −11 to −6 displayed a strong preference for the wild-type sequence GCCGUU; mutation to any other base reduced the ability for that substrate to be edited (Fig. 6; Table 1). At positions −5, −4, and +3 a strong preference for pyrimidines is evident. At the −2 position, a C nucleotide is preferred over the native A nucleotide. Transcripts with nucleotide changes to G residues in the regions −10/−9, −7/−4, and −2/−1 are extremely poor editing substrates. The editing apparatus appears to tolerate G residues in the positions 5′ of the editing site where there are G residues in the wild-type sequence.
FIGURE 6.
The nucleotide preference for the tobacco editing apparatus at important positions around the editing site. The editing percentages of RNA substrates with 1 nt mutated to each of the 3 nt at each important position were calculated relative to the nucleotide present in wild-type substrates containing tobacco sequences. The amount of editing in mutant substrates was then compared through generation of a sequence logo using the Weblogo program (Crooks et al. 2004). The size of the characters is based on the relative editing of each substrate. Larger characters represent nucleotides that are preferred at positions around the editing site for more efficient editing.
TABLE 1.
Relative editing in RNA substrates with one nucleotide mutated to each of the three alternate bases at important positions around the editing site
The edited nucleotide is determined by a molecular ruler from a 5′ element
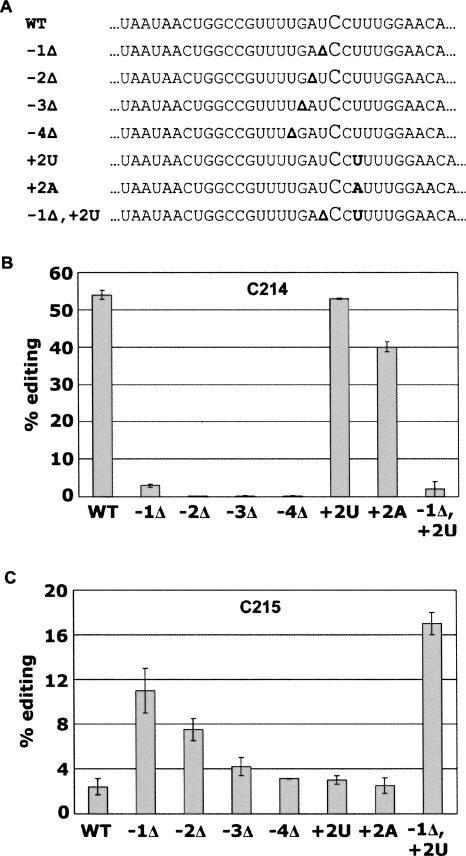
Although the editing site in psbE is flanked by a 3′ C, editing has been detected only at C214. This specificity could either be due to an editing mechanism that is processive, which edits the first C it encounters from a cis-acting element, or that utilizes a molecular ruler, where the editing site is a precise distance from a cis element and the edited C. It is possible that the critical sequences detected 5′ of the editing site in psbE (Fig. 7) could act as the “anchor” for a molecular ruler.
FIGURE 7.
Editing in C214 and C215 in substrates with deletions or insertions compared with wild-type tobacco sequence. (A) Alignment of mutant substrates constructed to examine spacing constraints for editing. Bolded characters represent deleted or inserted nucleotides. Gray bars depict the percentage of editing of mutant substrates at the normal editing site C214 (B) and the 3′ adjacent nucleotide C215 (C). (B,C) Error bars are calculated from duplicate reactions as 1 SD from the mean.
We created seven substrates to test the effects of altering the local spacing around the editing site. We deleted nucleotides 5′ of the editing site and inserted nucleotides 3′ to determine whether the native editing site C214 was invariably edited in the substrates in vitro or whether editing could occur at the 3′ nucleotide C215 (Fig. 7).
There was no editing at the native editing site C214 in substrates with nucleotides deleted immediately 5′ of the editing site. Editing was not significantly reduced when a U was inserted 3′ of C215. Editing was moderately reduced when an A was inserted at position +2. The A insertion influenced the spacing of the minor 3′ element UU at +2/+3, but UU at +2/+3 is maintained in the substrate with the U insertion. Shifting nucleotides 1 nt 3′ by insertion of a U at +2 had no affect on editing. Therefore the location of the +8/+9 nucleotides, the identity of which is important in editing efficiency (Fig. 2), can be shifted without affecting editing. Substrates −4Δ, −3Δ, −2Δ, and −1Δ were not edited in vitro at C214.
Substrates −1Δ, −2Δ, and −1Δ, +2U all exhibited significant editing at the 3′ adjacent nucleotide C215. Therefore, the editing site is determined by the distance of a C nucleotide from the 5′ cis element. The amount of editing at C215 is much lower than the wild-type editing at C214. This is likely to be due to the presence of a C immediately 5′ of C215, while the presence of A or U at −1 results in higher editing at C214 than a 5′ C (Miyamoto et al. 2004). Insertion of nucleotides 3′ of the editing site did not lead to a shift in the C target; therefore, the sequence 3′ of the editing site is not an important determinant for the location of editing. The −1Δ, +2U substrate was more efficiently edited at C215 than the substrate carrying only the −1Δ, suggesting a preference for greater distance between the C target and a downstream element.
DISCUSSION
The minimal size of the surrounding sequence sufficient for editing of a particular C has not been characterized for the vast majority of the 38 known tobacco editing sites in chloroplasts. The known minimal regions of surrounding sequence for editing in vivo are in close proximity to the editing sites: −16/+5 in NTpsbL C2, −12/+11 in NTndhB C683, −21/+2 in NTndhB C692, and −20/+6 in NTrpoB C473. The smallest region identified to be sufficient for correct selection of a C target in vitro is the −13/+10 region around NTpsbE C214.
We observed that the editing efficiency of smaller in vitro substrates is much lower compared to larger substrates with more native sequence around the editing site. The reduced editing efficiency in smaller editing substrates may preclude determination of the minimal region for editing for many of the uncharacterized editing sites. The lower editing efficiency in smaller substrates may result from the absence of auxiliary sequences that enhance editing in vitro, or a reduction in the affinity of the editing components for small RNAs.
By determining the nucleotide preference for editing at important positions, we have identified a 6-nt sequence, GCCGUU that is critical for editing of psbE transcripts in vitro. A 56-kDa protein has been found to undergo specific UV cross-linking to psbE transcripts (Miyamoto et al. 2004). PsbE transcripts with mutations in the −15/−6 region did not compete for binding of the 56-kDa protein to a wild-type psbE transcript, indicating that the −15/−6 region contains some critical sequences for factor binding. It seems likely that the actual region important for binding of this factor is at −11/−6, where the GCCGUU motif we have detected is located. Our studies have correlated a region critical for efficient editing with a region known to be important for binding of a protein.
The GCCGUU motif is found near the psbE editing site but not proximal to other known C targets in chloroplasts. The GCCGTTnnnnnC sequence is present only once within the known tobacco chloroplast protein-coding sequences. Therefore, the 6-nt motif may be sufficient to uniquely identify one cytidine within all chloroplast coding sequences. If a particular factor binds to this motif then its role in editing is likely to be specific to psbE rather than to multiple editing sites. This suggests that some editing sites are independently regulated by one specificity factor. A single factor also appears important in editing of NTndhD C2 because mutation in a single gene encoding a putative RNA-binding protein results in abolition of editing only at NTndhD C2 (Kotera et al. 2005). However, the reduction of editing observed at multiple sites following overexpression transcripts encoded by a chloroplast transgene carrying a similar editing site suggests that there also are groups of sites that share cis elements and factors (Chateigner-Boutin and Hanson 2002; Hayes et al. 2006). Perhaps some chloroplast editing sites are regulated individually and other editing sites are regulated by shared factors.
At present, our study is the only analysis of nucleotide preference surrounding a chloroplast editing site where substrates were tested in which the wild-type nucleotide was mutated to each of the other 3 nt. This thorough analysis of nucleotide preference for editing reveals that sequence requirements are more complex than would be detected by merely testing substrates in which the complementary nucleotide has replaced the native nucleotide. Although the 6-nt motif GCCGUU is critical for editing, other nucleotide preferences are less nucleotide specific. At positions −5, −4, and +3, there is a preference for pyrimidines, and at −2, a C or A will suffice. The increase in editing in a substrate with a C at −2 instead of the native A may be due to a general preference for the dipyrimidines CU at the −2/−1 positions by the editing mechanism. Although the dipyrimidine is probably favored, mutation of an A at −2 to a C may be constrained due to the requirement of maintaining an Asp codon. G nucleotides at many positions reduce editing efficiency; unexpectedly, a C at +8 completely abolishes editing. We have not been able to create models involving secondary structures that might explain these sequence preferences and requirements.
We have determined that the edited nucleotide in psbE is a precise distance from the discovered 5′ cis element. Therefore a molecular ruler is a more likely mechanism for the observed specificity of selection of a C target than a processive enzyme. Because NTndhB C692 also appears to be sensitive to distance (Hermann and Bock 1999), a measuring mechanism may be a feature common to all chloroplast editing sites. However, the impact of 5′ and 3′ nucleotide insertions or deletions has only been extensively studied for two sites in chloroplasts. More editing sites will have to be analyzed to make global conclusions about mechanisms for target selection.
MATERIALS AND METHODS
Preparation of tobacco chloroplast extracts
Tobacco chloroplast extracts were prepared as described by Hegeman et al. (2005). Tobacco (Nicotiana tabacum var. Petit Havana) was grown at 25°C for 4–6 wk under metal halide lights with a 16-h light, 8-h dark cycle. Plants were covered for 2 d before chloroplast isolation. Chloroplasts were isolated from fully expanded leaves as described by Miyamoto et al. (2002). Intact chloroplasts were separated on a continuous Percoll gradient and washed. Chloroplasts were lysed using a Triton X-100 and a hypertonic buffer to produce chloroplast extracts. Extracts were then dialyzed and contained ∼20 μg/μL of protein.
Creation of RNA substrates
Oligonucleotides (Integrated DNA Technologies) were used to produce DNA templates by PCR (Table 2). Templates containing Arabidopsis psbE sequences with different lengths of 5′ sequence from −150 to −13 were constructed as described by Hegeman et al. (2005). Templates with single nucleotide mutations were constructed using mutant primers.
TABLE 2.
Sequences of oligonucleotides (Integrated DNA Technologies) used
TABLE 2.
Continued
Synthetic RNA was created through in vitro transcription using the DNA template PCR product. The DNA template was removed by TurboDNAse digestion. The RNA was then cleaned up using the ZYMOcleanup-5 kit.
Editing in vitro
Reaction conditions for editing in vitro were similar to those described by Hegeman et al. (2005). Optimized reaction conditions included the elimination of magnesium acetate and an increase in ATP concentration from 10 mM to 100 mM. Poisoned primer extension (PPE) was carried out as by Hegeman et al. (2005) to quantify the percent editing in templates. Three different primers were used for PPE: NTpsbE_PPE_G was used to quantify editing at C214 of all substrates without mutations within the 5′ region; NTpsbE_PPE_C was used to quantify editing at the C214 editing site in substrates with mutations in the 3′ region; and NTpsbE_AltC_G was used to quantify editing at C215.
Competition experiments
Self-competitor RNAs were created identically to the psbE substrates except the flanking sequences were swapped. Primers were used to add the T7KS bacterial sequence 5′ of the psbE sequence, and the SK sequence formed the 3′ end. RNA competitors were then produced by in vitro transcription from the PCR product template. RNA competitors, therefore, could not be amplified by using the same SK and KS primers as used for the editing substrates. Different concentrations of RNA competitors were then added to typical reaction conditions immediately after the addition of 10 fmol of substrate.
ACKNOWLEDGMENTS
This work was supported by NSF grant MCB 0344007 to M.R.H and an NIH graduate traineeship to M.L.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.295607.
REFERENCES
- Bock, R., Hermann, M., Fuchs, M. Identification of critical nucleotide positions for plastid RNA editing site recognition. RNA. 1997;3:1194–1200. [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin, A.L., Hanson, M.R. Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol. Cell. Biol. 2002;22:8448–8456. doi: 10.1128/MCB.22.24.8448-8456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, S., Maliga, P. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 1996;15:5958–5964. [PMC free article] [PubMed] [Google Scholar]
- Corneille, S., Lutz, K., Maliga, P. Conservation of RNA editing between rice and maize plastids: Are most editing events dispensable? Mol. Gen. Genet. 2000;264:419–424. doi: 10.1007/s004380000295. [DOI] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer, R., Kiefer-Meyer, M.C., Kossel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. 1997;94:6285–6290. doi: 10.1073/pnas.94.12.6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, M.L., Reed, M.L., Hegeman, C.E., Hanson, M.R. Sequence elements critical for efficient RNA editing of a tobacco chloroplast transcript in vivo and in vitro. Nucleic Acids Res. 2006;34:3750–3762. doi: 10.1093/nar/gkl490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman, C.E., Hayes, M.L., Hanson, M.R. Substrate and cofactor requirements for RNA editing of chloroplast transcripts in Arabidopsis in vitro. Plant J. 2005;42:124–132. doi: 10.1111/j.1365-313X.2005.02360.x. [DOI] [PubMed] [Google Scholar]
- Hermann, M., Bock, R. Transfer of plastid RNA-editing activity to novel sites suggests a critical role for spacing in editing-site recognition. Proc. Natl. Acad. Sci. 1999;96:4856–4861. doi: 10.1073/pnas.96.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T., Sugiura, M. Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: Development of a chloroplast in vitro RNA editing system. EMBO J. 2001;20:1144–1152. doi: 10.1093/emboj/20.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch, B., Maier, R.M., Appel, K., Igloi, G.L., Kossel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991;353:178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- Inada, M., Sasaki, T., Yukawa, M., Tsudzuki, T., Sugiura, M. A systematic search for RNA editing sites in pea chloroplasts: An editing event causes diversification from the evolutionarily conserved amino acid sequence. Plant Cell Physiol. 2004;45:1615–1622. doi: 10.1093/pcp/pch191. [DOI] [PubMed] [Google Scholar]
- Kahlau, S., Aspinall, S., Gray, J.C., Bock, R. Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J. Mol. Evol. 2006;63:194–207. doi: 10.1007/s00239-005-0254-5. [DOI] [PubMed] [Google Scholar]
- Kotera, E., Tasaka, M., Shikanai, T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- Maier, R.M., Neckermann, K., Igloi, G.L., Kossel, H. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- Miyamoto, T., Obokata, J., Sugiura, M. Recognition of RNA editing sites is directed by unique proteins in chloroplasts: Biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol. Cell. Biol. 2002;22:6726–6734. doi: 10.1128/MCB.22.19.6726-6734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, T., Obokata, J., Sugiura, M. A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts. Proc. Natl. Acad. Sci. 2004;101:48–52. doi: 10.1073/pnas.0307163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N.M., Hanson, M.R. Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA. 2002;8:497–511. doi: 10.1017/s1355838202029424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T., Yukawa, Y., Wakasugi, T., Yamada, K., Sugiura, M. A simple in vitro RNA editing assay for chloroplast transcripts using fluorescent dideoxynucleotides: Distinct types of sequence elements required for editing of ndh transcripts. Plant J. 2006;47:802–810. doi: 10.1111/j.1365-313X.2006.02825.x. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber, C., Regel, R., Du, T.G., Hupfer, H., Herrmann, R.G., Maier, R.M. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: The role of RNA editing in generating divergence in the process of plant speciation. Mol. Biol. Evol. 2002;19:1602–1612. doi: 10.1093/oxfordjournals.molbev.a004222. [DOI] [PubMed] [Google Scholar]
- Tillich, M., Funk, H.T., Schmitz-Linneweber, C., Poltnigg, P., Sabater, B., Martin, M., Maier, R.M. Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 2005;43:708–715. doi: 10.1111/j.1365-313X.2005.02484.x. [DOI] [PubMed] [Google Scholar]
- Tsudzuki, T., Wakasugi, T., Sugiura, M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 2001;53:327–332. doi: 10.1007/s002390010222. [DOI] [PubMed] [Google Scholar]
- Wakasugi, T., Hirose, T., Horihata, M., Tsudzuki, T., Kossel, H., Sugiura, M. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: The pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl. Acad. Sci. 1996;93:8766–8770. doi: 10.1073/pnas.93.16.8766. [DOI] [PMC free article] [PubMed] [Google Scholar]