Abstract
Potexvirus genomes contain conserved terminal elements that are complementary to multiple internal octanucleotide elements. Both local sequences and structures at the 5′ terminus and long-distance interactions between this region and internal elements are important for accumulation of potato virus X (PVX) plus-strand RNA in vivo. In this study, the role of the conserved hexanucleotide motif within SL3 of the 3′ NTR and internal conserved octanucleotide elements in minus-strand RNA synthesis was analyzed using both a template-dependent, PVX RNA-dependent RNA polymerase (RdRp) extract and a protoplast replication system. Template analyses in vitro indicated that 3′ terminal templates of 850 nucleotides (nt), but not 200 nt, supported efficient, minus-strand RNA synthesis. Mutational analyses of the longer templates indicated that optimal transcription requires the hexanucleotide motif in SL3 within the 3′ NTR and the complementary CP octanucleotide element 747 nt upstream. Additional experiments to disrupt interactions between one or more internal conserved elements and the 3′ hexanucleotide element showed that long-distance interactions were necessary for minus-strand RNA synthesis both in vitro and in vivo. Additionally, multiple internal octanucleotide elements could serve as pairing partners with the hexanucleotide element in vivo. These cis-acting elements and interactions correlate in several ways to those previously observed for plus-strand RNA accumulation in vivo, suggesting that dynamic interactions between elements at both termini and the same subset of internal octanucleotide elements are required for both minus- and plus-strand RNA synthesis and potentially other aspects of PVX replication.
Keywords: RNA, replication, PVX, virus, interactions
INTRODUCTION
Both in vitro and in vivo studies on a variety of plus-strand RNA viruses indicate that replication is dependent on a diverse array of sequence and RNA secondary structural elements located throughout the genomes (Buck 1996; Dreher 1999; Kao et al. 2001; Brian and Baric 2005). Synthesis of minus- and plus-strand RNA has been shown to involve local promoter elements near the termini and upstream of subgenomic RNA (sgRNA) species. In some instances, these processes have also been shown to involve long-distance cis interactions among elements throughout the genomic RNA or between termini (Klovins et al. 1998; Kim and Hemenway 1999; Zhang et al. 1999; Frolov et al. 2001; Herold and Andino 2001; You et al. 2001; Lindenbach et al. 2002; Alvarez et al. 2005; Fabian and White 2006; Miller and White 2006), or trans-interactions between different RNA species (Sit et al. 1998; Eckerle and Ball 2002). Depending on the system, these elements and/or interactions can function as part of core transcription promoter elements, enhancers (Sit et al. 1998; Klovins and van Duin 1999; Nagy et al. 1999, 2001; Ray and White 1999, 2003; Sullivan and Ahlquist 1999; Eckerle and Ball 2002; Panavas and Nagy 2003; Ranjith-Kumar et al. 2003; Zhang and Simon 2003; Murray et al. 2004), and/or repressors (Pogany et al. 2003; Zhang and Simon 2003; Zhang et al. 2004). Additionally, dynamic interactions among elements serve as riboswitches to regulate different phases of the replication pathway (Olsthoorn et al. 1999; Koev et al. 2002; Pogany et al. 2003; Goebel et al. 2004; Zhang et al. 2004, 2006).
Requirements for viral minus- and plus-strand RNA synthesis have been extensively studied in our laboratory using potato virus X (PVX) as a model system. As the type member of Potexviruses, PVX is a rod-shaped virus containing a capped and polyadenylated 6438 nucleotides (nt), genome containing five open reading frames (ORFs) (for review, see Batten et al. 2003). ORF1 encodes the 165-kDa replicase-possessing RNA-dependent RNA polymerase (RdRp) activity, and is followed by overlapping reading frames for the triple gene block (TB) proteins of 25, 12, and 8 kDa. ORF5 corresponds to the 25-kDa coat protein (CP). In addition to viral minus- and plus-strand genomic length RNAs, two major sgRNA species are produced during infection—the 2.1- and 0.9-kb RNAs corresponding to TB and CP transcripts, respectively. A third sgRNA species of ~1.4 kb has been observed in some strains of PVX (Kohm et al. 1993), and has been shown to be a template for expression of the TB2 and TB3 proteins in transgenic plants (Verchot et al. 1998).
Our laboratory has investigated the requirements for PVX RNA accumulation in vivo (Kim and Hemenway 1996, 1997, 1999; Miller et al. 1998, 1999; Kim et al. 2002; Pillai-Nair et al. 2003) by quantifying minus- and plus-strand RNA levels in tobacco protoplasts inoculated with wild-type and mutant PVX transcripts. We have also developed a template-dependent in vitro replication system for PVX (Plante et al. 2000). Kim and Hemenway (1996) found that multiple elements in the 5′ NTR of the PVX genome are required for accumulation of both genomic and sgRNA plus-strand RNAs in protoplasts. Conserved elements located upstream of the TB and CP sgRNAs are also important for sgRNA synthesis (Kim and Hemenway 1997). Additionally, interactions between a conserved element in the 5′ NTR and the complementary, conserved octanucleotide elements upstream of TB or CP are needed for optimal accumulation of all plus-strand RNA species (Kim and Hemenway 1999). Thus, accumulation of PVX plus-strand RNAs in vivo is dependent on both local elements and long-distance interactions.
Similarly, Pillai-Nair et al. (2003) found that multiple local elements in the 3′ NTR are required for PVX minus-strand RNA accumulation, but that some of these elements differentially affect minus- and plus-strand RNA levels. The hexanucleotide element previously shown to be important for replication of PVX (Sriskanda et al. 1996) and other potexviruses (White et al. 1992; Chiu et al. 2002), the U-rich motif known to bind host protein in vitro (Sriskanda et al. 1996), and the sequence between these motifs, are important for minus-strand RNA accumulation in protoplasts (Pillai-Nair et al. 2003). In addition, formation of stem–loop 3 (SL3) RNA secondary structure containing the hexanucleotide motif in the terminal loop is also required for PVX minus-strand RNA accumulation. The very 3′ region containing overlapping, potential polyadenylation motifs referred to as near upstream elements (NUEs) is more important for PVX plus-strand RNA accumulation than for minus-strand RNA accumulation (Pillai-Nair et al. 2003) and poly(A) tail length (J. Batten, N. Pillai-Nair, and C. Hemenway, unpubl.). In contrast, 13 nt between the U-rich element and the NUEs are dispensable for minus-strand RNA accumulation. Although these studies suggest that several 3′ NTR elements directly affect PVX minus-strand RNA synthesis in vivo, mutations in these elements may also affect other aspects of the viral replication cycle, such as translation, and contribute to reduced minus-strand RNA levels.
Interpretation of viral RNA synthesis in vivo is usually complicated by other viral processes such as translation and assembly and, potentially, by cellular processes. Similarly, in vitro systems are limited by loss of host factors during RdRp purification, association of replicase complexes with membranes, and other limiting features of extracts. Thus, comparative studies between in vitro and in vivo systems are necessary to fully investigate basic requirements for viral RNA synthesis during viral infection. In this study, we further analyzed the role of the conserved hexanucleotide motif within SL3 of the 3′ NTR and internal conserved octanucleotide elements in minus-strand RNA synthesis using both the template-dependent, PVX RdRp extract system (Plante et al. 2000) and our protoplast replication system (Kim and Hemenway 1996). In assessing template requirements for minus-strand RNA synthesis in vitro, we discovered that, in addition to the 3′ NTR containing the hexanucleotide motif in SL3, the CP conserved octanucleotide element upstream was required for optimal transcription. Additional experiments to disrupt interactions between one or more internal conserved octanucleotide elements or the 3′ hexanucleotide element showed that long-distance interactions are necessary for minus-strand RNA synthesis both in vitro and in vivo. These data suggest that during PVX replication, dynamic interactions between sequences at both termini and the same subset of internal octanucleotide elements are required for both minus- and plus-strand RNA synthesis.
RESULTS
Templates with complementary elements are required for optimal minus-strand RNA synthesis in vitro
To begin to compare sequence and structural requirements for PVX minus-strand RNA synthesis in vitro and in vivo, we analyzed different plus-strand RNA templates from wild-type (wt) and mutant clones using our PVX RdRp in vitro assay (Plante et al. 2000). RCNMV RNA2 was used as a negative control to monitor the template specificity of the plant RdRp extract for PVX RNA. All PVX templates were designed to be co-terminal with the 3′ end of the PVX genome (Fig. 1A). They were obtained either by transcription from the wild-type PVX cDNA clone, p10 (txt 193; nt 6249–6452), which contains 193 nt from the 3′ end, or from PCR DNA fragments 850 nt (pcr 850; nt 5606–6452) or 200 nt (pcr 200; nt 6255–6452) in length. Corresponding RNA templates lacking 10 nt (nt 6368–6377) around the hexanucleotide element in SL3 (Δ10 mutation) were similarly synthesized (Fig. 1). This Δ10 mutation has been shown to exhibit reduced levels of PVX minus-strand RNA in vivo (Pillai-Nair et al. 2003).
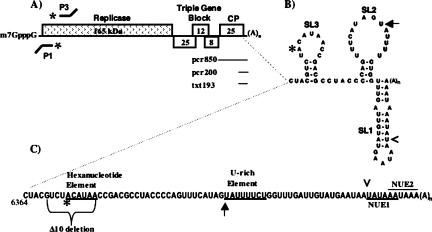
FIGURE 1.
PVX Genome organization and cis-acting elements in the 3′ NTR. (A) The five ORFs are depicted as boxes, with sizes noted in kilodaltons. Probes for S1 nuclease assays are shown above the genome (P3 for minus-strand RNA detection) and below the genome (P1 for plus-strand RNA detection). Templates used for RdRp assays (pcr850, pcr200, and txt 193) are listed below the 3′ region of the genome as horizontal lines. (B) The secondary structure for the PVX 3′ NTR RNA, as determined by Pillai-Nair et al. (2003), is labeled to indicate the three stem–loops, SL1, SL2, and SL3. (C) The linear sequence for the 3′ NTR is shown, along with the hexanucleotide element (ACAUAA, starting with “*”), the U-rich element (UAUUUUCU, starting with the arrow), and the two near upstream elements (NUE1 and NUE2, starting with the caret). The region deleted in the Δ10 mutant is denoted by a bracket below the sequence.
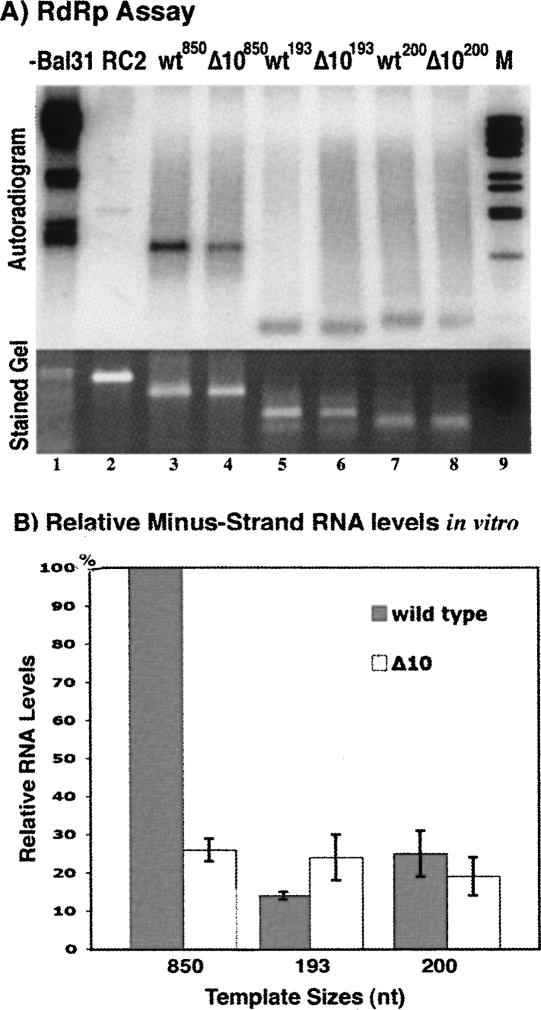
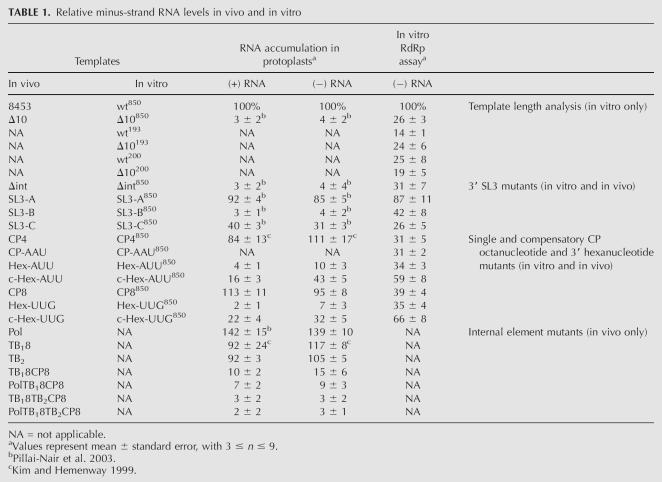
Of the different RNA transcripts added to the RdRp extracts, the wt850 templates (Fig. 2, lane 3) resulted in greater minus-strand RNA synthesis than either the wt193 (Fig. 2, lane 5) or wt200 (Fig. 2, lane 7) templates, which produced 14% and 25% of the wt850 levels, respectively. In addition, only the 850-nt template exhibited sensitivity to the Δ10 deletion (Fig. 2, lane 4), with a reduction to 26% of the wild-type minus-strand RNA synthesis levels. Both Δ10193 and Δ10200 templates performed either as well as or better than the corresponding wild-type templates. These data indicate that the 850-nt-long, 3′ terminal PVX RNA transcript functions as a template for minus-strand RNA synthesis in a manner similar to that observed in vivo (Pillai-Nair et al. 2003). Hence, 850-nt templates were used in the following RdRp assays.
FIGURE 2.
Template selection for in vitro analyses of minus-strand RNA synthesis. (A) Templates derived by transcription of 3′ terminal 850, 200, or 193 nt from either wild-type or Δ10 mutant PVX cDNAs were added to PVX RdRp extract treated with nuclease Bal 31 (template-dependent extract), and reaction products were analyzed by agarose gel electrophoresis. The autoradiogram in the top gel shows products from RdRp assays; the corresponding stained gel below serves as control for input template levels. Lane 1 includes products derived from untreated PVX RdRp extract, and represents endogenous template activity. A double-stranded DNA marker (bacteriophage lambda digested with BstEII) is shown in lane 9. Lanes 2–8 contain products obtained upon addition of templates to nuclease Bal31 treated extracts. Lane 2 contains products obtained with an unrelated viral RNA template, red clover necrotic mosaic virus (RCNMV) RNA2. Other templates tested include 850-nt pcr transcripts from wild type (lane 3) and Δ10 mutant (lane 4), 193-nt transcripts from wild type (lane 5) and Δ10 (lane 6), and 200-nt pcr transcripts from wild type (lane 7) and Δ10 (lane 8). (B) Quantification of RNA levels for templates analyzed in panel A. Wild-type and mutant template values are indicated by gray and white bars, respectively. Error bars represent calculated stand error values for experiments repeated at least three and up to nine times (3 ≤ n ≤ 9).
The hexanucleotide element and SL3 secondary structure in the 3′ NTR are important for minus-strand RNA synthesis in vivo and in vitro
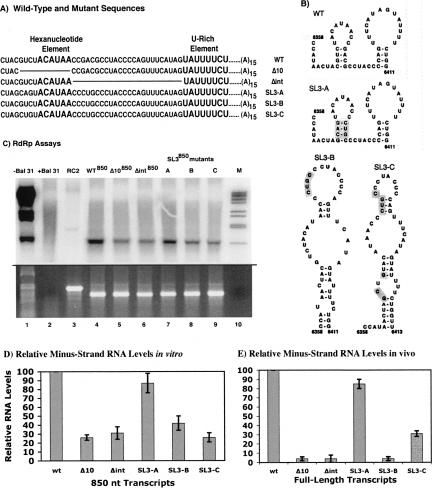
Pillai-Nair et al. (2003) demonstrated that formation of SL3 (nt 6367–6383), which contains the hexanucleotide sequence in the terminal loop, is required for minus-strand RNA accumulation in protoplasts. To further study the role of the conserved hexanucleotide element in minus-strand RNA synthesis in vitro, we tested four additional mutations previously described by Pillai-Nair et al. (2003) in the context of the 850-nt transcripts (Fig. 3A,B). The Δint mutation deletes 24 nt (nt 6378–6401) between the hexanucleotide element and the U-rich region, which is predicted to disrupt SL3 formation. As shown in Figure 3C,D, RNA synthesis from the Δ10 (Fig. 3C, lane 5) and Δint (Fig. 3C, lane 6) templates in the RdRp assays was decreased to 27% and 31% of wild-type levels, respectively. These data are consistent with the effect of these mutations in the protoplast system (Fig. 3E), except the reduction in minus-strand RNA accumulation in vivo was greater, where protoplasts inoculated with Δ10 and Δint mutations within the full-length transcripts exhibited RNA levels decreased to 4% of wild-type levels (Pillai-Nair et al. 2003).
FIGURE 3.
Effects of mutations in the hexanucleotide region on minus-strand RNA synthesis. (A) Sequences extending from just upstream of the hexanucleotide element and through the U-rich element of the PVX 3′ NTR are shown for wild-type and mutant templates. The regions of sequence deleted in mutants Δ10 and Δint are indicated by lines within the sequence. Site-directed mutations in SL3 (SL3A, SL3B, SL3C) are indicated by text that is underlined. (B) Corresponding predicted structures for wild-type and SL3 mutants (Pillai-Nair et al. 2003) are illustrated, with shading of altered sequences. (C) Products obtained in RdRp assays with 850-nt templates corresponding to the wild-type and mutant sequences. Lane 1 contains products from endogenous templates in the untreated RdRp extracts. Lane 2 represents a control for nuclease Bal 31 treatment, indicating that endogenous templates have been removed and no specific products are obtained. Lane 10 contains the double-stranded DNA marker described in Figure 2. Lanes 3–9 include products obtained when wild-type and mutant templates were added to nuclease Bal 31 treated extracts. (D) Quantification of the RdRp assay data. (E) Data obtained with corresponding mutant and wild-type full-length transcripts in protoplasts (Pillai-Nair et al. 2003). Error bars are as described in the Figure 2 legend.
Three other mutations tested for minus-strand RNA accumulation in vitro contained alterations to the stem sequence of SL3 (Fig. 3B). In mutant SL3-A, the sides of the stem were swapped without altering formation of the stem. Both SL3-B and SL3-C mutants, which contain changes to only one side or both sides of the stem, respectively, are predicted to form altered conformations that differ from SL3. The SL3-A mutation, previously shown in vivo to exhibit near-wild-type activity for minus-strand RNA accumulation, also exhibits near-wild-type minus-strand RNA synthesis in vitro (Fig. 3C, lane 7; Fig. 3D). Alterations in SL3 structure in mutants SL3-B (Fig. 3C, lane 8) and SL3-C (Fig. 3C, lane 9) resulted in decreased RNA synthesis (42% and 26%, respectively) in vitro, but these decreases were not as large as those observed in vivo (Fig. 3E). These data indicate that both the sequence of the hexanucleotide element and formation of SL3 are important for minus-strand PVX RNA synthesis both in vitro and in vivo.
Long-distance RNA–RNA interactions are required for optimal minus-strand RNA accumulation both in vitro and in vivo
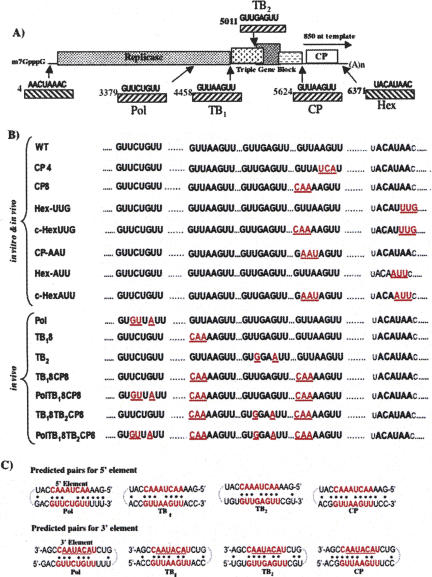
Our previous studies on PVX plus-strand RNA synthesis in protoplasts (Kim and Hemenway 1997, 1999) showed that long-range RNA–RNA interactions between the conserved element at the 5′ end of the PVX genome (nt 4–11; AACUAAAC) and the conserved octanucleotide sequences (GUUAAGUU) located upstream of either the triple gene block (nt 4458–4466) or CP (nt 5624–5632) are critical for optimal plus-strand RNA accumulation (Fig. 4A). Two other conserved sequences within the RdRp coding region (Pol at nt 3379–3386) and upstream of TB2 (TB2 at nt 5011–5018) were also identified as potential pairing partners. The predicted number of base pairs between the 5′ element and these internal elements is either 6 or 7 out of 8 nt (9–10 out of 11 nt if sequence extended beyond the octanucleotide element). Similarly, any of these internal octanucleotide sequences could potentially pair with the complementary hexanucleotide element (ACAUAA) in the terminal loop of SL3 in the 3′ NTR, and thus be important for minus-strand RNA synthesis (Fig. 4A,C).
FIGURE 4.
Mutations introduced into internal and terminal conserved elements for analyses of long-distance RNA–RNA interactions. (A) Positions within the PVX genome of conserved octanucleotide elements near the 5′ terminus (5′), upstream of the triple gene block (TB) and coat protein gene (CP). The conserved hexanucleotide element in the 3′ NTR (Hex) is also shown within an octanucleotide sequence that is also conserved. The 5′ and Hex elements are both complementary to the internal octanucleotide elements. Two other octanucleotide sequences (Pol and TB2) that are complementary to terminal elements are also illustrated. The position of the 850-nt template is shown above the CP gene as a horizontal arrow. (B) Mutations within the conserved elements are noted in red text. The top group of mutants was used to investigate minus-strand RNA synthesis both in vitro and in vivo because they could be tested in the context of both the 850-nt templates and in full-length transcripts. The lower group of mutants was only tested in vivo in the context of the full-length transcripts. (C) Predicted base pairs between the 5′ octanucleotide element or the hexanucleotide element (within the octanucleotide context) and internal elements.
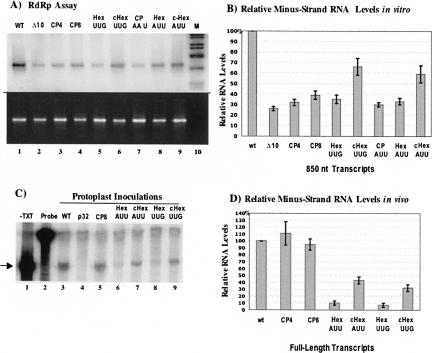
To determine if potential RNA–RNA interactions are required for minus-strand RNA synthesis, we first analyzed mutations (Fig. 4B) in the hexanucleotide element and in CP octanucleotide sequences using the 850-nt templates for in vitro RdRp assays and the full-length templates in protoplasts. Mutants CP4 and CP8 disrupt three out of six possible base pairs with the complementary hexanucleotide motif sequence. These mutants were previously found (Kim and Hemenway 1997) to exhibit decreased plus-strand RNA accumulation in protoplasts, but no significant difference in minus-strand RNA accumulation in the context of the full-length PVX RNA (Kim and Hemenway 1997). The CP-AAU mutant (nt 5625–2657; UUA to AAU) also contains 3 nt changes in the CP octanucleotide, disrupting three out of six potential pairs between the CP and hexanucleotide elements. The Hex-UUG mutant (nt 6376–6378; AAC to UUG) and Hex-AUU mutant (nt 6375–6377; UAA to AUU) were made to change 3 nt of the hexanucleotide element and eliminate three out of six potential base pairs with the upstream CP octanucleotide sequence. The corresponding compensatory mutants, c-HexUUG and c-HexAUU, were designed to restore complementarity and potential base-pairings between CP8/Hex-UUG and CP-AAU/Hex-AUU, respectively.
As shown in Figure 5 and Table 1, the CP4850 (Fig. 5A, lane 3), CP8850 (Fig. 5A, lane 4), and CP-AAU850 (Fig. 5A, lane 7) mutants exhibited reduced minus-strand RNA synthesis in vitro, to 31%, 39%, and 31% of wt850 levels, respectively. This was in contrast to near-wild-type levels produced when similar mutations were analyzed in the context of the full-length PVX transcripts in protoplasts (Fig. 5C,D). These differences suggest that the hexanucleotide element may pair with the other conserved octanucleotide sequences available in the full-length PVX RNA, which are not present in the 850-nt templates analyzed in vitro. In contrast, the hexanucleotide element mutants, Hex-UUG (Fig. 5A, lane 5; Fig. 5B) and Hex-AUU (Fig. 5A, lane 8; Fig. 5B) exhibited decreases in minus-strand RNA synthesis to 35% and 34%, respectively, in vitro, and to 7% and 10%, respectively, in vivo (Fig. 5C,D). These data, further summarized in Table 1, indicate that the sequence of the hexanucleotide element in the 3′ NTR is absolutely required for minus-strand RNA synthesis in vitro and in vivo.
FIGURE 5.
Analyses of single element mutants and compensatory mutants in vitro and in vivo. (A) RdRp assays with 850-nt templates from wild-type and mutant templates. Products obtained upon addition of templates to treated extracts are shown in lanes 1–9. The products from wild-type and Δ10 mutant are shown in lanes 1 and 2. Lanes 3, 4, and 7 contain products from addition of single CP element mutants, CP4, C8, and CP-AAU, respectively. Products from hexanucleotide element mutants, HexUUG and HexAUU, are shown in lanes 5 and 8, respectively. Products from compensatory mutant templates, cHexUUG and cHexAUU, are shown in lanes 6 and 9. Lane 10 contains the double-stranded DNA marker. (B) Quantitative comparison of the relative minus-strand RNA levels from the in vitro data. Error bars are as described in the Figure 2 legend. (C) Minus-strand RNA accumulation in protoplasts electroporated with wild-type and mutant full-length transcripts. Lane 1 serves as a control to indicate the correct protected fragment (noted by the arrow) upon nuclease S1 treatment of the P3 probe (Fig. 1) hybridized to wild-type PVX minus-strand RNA transcript. Lane 2 contains the untreated input P3 probe. Minus-strand RNA products detected by nuclease S1 assays in protoplasts inoculated with wild-type, p32 replication defective control, CP8, HexAUU, cHexAUU, HexUUG, and cHexUUG appear in lanes 3–9, respectively. (D) Quantitative comparison of protoplast data. Error bars are as described in the Figure 2 legend.
TABLE 1.
Relative minus-strand RNA levels in vivo and in vitro
Analyses of the compensatory mutants, c-HexUUG and c-HexAUU, showed that minus-strand RNA accumulation was partially restored both in vitro and in vivo when the mutant hexanucleotide had a complementary partner upstream (Fig. 5; Table 1). Minus-strand RNA levels increased from 34% in RdRp assays with the Hex-AUU mutant templates (Fig. 5A, lane 8) to 59% with the cHex-AUU mutant transcripts (Fig. 5A, lane 9); an increase from 35% to 66% was observed for Hex-UUG and cHex-UUG mutant templates, respectively (Fig. 5A, lanes 5,6; Fig. 5B). Similarly, RNA levels in protoplasts increased from 7% to 10% when inoculated with hexa mutants to 30%–40% of wild-type levels after inoculation with compensatory mutants (Fig. 5C, lanes 6–9; Fig. 5D). These data indicate that in the context of the full-length cDNA clone, the wild-type hexanucleotide element can pair with more than one octanucleotide sequence to initiate minus-strand RNA synthesis, but a mutant hexanucleotide element prevents initiation altogether. In contrast, in the context of the 850-nt template analyses in vitro, mutation in either element results in decreased minus-strand RNA synthesis because the sequence of the hexanucleotide element is required for RNA synthesis and there are no additional complementary partners to supply the required pairing interaction.
Minus-strand RNA synthesis can be inhibited by mutation of multiple, conserved internal elements
Additional mutations were made to further study the potential for the hexanucleotide element in the 3′ NTR to interact with one or more of several conserved octanucleotide sequences upstream. In addition to the TB1 and CP octanucleotide sequences that have been shown to be important in interactions for plus-strand RNA synthesis (Kim and Hemenway 1997), two other elements have been observed (Kim and Hemenway 1999). As shown in Figure 4A, one starts at nt 3379 in the RdRp coding region (Pol) and the other is at nt 5011 upstream of the second TB coding region (TB2). Aside from the complementarity between these two elements and terminal sequences, no other functional significance has been reported for either of them. Consequently, in addition to the single CP octanucleotide mutants (CP4 and CP8) that were made previously (Kim and Hemenway 1997), five new mutants were constructed—single Pol octanucleotide mutant (Pol), double TB1-CP octanucleotide mutant (TB18CP8), two forms of triple octanucleotide mutants (PolTB18CP8 and TB18TB2CP8), and a quadruple mutant (PolTB18TB2CP8). For each element mutated, three nucleotide changes were made to eliminate three potential base-pairings between the conserved octanucleotide element and the hexanucleotide sequence. Changes to the Pol element, which lies within the replicase coding sequence, did not alter the coding sequence. Similarly, for the conserved CP element that lies within the C-terminal part of the third TB coding sequence, mutations in the CP element did not alter this coding sequence.
As shown in Figure 6A,C and Table 1, mutation of the single Pol element resulted in 120% (Fig. 6A, lane 4) of the wild-type minus-strand RNA levels in protoplasts, indicating that this mutation did not affect polymerase activity or minus-strand RNA accumulation. Similarly, mutations of only the CP8 element (Fig. 6A, lane 6; Fig. 6C) or the TB2 element (Fig. 6B, lane 5; Fig. 6C) did not alter levels of minus-strand RNA accumulation. Kim and Hemenway (1997) previously found that both of the single CP8 and TB18 mutants supported wild-type levels of minus-strand RNA accumulation in protoplasts. These data indicate that when one of these internal elements is mutated, others can serve to potentially pair with the hexanucleotide element and facilitate minus-strand RNA synthesis.
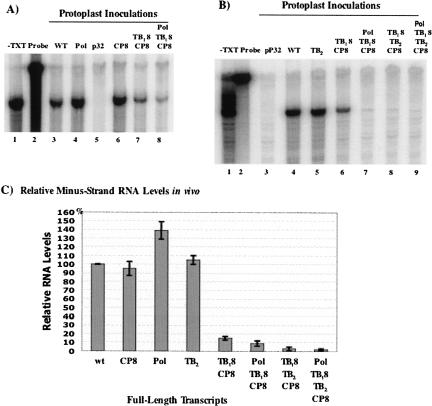
FIGURE 6.
Analyses of interactions between the hexanucleotide element and multiple internal elements in vivo. (A,B) Accumulation of minus-strand RNA in protoplasts inoculated with full-length transcripts from wild-type, single element mutants, or multiple element mutants. Control reactions in lanes 1 and 2 are as described in the Figure 5 legend (panel C). Lanes 3 and 5 in panel A contain protected minus-strand RNA fragments from protoplasts inoculated with wild-type or p32 (replicase defective control), respectively. Products from protoplasts inoculated with single element mutants, Pol and CP8, are in lanes 4 and 6, respectively. Products from the TB8CP8 double element mutant and the PolTB8CP8 triple element mutant are in lanes 7 and 8. Similarly, panel B includes products from protoplasts inoculated with the single element mutant, TB2, (lane 5), double element mutant TB8CP8 (lane 6), and additional multiple element mutants, PolTB8CP8, TB8TB2CP8, and PolTB9TB2CP9 (lanes 7–9). (C) Quantitative representation of data in panels A and B.
In contrast, minus-strand RNA accumulation was significantly decreased to 15% of wild-type levels when the double octanucleotide mutant, TB18CP8 (Fig. 6A, lane 7; Fig. 6C), was tested in protoplasts. When three upstream octanucleotide elements were mutated in PolTB8CP8 (Fig. 6A, lane 8; Fig. 6B, lane 7; Fig. 6C) and in TB18TB2CP8 (Fig. 6B, lane 8; Fig. 6C), the minus-strand RNA levels were further decreased to 9% and 3% of wild-type levels, respectively. When the mutant containing modifications to all four conserved elements, PolTB18TB2CP8, was inoculated into protoplasts, the minus-strand RNA accumulation was severely reduced to 2% (Fig. 6B, lane 9; Fig. 6C) of wild-type levels. As expected, plus-strand RNA levels in protoplasts inoculated with these templates mirrored the minus-strand RNA levels for all mutants tested (Table 1). Collectively, these data show that the hexanucleotide element in the 3′ NTR is important for minus-strand RNA synthesis in vitro and in vivo. This hexanucleotide element interacts with a complementary octanucleotide sequence for optimal RNA synthesis, and any of several octanucleotide elements located upstream at distances of 800 nt or greater may serve as the partner in vivo.
DISCUSSION
Hexanucleotide-dependent minus-strand RNA synthesis in vitro occurs only with longer templates containing a complementary element
Our previous analyses of full-length PVX transcripts containing 3′ NTR mutations in protoplasts (Pillai-Nair et al. 2003) indicated that deletion of the hexanucleotide element and disruption of its placement in the terminal loop of SL3 severely reduced minus-strand RNA levels. Although studies on several other viruses indicated that 3′-terminal elements were sufficient to study template requirements for minus-strand RNA synthesis in vitro (Duggal et al. 1994; Song and Simon 1995; Deiman et al. 1997; van Rossum et al. 1997; Singh and Dreher 1998; Chapman and Kao 1999; Dreher 1999; Turner and Buck 1999; Osman et al. 2000; Sivakumaran et al. 2000; Olsthoorn and Bol 2002; Panavas et al. 2002), we found that templates containing ~200 nt from the 3′ end (including the 72-nt 3′ NTR) were not transcribed well and did not exhibit dependence on the hexanucleotide element within SL3.
Based on our previous studies (Kim and Hemenway 1996, 1997, 1999; Miller et al. 1998) on cis-acting elements required for PVX plus-strand RNA accumulation in vivo, we reasoned that minus-strand RNA synthesis in vitro may also require interactions between complementary elements. Upon testing a PVX transcript extending just upstream of the conserved octanucleotide element in the promoter region for CP sgRNA, we found better transcription in vitro and a dependence on the complementary hexanucleotide element and SL3. In addition, these 850-nt templates were also useful for testing other elements in the 3′ NTR required for minus-strand RNA synthesis in vitro (B. Hu, S, Yoshinari, and C. Hemeway, unpubl.). Although core promoter elements may reside in the PVX 3′-terminal region, interaction between the conserved hexanucleotide element and the complementary conserved element upstream of the CP sgRNA is necessary for efficient transcription of the 850-nt template in vitro. Perhaps this interaction serves an enhancer-like function. For BMV, a replication enhancer-like element for minus-strand RNA initiation is located close to the transcription start site (Ranjith-Kumar et al. 2003). Internal enhancer elements have been shown to be important for replication of several different viruses, with most of these elements functioning in cis (Klovins and van Duin 1999; Nagy et al. 1999, 2001; Ray and White 1999, 2003; Panavas and Nagy 2003; Zhang and Simon 2003; Murray et al. 2004) and a few in trans (Sit et al. 1998; Eckerle and Ball 2002).
Repressor elements positioned upstream of core promoters have been identified in the 3′ regions of tombusvirus genomic and DI-72 RNA (Pogany et al. 2003; Na and White 2006) and the sat-C RNA of turnip crinkle virus (Zhang et al. 2004; Zhang and Simon 2005). These elements and interactions with other regulatory sequences serve to repress transcription in vitro. Although they also appear to function in vivo, they may not act to repress transcription or to the extent observed in vitro. In our system, the interaction occurs over a much longer distance and we observe similar effects upon disruption of interactions in vitro and in vivo, suggesting that interactions between elements are important for optimal RNA synthesis. Also, within the context of the 200 nt PVX transcript used as a template in vitro, the Δ10 mutation was not different from the wild-type template, indicating that the hexanucleotide element is not serving as a repressor in this system. Although we cannot rule out that repressor elements may reside in the smaller PVX templates that we tested or that inclusion of sequences upstream may relieve repression, our data indicate that interactions between the hexanucleotide element and an internal element function similarly in vitro and in vivo during minus-strand RNA synthesis.
Inhibitory effects of mutations on RNA synthesis in vitro are not as great as those observed in vivo
As summarized in Table 1, trends between in vivo and in vitro data were similar in terms of requirements for the hexanucleotide element, SL3 formation, and long-distance interactions between complementary elements, but the decreases in minus-strand RNA levels in vitro were not as great as those observed in vivo when testing mutants. This type of response, with similarity in trend and difference in extent, has also been reported for comparative in vitro and in vivo studies with BMV (Sivakumaran et al. 2003; Hema and Kao 2004). Also, we have never observed a complete inhibition of minus-strand RNA synthesis using our in vitro RdRp system, even with deletion of 60 nt from the 3′ end or when using short templates within the CP coding sequence (B. Hu, S, Yoshinari, and C. Hemeway, unpubl.); there is always some low level of product. This low level of product always corresponds to the size of the input template and is not observed in the absence of added templates; thus, it is not due to residual activity from endogenous templates not removed during extract preparation. This low level of product may reflect that preparation of templates by run-off transcription with bacteriophage T7 polymerase results in a fraction of templates that have not assumed folded structures that allow for interaction between complementary elements. Thus, the percentage of templates that remain unpaired could produce a level of background similar to the lower levels of transcription that we observed with the 200-nt templates.
It may be that preparation of the RdRp extract and removal of the endogenous template to obtain the template-dependent version resulted in a low level of nonspecific initiation by the RdRp complex due to alteration of a fraction of the complexes that renders them less specific. Conversion of the RdRp complexes from their usual cis-preferential form to a template-dependent form that then has to function in trans may account for such altered complexes. Cis-preferential replication of PVX in vivo is supported by data indicating that a replication-defective PVX RNA cannot be complemented in trans by a replication-competent transcript after co-inoculation of these plus-strand templates into protoplasts (K.-H. Kim and C. Hemenway, unpubl. data). In addition, defective RNAs that would be replicated in trans by viral RdRp have been observed for some potexviruses (White et al. 1991; Lin and Hsu 1994), but not for PVX. Thus, it is possible that PVX template requirements using the trans-functioning RdRp in vitro preparation may differ from those observed in vivo.
In contrast to the in vitro system where only transcriptional effects are observed, the greater reductions observed in minus-strand RNA accumulation in protoplasts may be due to a combination of effects on viral transcription, translation, and/or other events. Also, it may be that RNA elements and interactions play a more critical role in setting up replication complexes in vivo than in vitro. Requirements for such RNA elements in this important process have been found for brome mosaic virus (Quadt et al. 1995), alfalfa mosaic virus (Vlot et al. 2001), and cucumber necrosis virus (CNV) (Panaviene et al. 2004, 2005). For the CNV studies, replication complexes were isolated from yeast transformed with replicase proteins, p33 and p92, and either wild-type DI-72 RNA or mutant versions of the DI-72 RNA; these complexes were then used for transcription of added templates. They found that specific regions of the DI were important for efficient transcription of exogenous templates, indicating their involvement in setting up replication complexes. Our in vitro system differs in that replication complexes isolated from infected plants are treated to remove the endogenous template, which is likely to have some impact on the assembled complex. Therefore, although there may still be a need to set up or remodel replication complexes in the PVX RdRp preparations, this process may have less stringent requirements in vitro than in vivo for specific elements or interactions.
Long-distance interactions among conserved elements is a central mechanism for PVX replication
The results presented here indicate that requirements for minus-strand RNA synthesis in vivo and in vitro parallel those reported in our previous plus-strand RNA studies (Kim and Hemenway 1996, 1997, 1999) in several ways. First, the data indicate that interactions between a terminal sequence, the 5′-terminal octanucleotide sequence (nt 4–11) or 3′ hexanucleotide element, and one of the internal conserved elements is necessary for RNA accumulation. Second, the conserved elements at the termini are accessible for pairing. The 5′ octanucleotide element lies near the terminus of the PVX genomic RNA and is part of a region (nt 1–31) that is predicted to be unstructured, both by computer prediction and by solution structural analyses (Miller et al. 1998), which allows for this element to be accessible for interactions elsewhere. Similarly, the 3′ hexanucleotide sequence is in the terminal loop of SL3 in the 3′ NTR of PVX plus-strand RNA (Pillai-Nair et al. 2003). When mutations affect SL3 formation such that the hexanucleotide is no longer predicted to reside in a terminal loop, minus-strand RNA synthesis is inhibited, suggesting that the hexanucleotide element has to be accessible for participation in long-distance RNA–RNA interactions. The local secondary structures of internal conserved octanucleotide elements have not yet been defined.
Third, the multiple internal conserved elements are interchangeable for interactions in vivo. If one of the internal octanucleotide elements is mutated and affects corresponding sgRNA synthesis, it does not preclude the 5′ terminal sequence from pairing with another octanucleotide element elsewhere to support genomic RNA synthesis (Kim and Hemenway 1999). Similarly, several of the internal octanucleotide elements can serve to pair with the hexanucleotide element for initiation of minus-strand RNA synthesis. Mutation of only the Pol, TB1, TB2, or CP elements did not affect minus-strand RNA synthesis in vivo, indicating that they are interchangeable. When both TB1 and CP elements were mutated, minus-strand RNA levels were severely reduced to 15% of wild-type levels. Although these levels were further compromised in the triple and quadruple mutants to 3%–9% of wild-type levels, these data indicate that TB1 and CP elements are more optimal partners for the hexanucleotide motif than the Pol and TB2 elements during initial stages of minus-strand RNA synthesis. However, it is possible that mutations in the TB1 and CP elements affect secondary and/or tertiary structures that might facilitate interactions with the other elements. There may also be additional RNA–RNA contacts that restrict interactions with different elements because of their physical distance from each other on the genome or because of secondary and/or tertiary conformational constraints. Alternatively, there may be a requirement for several pairs of interchangeable partners to allow for dynamic changes during replication; if one pair is removed, the other cannot fully support replication. Replication may also be optimized (potentially by RNA conformation) for utilization of different elements during minus- and plus-strand RNA synthesis and help to regulate levels of each RNA species. Additional templates containing multiple upstream elements will need to be tested in vitro to determine if the element redundancy observed in vivo functions in vitro as well.
Fourth, conserved terminal and internal elements have multiple functions during replication. We have shown that the sequences of the conserved elements, in addition to complementarity, are important for RNA synthesis. This is supported by the observation that when compensatory mutations are made in one of the sg octanucleotide elements and the 5′-terminal sequence, gRNA levels are fully restored, but the corresponding sg plus-strand RNA accumulation is only restored to ~40%–50% of that observed with a wild-type octanucleotide sequence (Kim and Hemenway 1999). Similarly, the hexanucleotide sequence is required for initiation of minus-strand RNA synthesis. Whenever the hexanucleotide element in SL3 is mutated to disrupt interactions with internal octanucleotide elements, minus-strand RNA levels are reduced to near zero in protoplasts and to 20%–30% of wild-type levels in vitro. These levels are only partially restored by compensatory changes in one of the octanucleotide elements. This partial restoration phenotype has also been observed when compensatory mutations were used to analyze interactions associated with transcription of TBSV (Pogany et al. 2003) and TCV sat-C (Zhang et al. 2004). Binding of a recombinant RdRp from another potexvirus, bamboo mosaic virus (BaMV) to the hexanucleotide sequence of BaMV RNA (Huang et al. 2001), further supports the role of this element in transcription. In addition, we know that some mutations in the TB and CP octanucleotide elements that do not affect transcription of sgRNAs still prevent infection in plants, suggesting multiple functions for these elements (Kim and Hemenway 1997). Zhang and Simon (2005) also found phenotypic differences between protoplast and plant infection experiments when analyzing transcription regulatory elements for TCV sat-C. Taken together, these studies indicate that the conserved elements serve to function as promoter sequences, interaction partners, and in some other capacity (possibly translation and/or assembly) during replication. This multifunctionality of cis-acting elements has been reported for other viral systems (Panaviene et al. 2005: Zhang and Simon 2005), reflecting optimization of elements within viral genomes.
Dynamic interactions during PVX replication
Collectively, our data suggest a mechanism for PVX replication in which both termini pair with one or more of the internal octanucleotide elements during replication. As depicted in Figure 7, pairing of the terminal elements with internal elements serves to bring the termini closer together. Shown are interactions between the 5′ element and TB1 octanucleotide and between the 3′ hexanucleotide element and CP octanucleotide. These particular interactions would result in complementary pairs separated by 1159 nt of internal sequence. Pairing of the 5′ element with the TB octanucleotide and the 3′ hexanucleotide with the TB2 element would shorten the internal sequence to 546 nt. Regardless of the actual nucleotide distance between different pairs, the physical distance between them is likely less because of secondary and tertiary folding of the internal sequence. Because our data indicate that disruption of one element does not preclude an interaction elsewhere, there are several potential arrangements that could be achieved depending on which internal elements are involved in the pairing. In addition to the complementary conserved elements already described here, there may be additional sequences and/or structures that interact to facilitate particular interactions during different stages of the replication process.
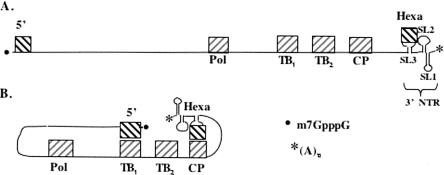
FIGURE 7.
Model for long-distance interactions required for PVX replication. (A) Relative locations of all conserved octanucleotide elements in the PVX genome are illustrated as hatched boxes. The direction of hatch marks in terminal elements are opposite to internal elements to denote complementarity for pairing. The 5′ m7GpppG cap structure and poly(A) tail mark the termini by a black dot and asterisk, respectively. Although not to scale, the three stem–loop structures (SL1, SL2, and SL3) are illustrated in the 3′ NTR, with the hexanucleotide element located in the loop of SL3. (B) One possible paired configuration. Other combinations of both terminal elements pairing with different internal elements are also possible and may be dynamic during replication.
One advantage of the pair illustrated in Figure 7, or other potential pairs, would be to bring the 3′ terminus closer to the polymerase as it is newly translated from this template, with pairings forming during or after translation. Although certain pairs may be more important for initiation of transcription and/or replication complex formation, other pairs may serve to facilitate RNA conformations relevant to the dynamic transcription process. Alternatively, pairing may form prior to translation and facilitate translation-factor-mediated interactions between the capped and polyadenylated termini as a variation on the closed-loop translation mechanism (Kahvejian et al. 2001; Merrick 2004). The multiple possibilities for interactions among the different elements are advantageous in that concomitant processes such as transcription of minus- and plus-strand RNAs, and potentially translation and/or assembly, will undoubtedly require dynamic interactions among these elements. Dynamic RNA conformations or riboswitches have been reported as important for virus replication in other systems (Olsthoorn et al. 1999; Koev et al. 2002; Pogany et al. 2003; Goebel et al. 2004; Zhang et al. 2004, 2006). Zhang et al. (2006) recently utilized temperature gradient gel electrophoresis (TGGE) to establish that TCV sat-C RNA forms an initial conformation that serves as a “pre-active” structure and must undergo conformational change to a phylogenetically inferred conformation or “active form” that is required for transcription. They proposed that interchangeable conformations may also serve to regulate the translation/replication switch.
Although long-distance cis or trans interactions between complementary elements have been found to be important for replication in a variety of virus systems, the PVX system is unique in that interactions between terminal elements and internal elements involve the same subset of conserved internal elements for both minus- and plus-strand RNA synthesis. This arrangement has the potential to position termini dynamically for transcription, translation, assembly, and/or other processes required for replication. Future studies will define the functional significance of multiple pairing partners, relative efficiency and interchangeability of different pairs, the extent to which the Pol and TB2 elements function during PVX replication, and the roles of potential pairing configurations during different stages of replication.
MATERIALS AND METHODS
Materials
Nuclease Bal31 was purified in our laboratory (S. Yoshinari and C. Hemenway, unpubl.) from Pseudoalteromonas espejiana cultures that were obtained from the American Type Culture Collection. Cellulase “Onozuka” R-10 and Pectolyase Y23, enzymes used for plant cell digestions, were purchased from Yakult Honsha Co. Ltd. and Karlan Research Products, respectively. Isotopes [α-32P] CTP and [γ-32P] ATP were purchased from New England Nuclear. Ribonucleotides, deoxynucleotides, and nuclease S1 were products from Roche Applied Science. Custom oligonucleotides for PCR and site-directed mutagenesis were synthesized by Sigma-Genosys. Taq DNA polymerase was obtained from Promega, and T7 RNA polymerase and restriction enzyme and modification enzymes were ordered from New England Biolabs. The Red Clover Necrotic Mosaic Virus (RCNMV) RNA2 cDNA clone was a gift from Dr. Steve Lommel (North Carolina State University).
Construction of mutants
Sequence changes introduced by site-directed mutagenesis into the wild-type PVX cDNA clone, pMON8453 (Hemenway et al. 1990), are shown in Figure 4. Two of the single conserved element mutants upstream of the coat protein gene, CP4 and CP8, were previously constructed by Kim and Hemenway (Kim and Hemenway 1999). Similarly, site-directed mutagenesis was used to generate changes in the conserved hexanucleotide element in the 3′ NTR (mutants Hex-UUG and Hex-AUU), in the element upstream of CP (CP-AAU), and upstream of the second of the triple gene block (TB2), and starting at nt 3379 of a similar sequence in the replicase gene (Pro). Compensatory mutant cHex-UUG was made by resecting mutant Hex-UUG and CP8; compensatory mutant cHex-AAU was made by resecting Hex-AUU and CP-AAU. The double, triple, and quadruple mutants, TB18CP8, PolTB18CP8, and TB18TB2CP8, respectively, were obtained from resection of mutants CP8, TB18, TB2, and Pol. Briefly, CP8 was digested with restrict enzymes Bsu36I and SphI to isolate a DNA fragment containing the mutation. The 1.9-kb fragment from CP8 was inserted into a similarly digested TB8 clone to make TB18CP8. For the triple mutant, PolTB18CP8, the TB18CP8 double mutant was digested with AflII and SphI to isolate a 2.35-kb fragment containing the two mutations. This fragment was ligated into a similarly digested Pol mutant clone. The quadruple mutant, PolTB18TB2CP8, was made by resection of the triple mutant and the single TB2 mutant clones. As the Pol and TB elements occur within the coding sequences, changes made to generate these mutants were designed to maintain the wild-type codons. All mutant constructs were sequenced to confirm the nucleotide changes.
Synthesis of RNA transcripts
Transcripts to be used as templates in the RdRp assays were generated either by run-off transcription from linearized clones or by transcription of PCR products derived from the 3′ regions of wild-type or mutant PVX cDNA clones. Standard PCR DNA amplification was used to generate two sizes of DNA fragments with a T7 RNA polymerase promoter sequence at the 5′ end. DNA fragments were denatured at 94°C, hybridized at 55°C, and elongated at 72°C (1 min each) for 35 cycles. The 850-nt and 200-nt transcripts were derived from PCR fragments synthesized with the 5′ primers, 850Top (5′-TAATACGACTCACTATAGCCACTCTCCGTTGAACG-3′) and 200Top (5′-TAATACGACTCACTATAGGCCAGGGCACAATCC-3′), respectively, and the same 3′ PVX bottom primer (5′-TTTTTTTTTTTTTTTTTTATTTATATTATTCATAC-3′). The p10 PVX cDNA was linearized with SpeI and RCNMV RNA2 cDNA was linearized with HindIII prior to run-off transcription (Kim and Hemenway 1996). For protoplast experiments, SpeI linearized wild-type and mutant full-length PVX clones were used to generate transcripts. The quality and quantity of RNA transcripts were assessed by electrophoresis on 1.4% agarose gels, ethidium bromide (EthBr) staining, and visualization using a UV Transilluminator. Relative mass for each sample was approximated by comparison to RNA standards of known concentration by visual approximation or densitometry scanning of the stained gels. Transcript sample volumes were adjusted to ensure that similar mass amounts were added to RdRp reactions or protoplast assays.
Preparation of template-dependent RdRp extract
Extracts containing PVX RdRp activity were prepared from PVX infected N. benthamiana plants as described by Plante et al. (2000). Fraction 3 derived from the sucrose gradients was dialyzed against Buffer B (50 mM Tris-HCl at pH 8.2, 5 mM MgCl2, 5% glycerol, 1 mM DTT, 1 μM lepeptin, 1μM pepstatin) to remove sucrose prior to subsequent treatments. Removal of the endogenous PVX template from the fraction 3 samples involved treatment of 40 μL extract with 1.5 μL Bal31 (S. Yoshinari and C. Hemenway, unpubl.) in a buffer containing 0.5% (v/v) NP-40 (Sigma) and 5 mM Ca(OAC)2. The extract was incubated at 30°C for 30 min before adding EGTA to a final concentration of 25 mM to stop the reaction.
In vitro RdRp assay
Wild-type or mutant PVX RNA transcripts were dried and carefully resuspended in 10 μL 5× RdRp reaction buffer containing 5mM A, U, GTP, 20% glycerol, 50 mM Tris-HCl (pH 8.2), 5 mM DTT, 20 units RNasin, 10 μg Actinomycin D, 375 mM KCl, and 5 mM MgCl2. RNA templates were then added to 38 μL of nuclease Bal31-treated extracts, along with 20 μCi [α-32P] CTP (800 Ci/mmol). Samples were incubated at 30°C for 1 h, and reactions were stopped by addition of 10 μL of 10% SDS and 130 μL of 10 mM:1 mM Tris-HCl (pH 8.0):EDTA. These reaction conditions were determined to be optimal for all templates tested in this study. Samples were then extracted with phenol and chloroform:iso-amyl alcohol (24:1 v/v) three times, and supernatants were adjusted to 250 mM ammonium acetate (final concentration) prior to addition of 1 μL of 10 mg/mL glycogen and precipitation with 2.5 volumes of 100% cold ethanol. After centrifugation, pellets were resuspended in diethylpyrocarbonate-treated H2O and electrophoresed on 1.4% agarose gels containing 0.05% SDS and 1× Tris-Acetate buffer at 4°C. Gels were stained with ethidium bromide and visualized with a UV Transilluminator before drying to quantify the amount of input RNA transcripts remaining after the RdRp assay. Dried gels were exposed overnight and analyzed with a Molecular Dynamics PhosphorImager and ImageQuant software (Molecular Dynamics) to quantify the bands. Because the ethidium-bromide-stained gels reflect variations in input RNA template levels, calculation of the relative amounts of these input RNA templates was used to adjust values for the corresponding bands on the autoradiograms. This serves to normalize the reactions so product levels can be directly compared. Although transcription of the 850-nt templates would incorporate four times as much label as the 200-nt template, there would be four times as many molecules of the 200-nt template in a given reaction. If both templates worked with similar efficiencies, the product intensities would be equivalent; differences in product levels would reflect dissimilar transcription efficiencies.
Protoplast inoculations and analyses of RNA accumulation
Nicotiana tobaccum (NT-1) cells were cultured and converted to protoplasts as previously described by Pillai-Nair et al. (2003). For protoplast inoculations, 5 μg capped RNA transcripts were diluted into 800 μL electroporation buffer (0.8% [w/v] NaCl, 0.02% [w/v] KCl, 0.02% [w/v] KH2PO4, 0.11% [w/v] Na2HPO4, 0.4 M Mannitol at pH 6.5) and added to 800 μL protoplasts (5×106 cells/mL) using a Bio-Rad Gene Pusler (250 V and 500 μF). Cells were placed on ice for 15 min and then plated into 15 mL of NT-1 protoplast media (0.43% [w/v] MS salts, 88 mM sucrose, 55 mM myo-inositol, 0.3 mM thiamine, 440 mM KH2PO4, 0.4 mg/mL of 2,4-dichlorophenoxy-acetic acid, 0.4 M mannitol). Cultures were incubated at 25°C (14 h light/14 h dark) for 40 h. These protoplasts were harvested and total RNA was isolated as described by Pillai-Nair et al. (2003).
For analyses of viral RNA accumulation in inoculated protoplasts, nuclease S1 assays were carried out as described previously (Kim and Hemenway 1996; Pillai-Nair et al. 2003). The P1 and P3 single-stranded DNA probes (Fig. 1A) for detection of plus- and minus-strand PVX RNAs, respectively, were prepared as previously described (Kim and Hemenway 1996). Protected fragments were subsequently resolved on 8% polyacrylamide–7 M urea gels. Products were visualized on dried gels using a PhosphorImager and analyzed with ImageQuant software.
ACKNOWLEDGMENTS
We thank Dr. Paul Wollenzien, Dr. David Brian, and Dr. Anne Simon for critically reading the manuscript and Stephanie Barefoot for technical assistance. This research was supported by NIH grant GM049841 to C.H.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.243607.
REFERENCES
- Alvarez, D.E., Lodeiro, M.F., Luduena, S.J., Pietrasanta, L.I., Gamarnik, A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten, J., Yoshinari, S., Hemenway, C.L. Potato virus X: A model system for virus replication, movement and gene expression. Mol. Plant Pathol. 2003;4:125–131. doi: 10.1046/j.1364-3703.2003.00156.x. [DOI] [PubMed] [Google Scholar]
- Brian, D.A., Baric, R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, K.W. Comparison of the replication of postive-stranded RNA viruses of plants and animals. Adv. Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, M.R., Kao, C.C. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- Chiu, W.W., Hsu, Y.H., Tsai, C.H. Specificity analysis of the conserved hexanucleotides for the replication of bamboo mosaic potexvirus RNA. Virus Res. 2002;83:159–167. doi: 10.1016/s0168-1702(01)00433-6. [DOI] [PubMed] [Google Scholar]
- Deiman, B.A., Seron, K., Jaspars, E.M., Pleij, C.W. Efficient transcription of the tRNA-like structure of turnip yellow mosaic virus by a template-dependent and specific viral RNA polymerase obtained by a new procedure [corrected] [published erratum appears in J. Virol. Methods 1997 66: 165–166] J. Virol. Methods. 1997;64:181–195. doi: 10.1016/s0166-0934(96)02166-0. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- Duggal, R., Lahser, F.C., Hall, T.C. Cis-acting sequences in the replication of plant viruses with plus-sense RNA genomes. Annu. Rev. Phytopathol. 1994;32:287–309. [Google Scholar]
- Eckerle, L.D., Ball, L.A. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology. 2002;296:165–176. doi: 10.1006/viro.2002.1377. [DOI] [PubMed] [Google Scholar]
- Fabian, M.R., White, K.A. Analysis of a 3′-translation enhancer in a tombusvirus: A dynamic model for RNA–RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov, I., Hardy, R., Rice, C.M. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA. 2001;7:1638–1651. doi: 10.1017/s135583820101010x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel, S.J., Hsue, B., Dombrowski, T.F., Master, P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78:669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hema, M., Kao, C.C. Template sequence near the initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 2004;78:1169–1180. doi: 10.1128/JVI.78.3.1169-1180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway, C., Weiss, J., O'Connel, K., Tumer, N.E. Characterization of infectious transcripts from a potato virus X cDNA clone. Virology. 1990;175:365–371. doi: 10.1016/0042-6822(90)90421-m. [DOI] [PubMed] [Google Scholar]
- Herold, J., Andino, R. Poliovirus RNA replication requires genome circularization through a protein protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.Y., Huang, Y.L., Meng, M., Hsu, Y.H., Tsai, C.H. Sequences at the 3′ untranslated region of bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. J. Virol. 2001;75:2818–2824. doi: 10.1128/JVI.75.6.2818-2824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian, A., Roy, G., Sonenberg, N. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 2001;LXVI:293–300. doi: 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- Kao, C.C., Singh, P., Ecker, D.J. De novo initiation of viral RNA-depndent RNA synthesis. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- Kim, K.-H., Hemenway, C. The 5′ nontranslated region of potato virus X RNA affects both genomic and subgenomic RNA synthesis. J. Virol. 1996;70:5533–5540. doi: 10.1128/jvi.70.8.5533-5540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.-H., Hemenway, C. Mutations that alter a conserved element upstream of the potato virus X triple block and coat protein genes affect subgenomic RNA accumulation. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- Kim, K.H., Hemenway, C.L. Long-distance RNA–RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:636–645. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.-H., Kown, S.-J., Hemenway, C.L. Cellular protein binds to sequences near the 5′ terminus of potato virus X RNA that are important for virus replication. Virology. 2002;301:305–312. doi: 10.1006/viro.2002.1559. [DOI] [PubMed] [Google Scholar]
- Klovins, J., van Duin, J. A long-range pseudoknot in Qβ RNA is essential for replication. J. Mol. Biol. 1999;294:875–884. doi: 10.1006/jmbi.1999.3274. [DOI] [PubMed] [Google Scholar]
- Klovins, J., Berzins, V., van Duin, J. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA. 1998;4:948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev, G., Liu, S., Beckett, R., Miller, W.A. The 3′-terminal structure required for replication of barley yellow dwarf virus RNA contains an embedded 3′ end. Virology. 2002;292:114–126. doi: 10.1006/viro.2001.1268. [DOI] [PubMed] [Google Scholar]
- Kohm, B.A., Goulden, M.G., Gilbert, J.E., Kavanagh, T.A., Baulcombe, D.C. A potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell. 1993;5:913–920. doi: 10.1105/tpc.5.8.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, N.-S., Hsu, Y.-H. A satellite RNA associated with bamboo mosaic potexvirus. Virology. 1994;202:707–714. doi: 10.1006/viro.1994.1392. [DOI] [PubMed] [Google Scholar]
- Lindenbach, B.D., Sgro, J.-Y., Ahlquist, P. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 2002;76:3905–3919. doi: 10.1128/JVI.76.8.3905-3919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick, W.C. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Miller, W.A., White, K.A. Long-distance RNA–RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, E.D., Plante, C.A., Kim, K.-H., Hemenway, C. Stem–loop structure in the 5′ region of potato virus X genome required for plus-strand RNA accumulation. J. Mol. Biol. 1998;284:591–608. doi: 10.1006/jmbi.1998.2174. [DOI] [PubMed] [Google Scholar]
- Miller, E.D., Kim, K.-H., Hemenway, C. Restoration of a stem–loop structure required for potato virus X accumulation indicates selection for a mismatch and a GNRA tetraloop. Virology. 1999;260:342–353. doi: 10.1006/viro.1999.9843. [DOI] [PubMed] [Google Scholar]
- Murray, K.E., Steil, B.P., Roberts, A.W., Barton, D.J. Replication of poliovirus RNA with complete internal ribosome entry site deletions. J. Virol. 2004;78:1393–1402. doi: 10.1128/JVI.78.3.1393-1402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na, H., White, K.A. Structure and prevalence of replication silencer–3′-terminus RNA interactions in Tombusviridae. Virology. 2006;345:305–316. doi: 10.1016/j.virol.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Nagy, P.D., Pogany, J., Simon, A.E. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus [in process citation] EMBO J. 1999;18:5653–5665. doi: 10.1093/emboj/18.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, P.D., Pogany, J., Simon, A.E. In vivo and in vitro characterization of an RNA replication enhancer in a satellite RNA associated with turnip crinkle virus. Virology. 2001;288:315–324. doi: 10.1006/viro.2001.1099. [DOI] [PubMed] [Google Scholar]
- Olsthoorn, R.C., Bol, J.F. Role of an essential triloop hairpin and flanking structures in the 3′-untranslated region of Alfalfa mosaic virus RNA in in vitro transcription. J. Virol. 2002;76:8747–8756. doi: 10.1128/JVI.76.17.8747-8756.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn, R.C., Mertens, S., Brederode, F.T., Bol, J.F. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, T.A.M., Hemenway, C.L., Buck, K.W. Role of the 3′ tRNA-like structure in tobacco mosaic virus minus-strand RNA synthesis by the viral RNA-dependent RNA polymerase in vitro . J. Virol. 2000;74:11671–11680. doi: 10.1128/jvi.74.24.11671-11680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas, T., Nagy, P.D. The RNA replication enhancer element of tombusvirus contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 2003;77:258–269. doi: 10.1128/JVI.77.1.258-269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas, T., Pogany, J., Nagy, P.D. Analysis of minimal promoter sequences for plus-strand synthesis by the cucumber necrosis virus RNA-dependent RNA polymerase. Virology. 2002;296:263–274. doi: 10.1006/viro.2002.1423. [DOI] [PubMed] [Google Scholar]
- Panaviene, Z., Panavas, T., Serva, S., Nagy, P.D. Purification of the cucumber necrosis virus replicase from yeast cells: Role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 2004;78:8254–8263. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene, Z., Panavas, T., Nagy, P.D. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 2005;79:10608–10618. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai-Nair, N., Kim, K.H., Hemenway, C.L. Cis-acting regulatory elements in the potato virus X 3′-nontranslated region differentially affect minus-strand and plus-strand RNA accumulation. J. Mol. Biol. 2003;386:701–720. doi: 10.1016/S0022-2836(02)01369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante, C.A., Kim, K.H., Pillai-Nair, N., Osman, T.A., Buck, K.W., Hemenway, C.L. Soluble, template-dependent extracts from Nicotiana benthamiana plants infected with potato virus X transcribe both plus- and minus-strand RNA templates. Virology. 2000;275:444–451. doi: 10.1006/viro.2000.0512. [DOI] [PubMed] [Google Scholar]
- Pogany, J., Fabian, M.R., White, K.A., Nagy, P.D. A replication silencer element in a plus-strand RNA virus. EMBO J. 2003;22:5602–5611. doi: 10.1093/emboj/cdg523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadt, R., Ishikawa, M., Janda, M., Ahlquist, P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar, C.T., Zhang, X., Kao, C.C. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 2003;77:1830–1839. doi: 10.1128/JVI.77.3.1830-1839.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, D., White, K.A. Enhancer-like properties of an RNA element that modulates tombusvirus RNA accumulation. Virology. 1999;256:162–171. doi: 10.1006/viro.1999.9630. [DOI] [PubMed] [Google Scholar]
- Ray, D., White, K.A. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 2003;77:245–257. doi: 10.1128/JVI.77.1.245-257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R.N., Dreher, T.W. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and -CCR- boxes: Initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998;4:1083–1095. doi: 10.1017/s1355838298980694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit, T.L., Vaewhongs, A.A., Lommel, S.A. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- Sivakumaran, K., Bao, Y., Roossinck, M.J., Kao, C.C. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 2000;74:10323–10331. doi: 10.1128/jvi.74.22.10323-10331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran, K., Hema, M., Kao, C.C. Brome mosaic virus RNA syntheses in vitro and in barley protoplasts. J. Virol. 2003;77:5703–5711. doi: 10.1128/JVI.77.10.5703-5711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C., Simon, A.E. Requirement of a 3′-terminal stem–loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- Sriskanda, V.S., Pruss, G., Ge, X., Vance, V.B. An eight-nucleotide sequence in the potato virus X 3′-untranslated region is required for both host protein binding and viral multiplication. J. Virol. 1996;70:5266–5271. doi: 10.1128/jvi.70.8.5266-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.L., Ahlquist, P. A brome mosaic virus intergenic RNA 3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 1999;73:198–204. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, R.L., Buck, K.W. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology. 1999;253:115–124. doi: 10.1006/viro.1998.9495. [DOI] [PubMed] [Google Scholar]
- van Rossum, C.M., Brederode, F.T., Neeleman, L., Bol, J.F. Functional equivalence of common and unique sequences in the 3′-untranslated regions of alfalfa mosaic virus RNAs 1, 2, and 3. J. Virol. 1997;71:3811–3816. doi: 10.1128/jvi.71.5.3811-3816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot, J., Angell, S.M., Baulcombe, D.C. In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. J. Virol. 1998;72:8316–8320. doi: 10.1128/jvi.72.10.8316-8320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C., Neeleman, L., Linthorst, H.J., Bol, J.F. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 2001;75:6440–6449. doi: 10.1128/JVI.75.14.6440-6449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, K.A., Bancroft, J.B., Mackie, G.A. Defective RNAs of clover yellow mosaic virus encode nonstructural/coat protein fusion products. Virology. 1991;183:479–486. doi: 10.1016/0042-6822(91)90977-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, K.A., Bancroft, J.B., Mackie, G.A. Mutagenesis of a hexanucleotide sequence conserved in potexvirus RNAs. Virology. 1992;189:817–820. doi: 10.1016/0042-6822(92)90614-u. [DOI] [PubMed] [Google Scholar]
- You, S., Falgout, B., Markoff, L., Padmanabhan, R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001;276:15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- Zhang, G., Simon, A.E. A multifunctional turnip crinkle virus replication enhancer revealed by in vivo functional SELEX. J. Mol. Biol. 2003;326:35–48. doi: 10.1016/s0022-2836(02)01366-9. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Simon, A.E. Importance of sequence and structural elements within a viral replication enhancer. Virology. 2005;333:301–315. doi: 10.1016/j.virol.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Zhang, G., Slowinski, V., White, K.A. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. J. Virol. 1999;73:550–561. doi: 10.1017/s1355838299982080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G., Zhang, J., Simon, A.E. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 2004;78:7619–7633. doi: 10.1128/JVI.78.14.7619-7633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G., Zhang, J., George, A.T., Baumstark, T., Simon, A.E. Conformational changes involved in initiation of minus-strand synthesis of a virus-associated RNA. RNA. 2006;12:147–162. doi: 10.1261/rna.2166706. [DOI] [PMC free article] [PubMed] [Google Scholar]