Abstract
The I-AniI maturase facilitates self-splicing of a mitochondrial group I intron in Aspergillus nidulans. Binding occurs in at least two steps: first, a specific but labile encounter complex rapidly forms and then this intermediate is slowly resolved into a native, catalytically active RNA/protein complex. Here we probe the structure of the RNA throughout the assembly pathway. Although inherently unstable, the intron core, when bound by I-AniI, undergoes rapid folding to a near-native state in the encounter complex. The next transition includes the slow destabilization and docking into the core of the peripheral stacked helix that contains the 5′ splice site. Mutational analyses confirm that both transitions are important for native complex formation. We propose that protein-driven destabilization and docking of the peripheral stacked helix lead to subtle changes in the I-AniI binding site that facilitate native complex formation. These results support an allosteric-feedback mechanism of RNA–protein recognition in which proteins engaged in an intermediate complex can influence RNA structure far from their binding sites. The linkage of these changes to stable binding ensures that the protein and RNA do not get sequestered in nonfunctional complexes.
Keywords: group I introns, RNA folding, RNA–protein interactions, RNP assembly
INTRODUCTION
RNA/protein complexes are essential for many aspects of post-transcriptional gene regulation; however, the mechanisms that govern their assembly are, for the most part, undefined. It is clear that recognition of RNA by proteins is a complex process that can include conformational changes in one or both partners. For some proteins (e.g., U1A protein), separate domains adopt a defined orientation only when bound to their substrate RNAs (Allain et al. 1996; Avis et al. 1996), whereas other proteins undergo a disorder-to-order transition upon RNA binding (e.g., L5 ribosomal protein from Xenopus) (DiNitto and Huber 2003). In many cases, RNAs also change conformation in subtle ways such as flipping out of bases from a loop (U1A RNA) (Gubser and Varani 1996), or more dramatically, such as the reorientation of helices in a three-way junction (Batey and Williamson 1998). This structural “flexibility” has been hypothesized to allow optimal juxtaposition of interacting groups through conformational adaptation (Leulliot and Varani 2001). The existence of conformational changes suggests that RNA/protein complex formation is a multistep assembly process populated with intermediates. However, the pathways by which RNA and protein partners bind one another are not well understood, since intermediates have been difficult to isolate and study experimentally. Moreover, how such intermediates contribute to the structure and thermodynamic stability of the final, native complex is largely unexplored.
Group I introns are self-splicing RNAs that, in some cases, require proteins for proper folding. Group I introns contain a conserved catalytic core composed of two extended helical domains. The P4–P6 domain includes the P5, P4, P6, and P6a helices and the P3–P9 domain is made from the P3, P7, P8, and P9 helices. The P4–P6 domain is engaged in multiple tertiary interactions with the catalytic P3–P9 domain to facilitate its proper folding (Woodson 2005). The 5′ and 3′ splice sites (SS) are present on another domain called P1–P2 that docks into the folded catalytic core prior to splicing. In general, protein cofactors facilitate RNA folding required for catalytic activity, but do not participate in the chemistry of the splicing reaction. Some cofactors bind with high specificity and only promote splicing of a single group I intron (Lambowitz et al. 1999). One member of this group is the I-AniI maturase that is required for splicing the A.n. COB group I intron in the mitochondrial (mt) cytochrome b (COB) gene from Aspergillus nidulans (Ho et al. 1997). I-AniI binds to the A.n. COB pre-RNA with picomolar affinity, but does not bind tightly to or promote splicing of other group I introns in vitro (Solem et al. 2002).
I-AniI appears to bind the A.n. COB intron through at least one on-pathway intermediate. The kinetics of complex formation and splicing are consistent with the formation of a relatively weak encounter complex that is resolved into a native, splicing–competent complex (Solem et al. 2002). The equilibrium constant defining the encounter complex (∼13 nM) is ∼1000 times less than that for nonspecific binding (1 μM) (Chatterjee et al. 2003), which suggests that this intermediate may contain some native RNA/protein contacts. We had hypothesized that the intermediate functioned to facilitate structure formation of both the catalytic core and the remainder of the I-AniI binding site by resolving misfolded RNA structure(s) (Solem et al. 2002).
In order to define the I-AniI/A.n. COB pre-RNA assembly pathway more precisely, we have employed chemical and enzymatic probing, oligonucleotide accessibility, interference mapping, and mutagenesis experiments. We find that the secondary structure of the free RNA is largely formed with the exception of two stacked helices (P4/6 and P3/7) that make up the intron core. I-AniI binding results in rapid folding of the helices and intron core. This transition is followed by destabilization and docking of the P1–P2 stacked helices. Mutations that impede the rapid folding of the core or deletion of the P1–P2 helices negatively affect binding, suggesting that both transitions are required for native complex formation. These data suggest a novel mechanism of RNA/protein assembly in which high-affinity protein binding is achieved via RNA remodeling promoted by a specifically, but weakly, bound protein in an intermediate complex.
RESULTS
Experimental strategy for probing the structure of A.n. COB intron
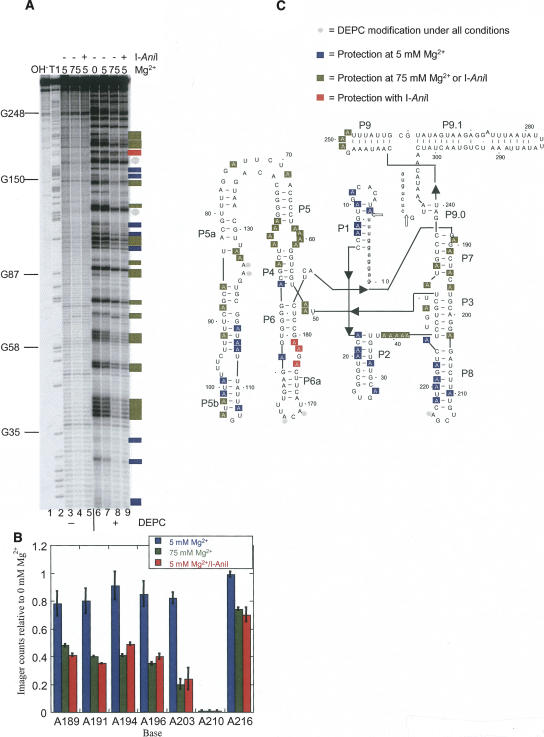
Chemical structure mapping and oligonucleotide hybridization were used to assess the secondary and tertiary structure of the A.n. COB pre-RNA. In these experiments splicing was prevented by withholding the essential group I intron splicing cofactor guanosine. The RNA was probed with diethyl pyrocarbonate (DEPC), which modifies the N7 of adenines that are accessible (i.e., not protected by base pairing, stacking, or tertiary interactions) (Ehresmann et al. 1987). Oligonucleotide hybridization followed by RNase H cleavage was used to monitor the accessibility of specific sequences throughout the RNA (Zarrinkar and Williamson 1994). Finally, iodine cleavage of phosphorothioate-substituted RNA was used to identify regions of A.n. COB pre-RNA that are solvent inaccessible due to interactions with the I-AniI protein or higher-order RNA folding (Caprara et al. 1996). The RNA was probed under various conditions that reflect different functional states of the intron. At 0 mM MgCl2, the intron does not self-splice and cannot bind I-AniI. At 5 mM MgCl2, the intron also does not self-splice but is competent to bind I-AniI. At 75 mM MgCl2 or 5 mM MgCl2 plus I-AniI protein, the intron undergoes self-splicing (i.e., the RNA is in an active conformation under both conditions) (Solem et al. 2002). The data were quantified and any position that showed a twofold or greater reduction in intensity relative to the absence of Mg2+ was scored as “protected” (see Material and Methods). The mapping data were evaluated based on similar biochemical data and crystal structures of related self-splicing group I introns. In this regard, the A.n. COB intron is classified as an IB4 intron containing an extra, peripheral subdomain, P5ab, that associates with the P4–P6 domain (Michel and Westhof 1990; Cate et al. 1996).
The A.n. COB catalytic core is unfolded at physiological conditions
Adenines in the helices of peripheral stem–loops (P1, P2, and P5b) of A.n. COB were protected from DEPC treatment in 5 mM Mg2+, providing evidence that the helices were base-paired (Fig. 1A,C). The lack of significant RNase H digestion with oligonucleotides that anneal to sequences throughout the peripheral P5ab extended helix, P5, and P2, was also consistent with stable secondary structure in these regions (Fig. 2, and see Fig. 5 below).
FIGURE 1.
Chemical structure mapping of the A.n. COB pre-RNA. (A) DEPC modification of end-labeled RNA. End-labeled A.n. COB was treated with DEPC in various concentrations of Mg2+ as well as in the presence of I-AniI. Sites of modification were cleaved by treatment with aniline and the products separated by denaturing gel electrophoresis. Lane 1, hydrolysis ladder; lane 2, G sequencing by partial T1 digestion; lanes 3–5, mock-treated RNA; lanes 6–9, DEPC-treated RNAs; lane 6, 0 mM Mg2+, lanes 3 and 7, 5 mM Mg2+; lanes 4 and 8, 75 mM Mg2+; lanes 5 and 9, 5 mM Mg2+ plus I-AniI. (B) Quantification of the DEPC mapping data encompassing the P7–P8 region. PhosophorImager counts in each lane were normalized and expressed as a ratio relative to 0 mM Mg2+. The histogram shows averaged ratios with the error bars representing standard deviations. (C) Secondary structure model of A.n. COB pre-RNA showing positions of DEPC modification or protection. Open arrows point to splice sites. Two independent DEPC mapping experiments were performed, and positions were scored as protected on the secondary structure model if a twofold or greater reduction in modification level relative to 0 mM Mg2+ was observed in both experiments. The state of modification for unmarked positions could not be assigned because of inconsistencies between experiments, degradation, or poor resolution in the gel. For the A.n. COB intron, adenines in single-stranded regions expected to make specific contacts to orient the four domains in a catalytically active conformation include (a) the A-rich bulge in P5a with the P4 helix; (b) the J3/4 and J6/7 junctions with P6 and P4 helices, respectively; (c) the P9 loop with the P5 helix; (d) the J4/5, J5/4, and J8/7 junctions with the 5′ SS containing P1 helix; and (e) the P2 loop with P8 helix.
FIGURE 2.
Oligonucleotide accessibility of the A.n. COB pre-RNA. (A) Locations of DNA oligonucleotide complementarity on a secondary structure of A.n. COB pre-RNA. (B) Accessibility experiments. Internally labeled RNAs were incubated in buffer containing 5 mM Mg2+ for 10 min and then a mixture of oligonucleotide and RNase H was added. After 1 min, the reaction was quenched and the products separated by denaturing gel electrophoresis. The region where each DNA oligonucleotide hybridizes is listed at the top of each lane in the gel image. 3E, DNA oligonucleotide targeted to the 3′ exon.
FIGURE 5.
Time-resolved mapping of I-AniI binding of A.n. COB pre-RNA. (A) Oligonucleotide accessibility. Internally labeled RNA was incubated with I-AniI for 10–300 sec and then probed by the addition of a DNA oligonucleotide and RNase H. Digestion occurred for 10 sec, the reaction quenched and products separated by denaturing polyacrylamide gel electrophoresis. The position of the oligonucleotide binding site is shown in Figure 2. SM, starting material; 0, no protein added. (B) Time-dependent RNA changes. The folding times for each region of the RNA were measured by plotting the fraction of RNA cleaved versus time. The time for RNase H digestion (10 sec) was added to each point. The data were fit to a single exponential. Folding of P7 and P4/6 were faster than the first time point (17 sec). Note that P1 and P2 become less accessible to the respective oligonucleotide probes at 10 min. This reproducible effect presumably reflects that P1 and P2 become stabilized in the native complex. (C) Accessibility of P1 and P2 using extended RNase H digestion. The data show that a significant fraction of RNA is cleaved at early (10–40 sec) times after protein addition when the pre-RNA was probed using a 1-min RNase H incubation time. (D) Iodine cleavage of GαS substituted A.n. COB RNA as a function of time. End-labeled RNA was incubated with I-AniI for 10–300 sec and then cleaved with iodine for 10 sec and the reaction quench with β-mercaptoethanol. The products were separated by denaturing gel electrophoresis. The regions of A.n. COB pre-RNA cleaved by iodine shown to the side of the gel image. All of the changes upon addition of I-AniI happened before the first time point (20 sec), and therefore were assigned a rate of >50 min−1. A similar rate of overall folding was also observed with RNase 1 and V1 (data not shown).
The mapping data also showed that the central core of the intron was unstable or misfolded in the absence of protein. All of the adenines that could be surveyed in the P7 and P3 helices were modified by DEPC suggesting that these coaxially stacked helices were unstable. Instability of P3, P7 as well as the stacked P4/P6 helices was also supported by strong RNaseH digestion with oligonucleotides that anneal in these regions (Fig. 2). Previously, the P7 and P3 helices were judged to be unfolded by their susceptibility to cleavage by the single-stranded specific Ribonuclease 1 (Downing et al. 2005). Thus, the four central helices that define the group I intron core are unstable in A.n. COB pre-RNA.
The DEPC modification patterns provided evidence that many tertiary interactions, which serve to correctly align the four extended helical domains, P3–P9, P4–P6, P5ab, and the splice sites containing P1–P2, were unstable. Based on strong conservation among group I introns, adenines in single-stranded regions within these domains are expected to engage in interactions that serve to orient the four domains in a catalytically active conformation (see legend to Fig. 1; Woodson 2005). Remarkably, adenines within almost all of these single-stranded regions were accessible to DEPC modification, suggesting that most of the predicted tertiary interactions were not formed at 5 mM Mg2+. Consistent with this interpretation, iodine cleavage of phosphorothioate-substituted A.n. COB pre-RNA occurred within regions expected to be within the solvent-inaccessible intron core (Fig. 3). Taken together, the probing data were consistent with a generally unfolded intron core for A.n. COB pre-RNA in the absence of protein.
FIGURE 3.
Phosphate backbone probing of A.n. COB pre-RNA. (A) Portion of a gel showing iodine cleavage of phosphorothioate substituted RNAs. A.n. COB pre-RNA was transcribed in the presence of one phosphorothioate analog and its 5′ end labeled. A.n. COB RNAs were incubated in various concentrations of Mg2+ as well as in the presence of I-AniI and then subjected to cleavage with iodine. The products were separated by denaturing gel electrophoresis and quantified. Nucleotide numbers are indicated at the side of the gel and the identity of the phosophorothioate substitution at the top. Lanes 1, 6, 11, and 16, mock-treated RNA; lanes 2, 7, 12, and 17, 0 mM Mg2+; lanes 3, 8, 13, and 18, 5 mM Mg2+; lanes 4, 9, 14, and 19, 75 mM Mg2+; lanes 5, 10, 15, and 20, 5 mM Mg2+ plus I-AniI. (B) Example of the quantification of the iodine mapping data. The histogram shows PhosophorImager counts in each lane that were normalized and expressed as a ratio relative to 0 mM Mg2+. (C) Summary of iodine cleavage data on the secondary structure of A.n. COB pre-RNA. Colored shading shows condition-specific protections, and positions that are cleaved under all conditions are demarcated by a gray oval. Three independent experiments were performed, and positions were scored as protected if a twofold or greater reduction in iodine cleavage relative to 0 mM Mg2+ was observed in each experiment. The relative amount of cleavage for unmarked positions could not be assigned because of inconsistencies between experiments, degradation, or poor resolution in the gel. Regions expected to be within a solvent inaccessible core include J3/4, J4/5, J5/4, J6/7, J8/7, the A-rich bulge in P5a, and the P7 helix. A portion of the P9.1 helix is shown schematically. (D) Position of putative I-AniI binding sites on the three-dimensional structure of the Tetrahymena intron. Red shading includes protected phosphate residues that lie on one side of the intron core. Other I-AniI-specific protections (blue shading) are on the opposite side of this surface and are likely due to interactions with the P1 helix.
I-AniI binding stabilizes secondary and tertiary structure
Binding of the I-AniI protein or high Mg2+ concentrations drove the folding of the A.n. COB intron into an active structure. Both DEPC modification and iodine cleavage patterns changed dramatically under splicing permissive conditions. Adenines in P3, P4, P6a, and P7 became protected in the presence of either 75 mM Mg2+ or I-AniI, which is consistent with formation and/or stabilization of these helices (Fig. 1). Furthermore, protection of adenines within those regions expected to form critical tertiary interactions (see above) is consistent with a folded intron structure. Phosphate backbone positions expected to be solvent inaccessible (see above) were also protected from iodine cleavage under both conditions (Fig. 3). Thus, the chemical protection data were consistent with the intron folding into an active structure.
Outlining the I-AniI binding site in A.n. COB pre-RNA
Differences between the modification patterns of A.n. COB pre-RNA in the presence of protein versus high Mg2+ concentrations reflect differences in RNA folding and protein contacts. In Figures 1C and 3C, these differences are shown schematically. Significantly, most of the idiosyncratic protein protections fell in one of two places (shown as red shading). One set of iodine protections was within the L5/5a loop and most likely reflected the formation of a specific RNA structure. By analogy with a similar region of the Tetrahymena self-splicing intron, P5a and P4 are expected to come in close contact via a sharp (∼150°) bend in the J5/5a region. The adenine-rich bulge in P5a makes hydrogen bond contacts with the minor groove of P4 to facilitate packing of the two helices (Cate et al. 1996). Other protein-specific protections were located in P4, P5a, P6, P6a, P9 (Fig. 3C), and J6a/6 (Fig. 1C), and most of which likely represent RNA binding sites (see below). Interestingly, enhanced iodine cleavage was observed adjacent to two assigned binding sites in P4 and P6, and this may reflect increased accessibility or strain introduced by protein binding. When mapped on the most recent three-dimensional structure of the Tetrahymena intron (Guo et al. 2004), the majority of I-AniI's phosphate backbone contacts form a wedge with the edges defined by residues in P4 (G54), P5a (G123, A124, and A125), and in P6 (G254, G253) and the tip by the loop in P9 (G248; Fig. 3D). Thus, I-AniI appears to contact three extended helical domains in the intron that are brought into close proximity by higher-order RNA folding.
Three I-AniI-dependent protections in P4 (C149) and J4/5 (A59, A60) lie on the side of P4 opposite the binding site described above (see blue shading in Fig. 3D). These protections are most likely due to docking of the 5′ SS containing P1 helix (Adams et al. 2004).
There are also a number of phosphate backbone protections observed only in 75 mM Mg2+ in P2, J2/3, P3, and P8 (Fig. 3C). Although not found in the Tetrahymena structure, the P2 and J2/3 elements are similar in size and structure to those in the bacteriophage Twort orf142–12 group I intron for which a crystal structure exists (Golden et al. 2005). When analogous sites in all four regions are mapped onto the three-dimensional structure, they cluster in a relatively small interface (data not shown). These protections may reflect an alternative structure of the intron in high Mg2+ or that binding of I-AniI may destabilize this region relative to high Mg2+ concentrations (see below).
Folding of the P7/P3 stacked helix is important for I-AniI binding
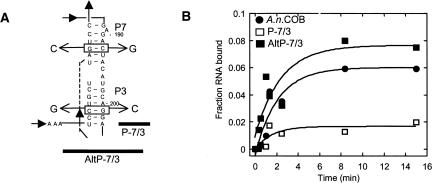
The structure probing experiments showed that binding of I-AniI is associated with extensive changes throughout the A.n. COB intron RNA that lead to a catalytically active structure. However, not all of these changes are necessarily important for encounter or native complex formation. One of the most dramatic changes outside of the I-AniI binding site involves folding of the P3 and P7 helices (Figs. 1, 2). To assess the effects of disrupting P7 and P3 on I-AniI binding, we created an intron RNA (P-7/3) containing mutations in both P7 (C192G) and P3 (G201C; Fig. 4A). To test if P7/P3 nucleotide changes affected I-AniI binding to the full-length A.n. COB pre-RNA, we separately measured the rates of association and dissociation. For two-step binding by I-AniI, the association rate for complex formation is described by the equation: k obs = K 1 k 2[I-AniI]/(K 1[I-AniI] +1), where K 1 is the equilibrium constant describing the formation of the encounter complex and k 2 is the rate constant that describes the transition from the encounter to the native complex (Solem et al. 2002). The encounter complex equilibrium constant (1/K 1) for the full-length RNA is ∼13 nM. Negative effects on binding such as an increase in 1/K 1 and/or decrease in k 2 can be determined by measuring the rate of complex formation at concentrations of I-AniI below 1/K 1. Therefore, we performed these experiments at 4 nM protein (Fig. 4B). The k obs for association at 4 nM I-AniI was around twofold slower for P-7/3 relative to A.n. COB pre-RNA (0.31 ± 0.1 versus 0.66 ± 0.2). The slower association rate of P-7/3 could reflect a change in the equilibrium constant for the encounter complex and/or in the isomerization rate of the encounter to the native complex (Solem et al. 2002). The amplitude of binding was also reduced for P-7/3 relative to A.n. COB pre-RNA (0.04 ± 0.03 versus 0.07 ± 0.03), but it is difficult to interpret the significance of this difference due to the large error associated with this measurement for the P-7/3 mutant. This considerable variability in amplitude between different preparations of P-7/3 RNAs may reflect a propensity for this mutant to mis-fold into a conformation that is unable to bind I-AniI. Compensatory mutations (AltP-7/3) restored the association rate and amplitude of RNA bound to that of the wild-type full-length A.n. COB pre-RNA (Fig. 4B). Interestingly, we observed no effect on the stability of the native complex as judged by rates of dissociation with the P-7/3 mutation (data not shown). Thus, it appears that the mutations primarily affect formation of the complex. In the following section we show that folding of both P7 and P3 precedes native complex formation providing evidence that the mutations most likely affect encounter complex formation.
FIGURE 4.
Mutations in P7 and P3 negatively affect binding. (A) Location of the mutations. Only the P7/3 helix is shown. P-7/3 has changes in P7 (C192G) and P3 (G201C) that disrupt base-pairing in the middle of both stems. Compensatory mutations in AltP-7/3 restore base-pairing in both helices. (B) Association kinetics for A.n. COB pre-RNA, P-7/3 and AltP-7/3. Internally labeled RNA was incubated with 4 nM protein, and at various times a molar excess of unlabeled A.n. COB pre-RNA was added to stop binding and the reactions filtered on nitrocellulose. The data were fit to a single exponential to obtain k obs. For three independent experiments, the k obs were: A.n. COB pre-RNA: 0.66 (±0.17) min−1, P-7/3: 0.31 (±0.1) min−1, and AltP-7/3: 0.56 (±0.17) min−1. The k obs at 4 nM I-AniI is essentially the same as measured previously [0.63 (±0.03) min−1; Solem et al. 2002].
Defining the encounter to native complex transition in the I-AniI/A.n. COB binding pathway
The structure mapping experiments defined the folded states of the A.n. COB pre-RNA prior to and after native complex formation. In order to further characterize the encounter complex and gain insight into the assembly pathway, we used a kinetic oligonucleotide hybridization technique (Zarrinkar and Williamson 1994). In these experiments, complex formation was initiated by the addition of I-AniI and the fraction of RNA accessible to oligonucleotide binding was determined by rapid (10 sec) RNase H cleavage of the RNA/oligonucleotide complex. By sampling the I-AniI/A.n. COB pre-RNA complex at various times after mixing, intermediate states along the binding pathway were defined. The assays were carried out with sufficient concentrations of protein to drive all of the RNA into the encounter complex. Thus, structural changes associated with the encounter complex are expected to occur rapidly while those associated with the transition to the native complex are expected to occur with a rate greater than or equal to that measured previously [2.7 min−1 (±0.2)] (Solem et al. 2002).
We tested a battery of oligonucleotides that hybridize along the length of the A.n. COB pre-RNA including the 3′ end of the pre-RNA, P7, P5ab, P5, P4/6, P3, P2, and P1 (Fig. 2). We observed no change in oligonucleotide hybridization in the 3′ end (accessible), P5ab, and P5 (both inaccessible) when I-AniI was added (data not shown, but see Fig. 2). In contrast, for P3, P4/6, and P7, oligonucleotide accessibility was reduced rapidly and dramatically after addition of I-AniI, suggesting that binding induced or stabilized helix formation in these elements (Fig. 5A, data not shown). The rapid formation of P3, P4/6, and P7 suggested that the A.n. COB pre-RNA in the encounter complex had undergone significant structural changes.
The transition to the native complex was reflected by changes in accessibility to the P1–P2 stacked helix. As shown in Figure 5A, P2 was minimally accessible to oligonucleotide hybridization in the absence of I-AniI (∼15% cleaved). However, addition of the protein induced a (relatively) slow increase in accessibility. A plot of the fraction RNA cleaved versus time revealed that the rate of induced accessibility was 3.0 (±0.4) min−1, which was very close to the rate for the transition from the encounter to the stable complex measured previously (Fig. 5B; Solem et al. 2002). We also observed increased accessibility to P1, although this helix was partially accessible in the absence of protein (∼25% cleaved; Fig. 5A,B). Again, the rate of increased accessibility was 3.0 (±0.3) min−1, which was close to the transition from the encounter to the stable complex. Accessibility to both P1 and P2 peaks after 1 min and then decreases.
In order to more accurately assess the fraction of I-AniI/RNA complexes with accessible P1 and P2 helices, we have probed the pre-RNA using a 1-min RNase H incubation time after binding proceeded for 10, 20, and 40 sec (Fig. 5C). These data show that a significant fraction of P1 and P2 is accessible in a time-dependent manner: specifically over 70% of RNA is cleaved at P2 and over 80% cleaved at P1 after binding is allowed to proceed for 40 sec. Thus, these observations support the idea that a pre-RNA with destabilized P1 and P2 helices likely represents an “on-pathway” intermediate in the binding pathway.
To address RNA folding on a more global scale, we used a kinetic iodine cleavage method. Complex formation with RNA substituted with guanosine phosphorothioates was initiated by the addition of I-AniI and accessible regions to iodine cleavage were assessed at various times after mixing. All of these changes occurred rapidly after the addition of I-AniI (Fig. 5D, data not shown). For example, protection from iodine cleavage in P3, P5a, and P5b occurred prior to the first time point (20 sec). Collectively, these data suggested that much of the A.n. COB pre-RNA folds rapidly after protein binding.
DISCUSSION
Binding of the I-AniI maturase to its cognate group I intron results in a hierarchical RNA folding pathway that ultimately leads to native complex formation and self-splicing activity. There are at least two folding transitions that occur upon addition of protein: one involving the formation of four central helices in the catalytic core, and another associated with destabilization of the splice site containing stacked helices. The finding that impeding or eliminating one of the folding steps by mutation negatively affects binding reveals the strong interdependence between RNA folding and I-AniI binding. When compared to other group I intron cofactors, folding of the intron by both stabilization and destabilization of specific RNA structures appears to be a unique feature of the I-AniI maturase.
Assembly of I-AniI/A.n. COB pre-RNA complex
Prior to binding of the protein, the A.n. COB intron is incompletely folded, since only helices peripheral to the catalytic core appear to be stable. The two stacked helices that make up the ribozyme's central core, P4/P6 and P3/P7, are unstable and as a result the intron RNA appears to lack significant tertiary structure. However, as shown for other group I intron RNAs, under our experimental conditions (5 mM Mg2+), interactions between metal ions and the polynucleotide chain likely drive the A.n. COB pre-RNA into a collapsed state that globally resembles the native state but lacks some secondary structure and stable tertiary structure (Buchmueller et al. 2000; Russell et al. 2000; Woodson 2000). Indeed, in 5 mM Mg2+, most of the intron peripheral helices are not accessible to various probes, suggesting a partially folded structure (Figs. 1, 2). However, although the intron is likely compact, it is still largely accessible to solvent (Fig. 3), as expected for collapsed, fluid RNA structures (Buchmueller et al. 2000).
Two RNA folding transitions result as I-AniI engages the collapsed A.n. COB intron (Fig. 5). In the first, the P4/6 and P3/7 stacked helices are rapidly stabilized and much of the global tertiary structure appears to be established (Fig. 5). These events occur on the time scale of encounter complex formation as measured previously (>10 sec) (Solem et al. 2002). This intermediate is not active in splicing, although the iodine cleavage patterns are consistent with tertiary structure formation (Fig. 5D; Solem et al. 2002). Presumably more subtle rearrangements in the catalytic core, not accessible by our current experiments, must occur prior to splicing activation and native complex formation (see below). Nonetheless, this transition is important for binding since mutations that destabilize P3 and P7 negatively affect binding (Fig. 4).
The second RNA folding transition is characterized by weakening of the P1 and P2 helices. Accessibility to oligonucleotide probes occurs at a rate of 3 min−1, which is essentially the same rate as the transition from encounter to native splicing active complex measured previously (Solem et al. 2002). These changes likely precede or are a part of this transition. The increase in accessibility presumably reflects a more dynamic structure of both P1 and P2. The importance of both helices to I-AniI binding is supported by the observation that deletion of P1/P2 decreases binding affinity by 4700-fold (Solem et al. 2002).
Stabilization of RNA structure is a hallmark of all group I intron splicing cofactors. High-affinity cofactors bind to correctly folded RNAs to stabilize structure, whereas RNA chaperones destabilize misfolded RNA structures to facilitate splicing (Lambowitz et al. 1999). I-AniI is the first high-affinity splicing cofactor to show both stabilizing and destabilizing effects on specific intron structures. I-AniI appears to stabilize intron core structure by binding directly to P5a, P4/P6, and P9 that are brought into close proximity by higher-order RNA folding.
It is more difficult to understand how binding of I-AniI in the encounter complex destabilizes P1 and P2 structure. One possibility is that I-AniI transiently interacts with P1 and P2 and thereby promotes destabilization. Protein-induced helix destabilization has been demonstrated in the case of the human pancreatic ribonuclease, the Cbp2p group I intron splicing cofactor, as well as many RNA chaperones (Sorrentino et al. 2003; Rajkowitsch et al. 2005; Bokinsky et al. 2006). Nonspecific, electrostatic interactions with the stacked helix may cause destabilization and subsequent refolding.
Protein-induced intron RNA folding events could also “force” a weakening of P1 and P2. Support for such a scenario comes from the observation that Mg2+-dependent tertiary interactions facilitate substantial rearrangements of secondary structure in the P5abc subdomain of the Tetrahymena intron (Wu and Tinoco 1998). It may be that the rapid folding of the intron core or binding itself generates torsional stress that is relieved by destabilization of P1 and P2. Another possible mechanism for weakening P1 and P2 comes from the observation that ligand-induced protein conformational changes have been implicated in transiently destabilizing RNA structure (Tompa and Csermely 2004). In this regard we have detected a specific conformational change in the C-terminal region of I-AniI upon binding the A.n. COB pre-RNA (A. Shumaker and M.G. Caprara, in prep.). The functional significance of this change with regard to RNA folding is currently under investigation.
RNA folding dynamics in an RNP intermediate
What is the functional relevance of destabilizing P1 and P2 to I-AniI binding? It was previously noted that deletion of P1 or P1 and P2 together significantly reduced the fraction of RNA in the native complex, suggesting that these elements were important for formation of the I-AniI binding site (Solem et al. 2002). The P1–P2 stacked helix and I-AniI bind on opposite surfaces of P4/P6/P6a, and therefore, the P1–P2 contribution to protein binding is most likely indirect (see Fig. 3D). Given the importance of these elements to native I-AniI binding, the changes we observe upon protein association likely reflect conformational changes that are necessary for native complex formation.
Destabilization of P1–P2 may facilitate docking of the domain that leads to further structural rearrangements in the core. In this regard, it is important to note that the rates of protein-dependent splicing, destabilization of P1 and P2, as well as the formation of the native complex, are similar, and thus report the same “slow” step in assembly. Since docking of P1–P2 must precede splicing, the similarity in rates suggests that docking occurs rapidly after these changes in P1–P2 are introduced. One model takes into account the finding that P1 and P2 are moderately stable in the absence of protein. Once the first folding transition has occurred, P1 and P2 may become trapped in inappropriate positions along the catalytic core in the encounter complex (Fig. 6). Misfolded intermediates in group I introns and other RNAs are very common, and their resolution can be rate limiting for folding (for review, see Thirumalai and Hyeon 2005). Increased flexibility of the P1 and P2, possibly stimulated by torsional stress incurred by protein binding and/or intron core folding, may be important to properly position the helix in the catalytic core. Consistent with this hypothesis is the finding that decreasing the thermostability and increasing the degrees of freedom between P1 and the Tetrahymena intron core increases the rate of docking ∼10-fold (Bartley et al. 2003). Furthermore, it has been shown that docking of P1 requires, and may facilitate, conformational changes in the structure of the P4 helix that contains part of the I-AniI binding site (Bartley et al. 2003; Adams et al. 2004). Therefore, in this model, the initial binding of I-AniI propagates changes through the RNA that ultimately “feedback” to facilitate formation of its specific binding site (Fig. 6).
FIGURE 6.
Model for A.n. COB pre-RNA/I-AniI assembly. Although important for binding, the P3–P7 domain is not shown for simplicity. Dashed lines indicate single-stranded or destabilized double-stranded regions and the cylinders represent helices. Prior to binding, the P4 and P6 helices in the core are unstable and, as a consequence, P1 and P2 are not properly positioned in the core. In the first step of binding, I-AniI associates with the intron RNA in a weak complex and promotes folding of the catalytic core. Within this complex, P1 and P2 become destabilized and subsequently dock into its intron's catalytic site that leads to the first step in splicing. The P1/P2 docked structure is inferred, since I-AniI binding results in splicing activation (see Discussion). This docking event may cause subtle rearrangements in the I-AniI binding site in P4, resulting in tighter binding and formation of the native complex. The relative orientation of the helices in the free RNA and intermediate complexes is not intended to reflect a specific conformation. I-AniI may dissociate from the RNA in the encounter complexes because of their short half-lives, but once in the native complex, binding is essentially irreversible (Solem et al. 2002).
Since it appears that protein binding is linked to P1–P2 association with the core, this allosteric-feedback model assures that protein binding is tightly coupled to RNA splicing. I-AniI appears unique in this regard, as other group I intron protein cofactors do not require P1–P2 structures for high-affinity binding, and in such cases splicing can be slow relative to the rate of protein binding (Saldanha et al. 1995; Webb and Weeks 2001; Longo et al. 2005). For example, binding by the Neurospora crassa mt tyrosyl tRNA synthetase protein results in stable, nonnative complexes that require an RNA helicase protein for native complex formation in vivo (Mohr et al. 2002).
An essential feature of the allosteric feedback mechanism proposed here is that proteins can influence RNA folding events far from their binding sites. Although local changes in RNA structure near protein binding sites have been routinely documented, distant effects are now becoming appreciated. For example, the bacterial L36 and yeast L3 ribosomal proteins affect RNA structure 60–100 Å away from their respective binding sites (Petrov et al. 2004; Maeder and Draper 2005). In both cases these changes are important for ribosomal function. Also, the signal recognition particle SRP19 protein changes the folding of 7S RNA 40 Å from its binding site (Kuglstatter et al. 2002; Hainzl et al. 2005). This conformational change is essential for the subsequent binding of SRP54 protein and SRP function. Thus, it seems likely that propagation of a protein binding “signal” throughout RNA is a common feature of many complex RNA/protein assemblies, and this property can be tightly coupled to biological function.
Not all maturases are alike
Finally, group I intron maturases are derived from homing DNA endonucleases, which adapted to function in splicing introns that encode them (Caprara and Waring 2005). It was recently shown that a close relative of I-AniI, the bi3 maturase from yeast, binds an RNA containing only the P4–P6 and P5ab domains of the mt COB gene (Longo et al. 2005). In contrast, I-AniI is incapable of binding the comparable, isolated A.n. COB structure (data not shown; Solem et al. 2002). The intron-binding site for the yeast maturase was mapped in the P5b stem–loop, whose sequence and structure are quite different from that of A.n. COB intron. In fact, deletion of this structure from the A.n. COB intron does not affect protein-dependent splicing (Geese and Waring 2001). These observations are consistent with the idea that the mechanisms of maturase recognition and facilitation of RNA folding can differ significantly even among closely related maturases (Downing et al. 2005). This most likely is a reflection of the different evolutionary paths that each intron/maturase pair follows during the process of adaptation to protein-assisted splicing.
MATERIALS AND METHODS
DNA templates, RNA transcription, and protein purification
A.n. COB intron containing pre-RNAs were transcribed from BamHI digested pT7AnCOBme (Solem et al. 2002). The P-7/3 template was constructed by overlapping PCR (Ho et al. 1989) using two overlapping mutagenic primers that simultaneously replace P7 C192 with a G and P3 G201 with a C (P-7AGAAGCCTCTCAGAGAGTAGTACATGCACGAGATCTTATGTC; P-3ARGACATAAGATCTCGTGCATGTACTCTCTGAGAGGCTTC; substitutions are underlined) and COB5E and COB3E, which hybridize to the ends of the A.n. COB gene in pT7AnCOBme. PCR and cloning were done as described by Solem et al. (2002). AltP-7/3 was constructed using two mutagenic oligonucleotides that change P3 C46 to a G and P7 G234 to a C as well as COB5E and COB3E using the P-7/3 plasmid as a template. Mutant RNAs were transcribed from plasmid digested with BamHI.
Transcription and purification of 32P-internally labeled and end-labeled RNAs were carried out as described by Solem et al. (2002) and Downing et al. (2005). For iodine structure mapping experiments, RNAs were prepared by adding NTPαS to the transcription reaction. All NTPαS analogs were purchased from Glen Research.
RNA structure-mapping experiments
For DEPC modification reactions, 5′-end-labeled full-length A.n. COB pre-RNA (80 nM) was incubated in the presence of 25 mM HEPES, pH7.5, 100 mM NaCl, and supplemented with 5 or 75 mM Mg2+ in the absence of protein at 37°C for 20 min. Protein was added (160 nM) to the appropriate reactions and incubated for 5 min followed by addition of 1 μL DEPC. The modification reactions were terminated by addition of 50 mM EDTA and 10 μg of Escherichia coli tRNA. The RNAs were extracted with phenol-chloroform-isoamyl alcohol (PCI; 25:24:1 v/v/v), ethanol precipitated twice and dried. The samples were treated with aniline/acetate buffer, pH 4.5, and incubated at 60°C for 15 min to cleave at the modified positions, and ethanol precipitated twice before being resuspended in water (Peattie 1979). The reaction products were separated on a series of denaturing 6% polyacrylamide/7 M urea gel, and the dried gels were quantified with a PhosphorImager (Molecular Dynamics). The data were analyzed by subtracting background (based on mock control samples) and band intensities were normalized for each lane with respect to the precursor RNA to account for any loading differences in each lane. Residues were scored protected if the band intensity was <50% of those under the 0 mM Mg2+ condition (Caprara et al. 1996).
For iodine footprinting experiments, 5′-end-labeled phosphorothioate-containing A.n. COB pre-RNA (30 nM) was incubated in the presence of 25 mM Tris-HCl, pH 7.5, 100 mM NaCl (TN), and supplemented with 5 or 75 mM Mg2+ in the absence of protein at 37°C for 10 min. Protein was added (150 nM) to the appropriate reactions and incubated for 5 min followed by addition of 2 μL of 1 mM iodine (Sigma Chemical Company) in ethanol. After 5 min, the cleavage reaction was stopped by adding 2 μL of β-mercaptoethanol and 10 μg of E. coli tRNA followed by organic extraction and ethanol precipitation. The reaction products were separated on a series of denaturing 6% polyacrylamide/7 M urea gels and the dried gels were quantified with a PhosphorImager. The data were analyzed as described above for the DEPC experiments.
For oligonucleotide accessibility experiments, internally labeled RNA (0.5 nM) was incubated in 15 μL TN containing 5 mM MgCl2 for 10 min 37°C. A mixture containing 1 μL of oligonucleotide (50 μM) and 0.5 μL of RNase H (5 U; USB Corporation) was added and the reaction incubated for 1 min. Aurin tricarboxylic acid (2 μL of a 5 mg/mL) was added to stop the reaction, and the reaction products were separated on a denaturing 6% polyacrylamide/7 M urea gel and visualized with a PhosphorImager.
Time-resolved structure mapping
Oligonucleotide accessibility time courses were performed with 0.5 nM internally labeled RNA incubated in 13.5 μL TN containing 5 mM MgCl2 for 10 min at 37°C. Reactions were initiated by mixing 13.5 μL of the RNA mixture with 1.5 μL of a 5 μM concentration of I-AniI prewarmed at 37°C. The reaction was incubated for 10–300 sec at 37°C. At each time point, 1 μL of oligonucleotide (50 μM) and 2 μL of RNase H (5 U) were added and the reaction incubated for 10 sec. Aurin tricarboxylic acid (2 μL of a 5 mg/mL solution) was added to stop the reaction, and the reaction products were separated on a denaturing 6% polyacrylamide/7 M urea gel and the dried gels were quantified with a PhosphorImager.
For time-resolved iodine cleavage experiments, 25 nM 5′-end-labeled GαS containing RNA was incubated in 13.5 μL TN containing 5 mM MgCl2 for 10 min at 37°C. Reactions were initiated by mixing 13.5 μL of the RNA mixture with 1.5 μL of a 2.6 μM concentration of I-AniI prewarmed at 37°C. The reaction was incubated for 10–300 sec at 37°C. At each time point, 2 μL of 1 mM iodine in ethanol was added and the reaction incubated for 10 sec. β-Mercaptoethanol (2 μL of a 100% solution) was added to stop the reaction, and the reaction products were separated on a denaturing 6% polyacrylamide/7 M urea gel and the dried gels were quantified with a PhosphorImager.
RNA binding experiments
Dissociation kinetics were performed and analyzed as described by Solem et al. (2002) and Downing et al. (2005). The association kinetic experiments were performed by preincubating 50 pM 32P-labeled RNA in 50 mM Tris-HCl, pH-7.5, 100 mM NaCl, 5 mM Mg2+, 10 mM DTT, 10% glycerol, and 0.1 μg/μL BSA at 37°C for 20 min. Reactions were initiated by mixing 100 μL of the RNA mixture with 2 μL of a 200 nM concentration of I-AniI prewarmed at 37°C. The reaction was incubated for 10–900 sec at 37°C. Complex formation was stopped by the addition of unlabeled A.n. COB pre-RNA to a concentration of 100 nM and subsequently filtered through nitrocellulose. The filters were washed with 3 mL of TN, dried, and counted using a scintillation counter. The data were fit to a first-order equation: fraction RNA bound = A(1 − e −kt), where A is the amplitude of RNA bound and k represents k obs for association.
ACKNOWLEDGMENTS
We thank Drs. Timothy Nilsen, Pieter deHaseth, and Edward Turk (CWRU) for critical reading of the manuscript, and Min Zhu for help with the manuscript preparation. B.J.K. was supported in part by a Cell and Molecular Biology Training Grant awarded through NIGMS. National Institutes of Health Grant GM-62853 to M.G.C. supported this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.307907.
REFERENCES
- Adams, P.L., Stahley, M.R., Gill, M.L., Kosek, A.B., Wang, J., Strobel, S.A. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain, F.H., Gubser, C.C., Howe, P.W., Nagai, K., Neuhaus, D., Varani, G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- Avis, J.M., Allain, F.H., Howe, P.W., Varani, G., Nagai, K., Neuhaus, D. Solution structure of the N-terminal RNP domain of U1A protein: The role of C-terminal residues in structure stability and RNA binding. J. Mol. Biol. 1996;257:398–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- Bartley, L.E., Zhuang, X., Das, R., Chu, S., Herschlag, D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Williamson, J.R. Effects of polyvalent cations on the folding of an rRNA three-way junction and binding of ribosomal protein S15. RNA. 1998;4:984–997. doi: 10.1017/s1355838298980426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky, G., Nivon, L.G., Liu, S., Chai, G., Hong, M., Weeks, K.M., Zhuang, X. Two distinct binding modes of a protein cofactor with its target RNA. J. Mol. Biol. 2006;361:771–784. doi: 10.1016/j.jmb.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmueller, K.L., Webb, A.E., Richardson, D.A., Weeks, K.M. A collapsed nonnative RNA folding state. Nat. Struct. Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- Caprara, M.G., Waring, R.B. Group I Introns and their maturases: Uninvited, but welcome guests. In: Belfort M., et al., editors. Homing endonucleases and inteins. Springer-Verlag; Heidelberg: 2005. [Google Scholar]
- Caprara, M.G., Mohr, G., Lambowitz, A.M. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., Doudna, J.A. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Chatterjee, P., Brady, K.L., Solem, A., Ho, Y., Caprara, M.G. Functionally distinct nucleic acid binding sites for a group I intron encoded RNA maturase/DNA homing endonuclease. J. Mol. Biol. 2003;329:239–251. doi: 10.1016/s0022-2836(03)00426-1. [DOI] [PubMed] [Google Scholar]
- DiNitto, J.P., Huber, P.W. Mutual induced fit binding of Xenopus ribosomal protein L5 to 5S rRNA. J. Mol. Biol. 2003;330:979–992. doi: 10.1016/s0022-2836(03)00685-5. [DOI] [PubMed] [Google Scholar]
- Downing, M.E., Brady, K.L., Caprara, M.G. A C-terminal fragment of an intron-encoded maturase is sufficient for promoting group I intron splicing. RNA. 2005;11:437–446. doi: 10.1261/rna.7225205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann, C., Baudin, F., Mougel, M., Romby, P., Ebel, J.P., Ehresmann, B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geese, W.J., Waring, R.B. A comprehensive characterization of a group IB intron and its encoded maturase reveals that protein-assisted splicing requires an almost intact intron RNA. J. Mol. Biol. 2001;308:609–622. doi: 10.1006/jmbi.2001.4609. [DOI] [PubMed] [Google Scholar]
- Golden, B.L., Kim, H., Chase, E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- Gubser, C.C., Varani, G. Structure of the polyadenylation regulatory element of the human U1A pre-mRNA 3′-untranslated region and interaction with the U1A protein. Biochemistry. 1996;35:2253–2267. doi: 10.1021/bi952319f. [DOI] [PubMed] [Google Scholar]
- Guo, F., Gooding, A.R., Cech, T.R. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol. Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Hainzl, T., Huang, S., Sauer-Eriksson, A.E. Structural insights into SRP RNA: An induced fit mechanism for SRP assembly. RNA. 2005;11:1043–1045. doi: 10.1261/rna.2080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Ho, Y., Kim, S.J., Waring, R.B. A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. Proc. Natl. Acad. Sci. 1997;94:8994–8999. doi: 10.1073/pnas.94.17.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuglstatter, A., Oubridge, C., Nagai, K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat. Struct. Biol. 2002;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- Lambowitz, A.M., Caprara, M.G., Zimmerly, S., Perlman, P.S. Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. In: Gesteland R.F., et al., editors. The RNA World II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [Google Scholar]
- Leulliot, N., Varani, G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- Longo, A., Leonard, C.W., Bassi, G.S., Berndt, D., Krahn, J.M., Hall, T.M., Weeks, K.M. Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase. Nat. Struct. Mol. Biol. 2005;12:779–787. doi: 10.1038/nsmb976. [DOI] [PubMed] [Google Scholar]
- Maeder, C., Draper, D.E. A small protein unique to bacteria organizes rRNA tertiary structure over an extensive region of the 50 S ribosomal subunit. J. Mol. Biol. 2005;354:436–446. doi: 10.1016/j.jmb.2005.09.072. [DOI] [PubMed] [Google Scholar]
- Michel, F., Westhof, E. Modeling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Mohr, S., Stryker, J.M., Lambowitz, A.M. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- Peattie, D.A. Direct chemical method for sequencing RNA. Proc. Natl. Acad. Sci. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, A., Meskauskas, A., Dinman, J.D. Ribosomal protein L3: Influence on ribosome structure and function. RNA Biol. 2004;1:59–65. doi: 10.4161/rna.1.1.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch, L., Semrad, K., Mayer, O., Schroeder, R. Assays for the RNA chaperone activity of proteins. Biochem. Soc. Trans. 2005;33:450–456. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- Russell, R., Millett, I.S., Doniach, S., Herschlag, D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat. Struct. Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- Saldanha, R.J., Patel, S.S., Surendran, R., Lee, J.C., Lambowitz, A.M. Involvement of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing. A new method for purifying the protein and characterization of physical and enzymatic properties pertinent to splicing. Biochemistry. 1995;34:1275–1287. doi: 10.1021/bi00004a022. [DOI] [PubMed] [Google Scholar]
- Solem, A., Chatterjee, P., Caprara, M.G. A novel mechanism for protein-assisted group I intron splicing. RNA. 2002;8:412–425. doi: 10.1017/s1355838202029321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino, S., Naddeo, M., Russo, A., D'Alessio, G. Degradation of double-stranded RNA by human pancreatic ribonuclease: Crucial role of noncatalytic basic amino acid residues. Biochemistry. 2003;42:10182–10190. doi: 10.1021/bi030040q. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D., Hyeon, C. RNA and protein folding: Common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- Tompa, P., Csermely, P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Webb, A.E., Weeks, K.M. A collapsed state functions to self-chaperone RNA folding into a native ribonucleoprotein complex. Nat. Struct. Biol. 2001;8:135–140. doi: 10.1038/84124. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. Compact but disordered states of RNA. Nat. Struct. Biol. 2000;7:349–352. doi: 10.1038/75106. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. Structure and assembly of group I introns. Curr. Opin. Struct. Biol. 2005;15:324–330. doi: 10.1016/j.sbi.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Wu, M., Tinoco, I., Jr. RNA folding causes secondary structure rearrangement. Proc. Natl. Acad. Sci. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar, P.P., Williamson, J.R. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]