Abstract
Mitochondrial tRNAs (mtRNAs) often lack domains and posttranscriptional modifications that are found in cytoplasmic tRNAs. These structural and chemical elements normally stabilize the folding of cytoplasmic tRNAs into canonical structures that are competent for aminoacylation and translation. For example, the dihydrouridine (D) stem and loop domain is involved in the tertiary structure of cytoplasmic tRNAs through hydrogen bonds and a Mg2+ bridge to the ribothymidine (T) stem and loop domain. These interactions are often absent in mtRNA because the D-domain is truncated or missing. Using gel mobility shift analyses, UV, circular dichroism and NMR spectroscopies and aminoacylation assays, we have investigated the functional folding interactions of chemically synthesized and site-specifically modified mitochondrial and cytoplasmic tRNAs. We found that Mg2+ is critical for folding of the truncated D-domain of bovine mtRNAMet with the tRNA’s T-domain. Contrary to the expectation that Mg2+ stabilizes RNA folding, the mtRNAMet D-domain structure was unfolded and relaxed, rather than stabilized in the presence of Mg2+. Because the D-domain is transcribed prior to the T-domain, we conclude that Mg2+ prevents misfolding of the 5′-half of bovine mtRNAMet facilitating its correct interaction with the T-domain. The interaction of the mtRNAMet D-domain with the T-domain was enhanced by a pseudouridine located in either the D- or T-domains compared to that of the unmodified RNAs (Kd = 25.3, 24.6 and 44.4 μM, respectively). Mg2+ also affected the folding interaction of a yeast mtRNALeu1, but had minimal effect on the folding of an E. coli cytoplasmic tRNALeu. The D-domain modification, dihydrouridine, facilitated mtRNALeu folding. These data indicate that conserved modifications assist and stabilize the formation of the functional mtRNA tertiary structure.
Keywords: mitochondrial RNA folding, aminoacylation, tRNAMet, tRNALeu
Abbreviations used: AAS, amino acid accepting stem; ASL, anticodon stem and loop domain; CD, circular dichroism spectropolarimetry; DSL, dihydrouridine (D) stem and loop domain; D:T complex, folding interaction of the D-half molecule with the T-half molecule; LeuRS, E. coli leucyl-tRNA synthetase; Kd, dissociation constant; LeuRSmt, mitochondrial leucyl-tRNA synthetase; bmtRNAMet, bovine mitochondrial methionine tRNA; ymtRNALeu-ΔACVL, yeast mitochondrial leucine tRNA lacking the anticodon and variable loop domains; tRNALeu1-ΔACVL, E. coli leucine tRNA lacking the anticodon and variable loop domains; MetRSmt, mitochondrial methionyl-tRNA synthetase; NMR, nuclear magnetic resonance spectrometry; PAGE, polyacrylamide gel electrophoresis; Ψ, pseudouridine; TSL, ribothymidine stem and loop domain; VSL, variable stem and loop domain
Introduction
tRNA gene transcription followed by nucleolytic cleavage, modification and folding generates the mature, functional form of the tRNA whether encoded in the bacterial cytoplasm, the eukaryotic nucleus or the mitochondrion. In protein synthesis, both cytoplasmic and mitochondrial tRNAs (mtRNA) are recognized by their respective aminoacyl-tRNA synthetase and translational factors, and bind to their cognate and wobble codons within the ribosomal A- and P-sites. The structural and functional domains of the aminoacyl stem (AAS) and the anticodon stem and loop (ASL) are conserved in both mitochondrial and cytoplasmic tRNAs (Figure 1). The L-shaped tertiary structure of a cytoplasmic tRNA is produced and stabilized by hydrogen bonds and Mg2+ coordination between the dihydrouridine (D) stem and loop domain (DSL) and the ribothymidine (T) stem and loop domain (TSL) (Figure 1). However, many mtRNAs have truncated or missing DSLs, and/or TSLs. How these tRNAs fold into the traditional L-shaped, three dimensional structure remains to be resolved. Importantly, mitochondrial tRNA gene mutations result in congenital human disease1,2 (www.mitomap.org). Mutations in mtRNAs, particularly those with truncated or missing DSLs and/or TSLs, could result in misfolded, nonfunctional RNA.
Figure 1.
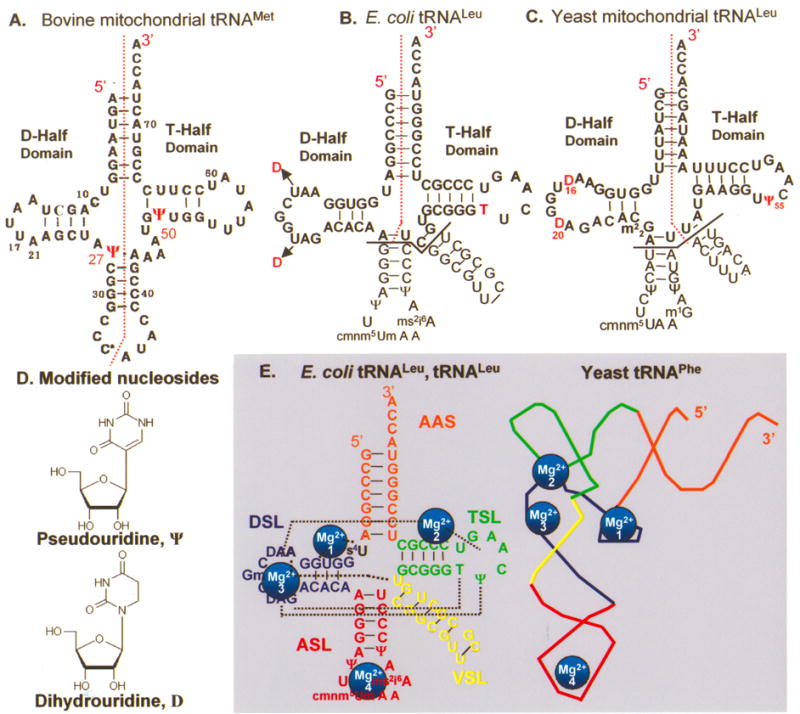
Primary and secondary structures of the native tRNAs. The tRNA D-domain and T-domain half molecules were chemically synthesized as indicated by the red dotted lines (••••), with and without modified nucleosides. A. D- and T-domain half molecules of bmtRNAMet were synthesized in their entirety with and without pseudouridine (Ψ) at positions 27 and 50. C* represents 5-formylcytidine and this position was left unmodified. B. The bolded sequences of the D- and T-domain half molecules of E. coli tRNALeu were synthesized without the anticodon stem and loop (ASL) and variable or extra stem and loop (VSL) domains. Only the unmodified E. coli tRNALeu D-half molecule was synthesized. However, the native tRNA has dihydrouridine (D) at positions 16 and 20. The T-half molecule was synthesized with and without T at position 54. C. The D- and T-domain half molecules of ymtRNALeu1 were chemically synthesized. The anticodon stem and loop (ASL) and variable or extra stem and loop (VSL) domains were not part of the chemical syntheses. The native tRNA has dihydrouridine (D) at positions 16 and 20. The ymtRNALeu1 D-half molecule was synthesized unmodified and with D at positions 16 and/or 20. The T-half molecule was synthesized with and without Ψ at position 55. D. The chemical structures of D and Ψ. E. The functional and structural domains common to cytoplasmic tRNAs are labeled for the E. coli tRNALeu, aminoacyl-stem (orange, AAS); dihydrouridine stem and loop (blue, DSL); anticodon stem and loop (red, ASL); variable (or extra) stem and loop (yellow, VSL); and ribothymidine stem and loop (green, TSL). The folding interactions that produce the three dimensional structure of the tRNA and its coordination of four Mg2+ ions (numbered light blue spheres) are shown. The black dotted lines indicate tertiary interactions that stabilize the L-shaped molecule and include those coordinating Mg2+.
In addition to the major nucleosides of A, U, G and C, both mitochondrial and cytoplasmic tRNAs have posttranscriptional modifications.3 Cytoplasmic tRNAs of all kingdoms have modified nucleosides that are conserved in chemistry and location within the tRNA sequence. The conserved modifications of cytoplasmic tRNAs appear to facilitate Mg2+ coordination and functional folding3,4. The T and pseudouridine (Ψ) modifications at nucleoside positions 54 and 55, respectively, in the T-domain are almost universal to cytoplasmic tRNAs (Figure 1).5 T54 and Ψ55 are also two of the first modifications to be synthesized in tRNA transcripts in vivo.6 Both modifications enhance the folding interaction between the yeast tRNAPhe DSL and the TSL.3 The D modification, the only known naturally-occurring, non-aromatic nucleoside, is also highly conserved (Figure 1).5
The functional folding of mtRNAs with truncated or missing DSL domains may be dependent on posttranscriptional modification. The native human mtRNALys with six modified nucleosides was found to fold into a form that could be aminoacylated in vitro. Yet, the in vitro transcript required N1-methylation of adenosine at residue 9 for functional folding to be detected.7,8 Although they are somewhat ubiquitous, little is known about the roles of other modifications in the folding of mtRNAs. Difficulty in the functional folding of mtRNA transcripts may also be affected by a predominance of A and U nucleotides. Mitochondrial tRNA gene products contain on average 35.9% A and 28.4% U, as compared to nuclear transcribed tRNAs, which are 19.6% A and 22.7% U.5,9
Because of the electrostatic repulsion of the negatively charged phosphates of the backbone, the tertiary folding interactions and the stability of tRNA molecules, as with other RNAs, is facilitated greatly by monovalent and divalent cations and/or polycations.10,11 With few exceptions, such as ribozymes, site-specific binding of Mg2+ does not appear to play an important role in RNAs functional folding.12 However, cytoplasmic tRNAs have Mg2+ ions bound loosely to the phosphate backbone. In addition, eleven distinct metal ions bind with high affinity at specific locations in the x-ray derived crystal structures.13 Two of the tightly bound Mg2+ found in cytoplasmic tRNAs are coordinated between the DSL and TSL (Figure 1).13,14 Should Mg2+ ions be coordinated at similar locations in truncated mtRNAs, they could be important to folding. Here, we report that conserved posttranscriptional modifications are important to mtRNA folding interactions, especially for bovine mtRNAMet that has a truncated D-loop. Unexpectedly, we found that Mg2+-dependent relaxation of D-domain structure was critical for functional folding of the bovine mtRNAMet (bmtRNAMet).
Results
Bimolecular mtRNA folding interactions: Effect of Mg2+
The L-shaped tertiary structure of the tRNA’s global fold is driven by interaction of the dihydrouridine (D-) domain with the ribothymidine (T-) domain (Figure 1). However, many mitochondrial tRNAs (mtRNA) have truncated D-stems and loops9,15 making it a challenge to compare their folding to cytoplasmic tRNAs. Therefore, we investigated the folding of two mtRNAs, bovine mtRNAMet and yeast mtRNALeu (bmtRNAMet and ymtRNALeu1) with a particular emphasis on the roles of modified nucleosides and Mg2+ and have compared these results to the folding of E. coli tRNALeu (Figure 1). Bovine mtRNAMet was investigated because its sequence is 92% identical to that of the human mtRNAMet and the identity and locations of the modified nucleosides are known16 whereas those for human mtRNAMet are inferred from the gene sequence (http://mamit-trna.u-strasbg.fr/table/Methionine.html).9
To investigate folding interactions, the tRNAs were synthesized chemically as half molecules with and without modifications (Figure 1). For instance, the bmtRNAMet molecule was synthesized as two half molecules, the D- and T-domain half molecules, with a break in the backbone of the anticodon loop (Figure 1). The D-domain and T-domain halves were 31 and 41 nucleotides, respectively. Half molecules of the bmtRNAMet were synthesized with pseudouridine at position 27 of the D-half (Ψ27) and at 50 in the T-half (Ψ50) (Figure 1). The yeast mitochondrial (ymtRNALeu1-ΔACVL) and E. coli tRNALeu (tRNALeu-ΔACVL)17 were chemically synthesized with partial anticodon stems and without their respective anticodon and variable loops (Figure 1). The shorter molecules were designed with the knowledge that in assessing biological activity, the cytoplasmic leucyl-tRNA synthetase did not require the anticodon stem and loop for aminoacylation of the tRNA.17 Our first objective was to confirm that the folding interactions of the various D-half molecules with their respective T-half molecules could be observed in a gel mobility shift assay.
The bimolecular folding interactions of the D- and T-half molecules of bmtRNAMet, ymtRNALeu1-ΔACVL and E. coli tRNALeu-ΔACVL could be observed by a gel mobility shift assay in the presence of 3 mM Mg2+ and at 4 °C (Figure 2A). The ethidium bromide stained complex of D- and T-half molecules (72mer) migrated more slowly than the individual D- and T-half molecules and comparably to that of unfractionated E. coli tRNAs, which ranged in length from 72–80 nucleotides (Figure 2A). Mg2+ concentrations higher than 3 mM did not increase the amount of complex formed (not shown). When the shifted band composed of the bmtRNAMet D:T complex was extracted from the gel and subjected to denaturing PAGE, both the 31 nucleotide D-half and the 41 nucleotide T-half molecules were recovered from the shifted band (not shown). Increasing amounts of complex were formed when the D-half molecule was titrated with increasing concentrations of the T-half molecule. With a molar excess of the T-half molecule, the bmtRNAMet D-half band in the gels disappeared (Figure 2B). Quantification of the formed bmtRNAMet complex at each concentration of added T-half molecules generated a binding curve (Figure 2C) from which the dissociation constant (Kd) of the D-half for the T-half molecule was determined to be 44.4 ± 4.3 μM. The Gibbs standard free energy (ΔG°) associated with the folding interaction was −6.8 kcal/mole. From gel mobility shift assays conducted at 4, 10, 20, 30 and 40 °C, the thermal stability of the reconstituted bmtRNAMet could be roughly estimated to be between 30 and 40 °C (not shown). As with yeast tRNAPhe,4 neither the T-half, nor the D-half molecules alone, in high concentrations generated the gel mobility shift observed when the two half molecules interacted in the presence of Mg2+ (data not shown).
Figure 2.
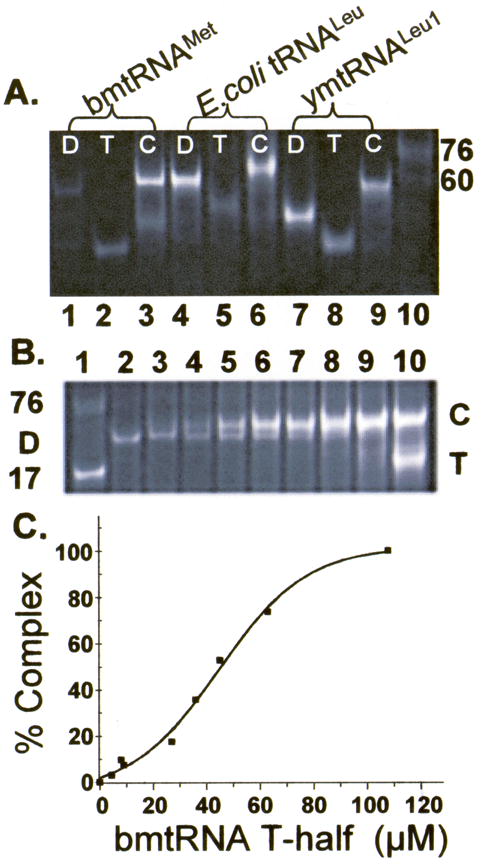
Gel mobility shift assay of tRNA folding interactions. A. The unmodified D- and T-domain half molecules of bmtRNAMet (lanes 1–3), E. coli tRNALeu-ΔACVL (E. coli tRNALeu, lanes 4–6) and ymtRNALeu1-ΔACVL (ymtRNALeu1, lanes 7–9) were subjected to polyacrylamide gel electrophoresis (4 °C) individually or together in the presence of 3 mM Mg2+. The individual half molecules, D (lanes 1, 4 and 7) and T (lanes 2, 5, and 8), and the D:T complex, C (lanes 3, 6, and 9), are denoted. The D- and T- half molecules of the three tRNAs migrate differently because they are of different lengths and nucleoside compositions. Two molecular size standards are also visible in the electrophoresis, a limited sample of unfractionated E. coli tRNA (3 μg) providing a broad band at ~76 nucleotides, and a 60mer RNA. B. Reconstitution of bmtRNAMet by formation of a complex between the unmodified D-domain half molecule titrated with the T-half molecule. The unmodified D-half molecule of bmtRNAMet (30 μM) was subjected to polyacrylamide gel electrophoresis (4 °C) in the presence of 3 mM Mg2+ and increasing concentrations of T-domain half molecule. Lane 1, standard 17 nucleotide ASL and unfractionated E. coli tRNA as markers; lane 2, 30 μM D-half molecule alone; lanes 3–9, 30 μM D-half molecule and increasing concentrations of the T-half molecule, lane 3, 4.8 μM, lane 4, 9.6 μM; lane 5, 19.3 μM; lane 6, 29 μM; lane 7, 39 μM; lane 8, 48.3 μM; lane 9, 68 μM; lane 10, 116 μM. Excess T-half molecule is evident in Lane 10. C. Binding of the unmodified bmtRNAMet D-half molecule to the unmodified T-half molecule in the presence of 3 mM Mg2+. Complex formed from titration of the D-half molecule with the T-half molecule and observed in PAGE (Fig. 2B) was quantified by analyzing the density of the bands. The percent of complex formed (relative to 100% in the presence of excess T-half molecule) is plotted as a function of T-half molecule concentration, generating a binding curve.
The folding interaction of the bmtRNAMet D- and T-domain half molecules observed in the presence of Mg2+ was difficult to detect in the absence of Mg2+ with only a faint band migrating at the location expected for complex formation (Figure 3). Though Mg2+ was a strong requirement for the folding interactions of the bmtRNAMet half molecules, Mg2+ was not strictly required for the D- and T-half molecules of E. coli tRNALeu-ΔACVL to form a complex. The folding interactions of the E. coli tRNALeu-ΔACVL half molecules were observable by PAGE mobility shift analysis in the absence of Mg2+ (Figure 3, lanes 4–6), albeit complex formation was enhanced in the presence of Mg2+. Migration of the E. coli tRNALeu-ΔACVL complex was slower than that of the 60mer standard (Figure 3). Interaction of the ymtRNALeu1-ΔACVL half-molecules also showed a significant dependence on Mg2+ (Figure 3, Lanes 7–9). The assessment of this interaction was difficult since the complex migrated very close to that of the free T-half molecule.
Figure 3.
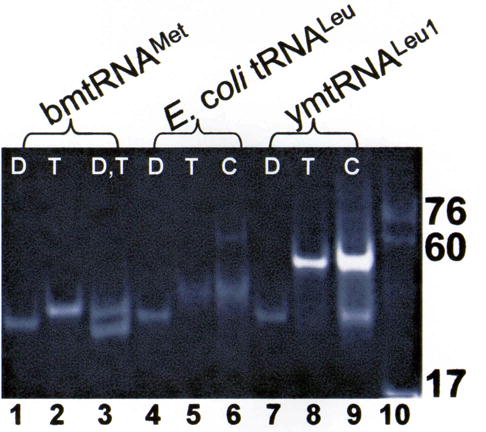
Differential effects of Mg2+ on the interaction of D- and T-domain half molecules of different tRNAs. The individual and combined, unmodified D- and T-half molecules of bmtRNAMet (lanes 1–3), E. coli tRNALeu-ΔACVL (E. coli tRNALeu, lanes 4–6) and ymtRNALeu1-ΔACVL (ymtRNALeu1, lanes 7–9) at a concentration of 30 μM each were subjected to PAGE in the absence of Mg2+ (4 °C). Only a faint band was seen to migrate at the location expected for complex formation by the mtRNAMet D- and T-half molecules. D:T complex formation was evident for E. coli tRNALeu-ΔACVL (C, lane 6) and as a band migrating slightly slower than that of the T-half molecule for mtRNALeu-ΔACVL (C, lane 9). Three molecular size standards are evident in lane 10, the ASL of yeast tRNAPhe (17 nucleotides), a 60mer RNA and unfractionated E. coli tRNA (~76 nucleotides).
Mg2+ affects the bmtRNAMet D- but not T-domain conformation
The need for Mg2+ in the folding of bmtRNAMet could be a general counter ion requirement for the phosphodiester backbone of both half molecules. Alternatively, the Mg2+ could have site-specific coordination that confers folding of one of the two half molecules. The electrophoretic mobility of the T-half molecule was not affected by Mg2+ (data not shown). However, the gel mobility of the D-half molecule was strongly affected by Mg2+. When Mg2+ was added to a concentration of 0.5 mM, the migration of the D-half molecule slowed considerably and two forms distinct from that in the absence of Mg2+ appeared (Da and Db; Figure 4) with Da migrating more slowly than Db. This observation suggests that Mg2+ binds to the D-half molecule in two different ways or with different stoichiometries (e.g. one and two Mg2+ per D-half molecule) leading to alternative conformational states. The distinctive migrations are indicative of Da, the slower migrating form being in an unfolded state, while Db, the faster migrating form was in a collapsed state. With increasing Mg2+ concentration, the slower migrating Da form predominated and at 3 mM Mg2+ was the only form observed (Figure 4). In comparison, the D-half molecules of E. coli tRNALeu-ΔACVL and ymtRNALeu1-ΔACVL were not as dramatically affected by Mg2+ concentrations, nor formed two distinct bands.
Figure 4.
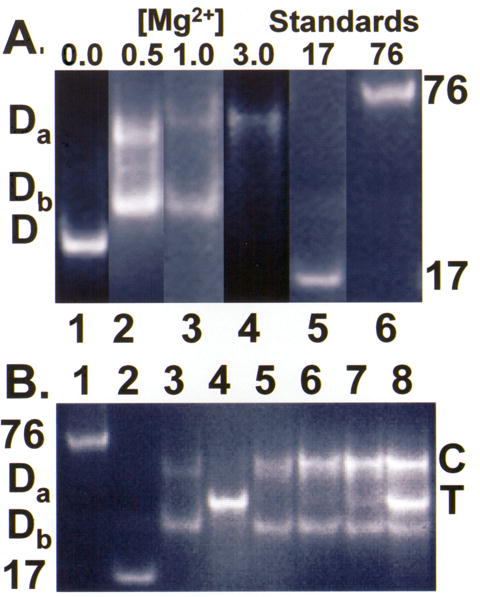
The effect of Mg2+ concentration on the folding interaction of the bmtRNAMet D- and T-domain half molecules. A. The unmodified bmtRNAMet D-domain half molecules at a concentration of 30 μM was subjected to PAGE (4 °C) in the absence of Mg2+, lane 1, and in the presence of different concentrations of Mg2+, lane 2, 0.5 mM; lane 3, 1.0 mM, and lane 4, 3.0 mM. Lanes 5 and 6 contained the molecular size standards of yeast tRNAPhe ASL, 17mer and unfractionated tRNA, 76mer, respectively, subjected to electrophoresis at 1 mM Mg2+. The lighter intensity of the bands observed at higher Mg2+ reflects the weaker staining of the unstructured molecule by ethidium bromide. Background coloration varies with magnesium concentration, and removal of excess stain. B. Titration of the unmodified bmtRNAMet D-domain half molecule (30 μM) with the unmodified T-domain at 1.0 mM Mg2+. The unmodified bmtRNAMet D-domain was titrated with increasing concentrations of T-domain half molecule. Lanes 1 and 2 are molecular weight standards of unfractionated tRNA, 76mer and yeast tRNAPhe ASL, 17mer, respectively. Lane 3 is comprised of the D-half molecule alone. At 1 mM Mg2+, two discrete bands are observed, Da and Db. Lane 4 is comprised of the T-half molecule alone (29 μM). The band for the D:T complex (C) increased in intensity with increasing amounts of T-domain half molecule (lane 5, 9.6 μM; lane 6, 19.3 μM; lane 7, 29 μM; lane 8, 56.3 μM). With increasing T-domain, the band for the slower moving D-half molecule conformation, Da, decreased in intensity, whereas the band for the faster moving D-half molecule conformation, Db, changed little. Excess T-half molecule is evident in lane 8.
The slower moving Da form of the D-half molecule observed in the presence of Mg2+ could be the result of dimer formation, as reported for a mutant of human mtRNALeu.18 In order to investigate this possibility, a [32P]-5′-end labeled D-half molecule was subjected to UV-photoactivated crosslinking at both 254 and 305 nm in the presence and absence of Mg2+. Whether UV-treated in the presence or absence of Mg2+, the D-half molecule migrated on a sequencing gel under denaturing conditions comparable to untreated samples, indicating that Mg2+ had not induced dimer formation (data not shown). In contrast, the intramolecular crosslink expected within yeast tRNAPhe, C48-U59, was observed.19
The abilities of the Da and Db forms of the D-half molecule to interact with the T-half molecule were examined using the gel shift assay at 1.0 mM Mg2+ and at increasing concentrations of the T-half molecule (Figure 4B). At 1.0 mM Mg2+, only the slower migrating Da form of the D-domain half molecule was titrated by the T-half into the complex, whereas the faster migrating Db form was not (Figure 4B). The minimal Mg2+ concentration needed to form and stabilize a complex of the bmtRNAMet half molecule was determined to be ~1.0 mM Mg2+. The 1.0 mM Mg2+ concentration was required in the reaction of the two half molecules, as well as during gel electrophoresis. Thermal denaturation and slow cooling of the D-half molecule did not alter its response to various Mg2+ concentrations as exhibited in gel electrophoresis. It also did not change the properties of the slower migrating Db form to bind the T-half molecule. Mitochondria accumulate Ca2+ to very high concentrations (500–800 μM).20 Though not reaching a concentration of 3 mM in vivo, 3 mM Ca2+ was equally effective as Mg2+ in promoting the folding interaction of the D- and T-half molecules (data not shown). Thus, reconstitution of bmtRNAMet from the D- and T-half molecules was dependent on a slower migrating form of the D-half molecule, and the amount of that conformer was in turn dependent on the Mg2+ concentration. The effect of Mg2+ on the properties of the D-half domain prompted study of the thermodynamic stability, and possible structural changes in this domain.
bmtRNAMet D-half molecule is unstructured in the presence of Mg2+
The UV-monitored thermal denaturation of RNAs assesses the thermal stability of the secondary structure in a molecule. The D-half molecule was denatured in Tris-Borate buffer, in the absence and presence of Mg2+ (Figure 5A). In the absence of Mg2+, the RNA displayed an obvious thermal transition with a high melting temperature (Tm) of 54.3 ± 1.4 °C (Table 1). This observation indicates that the D-half molecule has significant structure in the absence of Mg2+. The Tm of RNA is very dependent on the sequence length, number of stems and their length, and the extent of higher order structures. For instance, ASLs melt between 35–73 °C, depending on sequence. We found that the Tm of native E. coli tRNAMet was 53.4 °C under conditions used for the D-half molecule of bmtRNAMet. The Tm of an E. coli tRNAGlu minihelix (35 nucleosides) was 83 °C (unpublished data). In addition, the thermal denaturation of the D-half molecule of bmtRNAMet resulted in 18% hyperchromicity (Table 1). This is a significant value for a 31 nucleotide RNA with minimal intra-strand interactions (Table 1) when compared to 10% hyperchromicity observed for ASLs heptadecamers (unpublished data), and 18% and 26% hyperchromicity for the ymtRNALeu-ΔACVL and native E. coli tRNAMet, respectively, both of which have extensive secondary and tertiary structures. Under different conditions, RNA duplexes of the type of the Dickerson DNA dodecamer can exhibit as much as 30% hyperchromicity21 and an E. coli tRNAGlu minihelix (35 nucleosides) give a 30% hyperchromicity (unpublished data). The stability of the D-half in the absence of Mg2+ was also evident from the standard free energy change (ΔG°) for this transition of −12.3 ± 0.2 kcal/mole.
Figure 5.
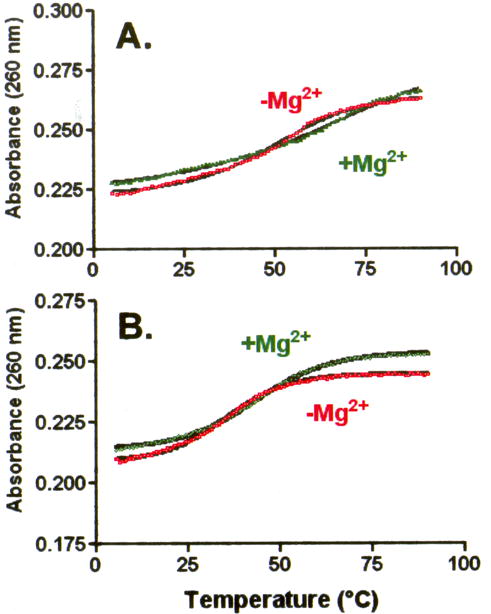
UV-monitored, thermal denaturations of the individual D- and T-half molecules with and without Mg2+. A. The unmodified bmtRNAMet D-domain (2.1 μM) was subjected to thermal denaturations in the absence (red □) and presence (green ▪) of 3.0 mM Mg2+. B. Thermal denaturation of the bmtRNAMet (2 μM) unmodified T-domain in the absence (red □) and presence (green Δ) of 3.0 mM Mg2+. Melts shown represent the averages of three separate experiments. In the presence of Mg2+, each experiment was conducted with a new sample due to metal catalyzed hydrolysis of the phosphodiester backbone at high temperatures.
Table 1.
Thermal stability of D- and T-half molecules and reconstituted bmtRNAMet
|
Tm (°C)a |
Hyperchromicity (%) |
||||
|---|---|---|---|---|---|
| bmtRNAMetHalf molecules and Reconstituted tRNA | No Mg2+ | Mg2+(3 mM) | ΔG°b(kcal/mole) | No Mg2+ | 3mM Mg2+ |
| D-half molecule | 54.3 | ------ | −12.3 | 18.0 | 11.0 |
| DΨ27-half molecule | 43.8 | ------ | −10.1 | 11.0 | 9.6 |
| T-half molecule | 36.1 | 44.6 | −9.1/−10.3 | 17.2 | 18.7 |
| TΨ50-half molecule | 28.6 | 42.1 | −7.6/−10.0 | 19.6 | 21.6 |
| D:T-reconstituted | ------ | 31.2 | −8.3 | ------ | 18.0 |
| DΨ27:T-reconstituted | ------ | 40.0 | −9.1 | ------ | 23.2 |
| D:TΨ50-reconstituted | ------ | 40.8 | −9.3 | ------ | 23.0 |
| D Ψ27:TΨ50-reconstituted | ------ | 41.4 | −9.2 | ------ | 18.2 |
Standard error for the Tm from three to five different transitions was ± 1.4 °C.
ΔG° for the D-half molecules in the absence of Mg2+, for the T-half molecules no Mg2+/3 mM Mg2+, and for the reconstituted bmtRNAMet in the presence of Mg2+. Standard error for ΔG° = ± 0.2 kcal/mole.
However, in the presence of 3 mM Mg2+ little to no transition in the absorbance of the D-half of bmtRNAMet was observed (Figure 5A) and the hyperchromicity was appreciably lower than that obtained in the absence of Mg2+ (11% vs. 18%, respectively; Table 1). Thus, Mg2+ caused some unfolding of the secondary structure of the D-half molecule, perhaps similar to that occurring during thermal denaturation. The presence of Mg2+, thus, resulted in what appears to be an unstructured or partially structured molecule.
The bmtRNAMet T-half RNA also exhibited a thermal transition in the absence of Mg2+. In the presence of Mg2+, the thermal stability of the bmtRNAMet T-half RNA increased (Figure 5B). The thermal transition and hyperchromicity exhibited by the T-half molecule, whether in the presence or absence of Mg2+, indicated the RNA possessed substantial secondary structure. As is expected of RNAs, the Tm was 8.5 °C higher in the presence of Mg2+ (Table 1). Interestingly, the Tm of the T-half molecule in the presence of Mg2+ was lower than the Tm of the D-half molecule in the absence of Mg2+ (44.6 ± 1.4 °C versus 54.3 ± 1.4 °C, respectively; Table 1). The Tm of the complex composed of the D- and T-half molecules was lower than that of the T-half alone (31.2 ± 2.0 °C).
Whereas temperature transitions measure thermal stability, circular dichroism spectrapolarimetry (CD) assesses the degree to which an RNA’s bases are stacked in loops, as well as in secondary structures.22,23 CD spectra of the bmtRNAMet D-domain half molecule were collected under conditions similar to those used during gel electrophoresis. In the absence of Mg2+, the CD spectrum exhibited a considerable positive ellipticity at 264 nm demonstrating that the majority of RNA was in a stable conformation (Figure 6A). However, the ellipticity of the D-domain half molecule dramatically decreased with the titration of Mg2+ (Figure 6A). Such a decrease in ellipticity at this wavelength is indicative of a loss in secondary structure, just as that observed with thermal denaturations. The change in ellipticity with increasing Mg2+ concentration also could reflect the Mg2+-induced change in equilibrium between the two D-half molecule conformers, from Db to Da, observed in PAGE. Whatever the conformational change produced by Mg2+, an apparent dissociation constant (Kd) of 2.5 mM could be extracted from the CD-derived binding curve (Figure 6B)
Figure 6.
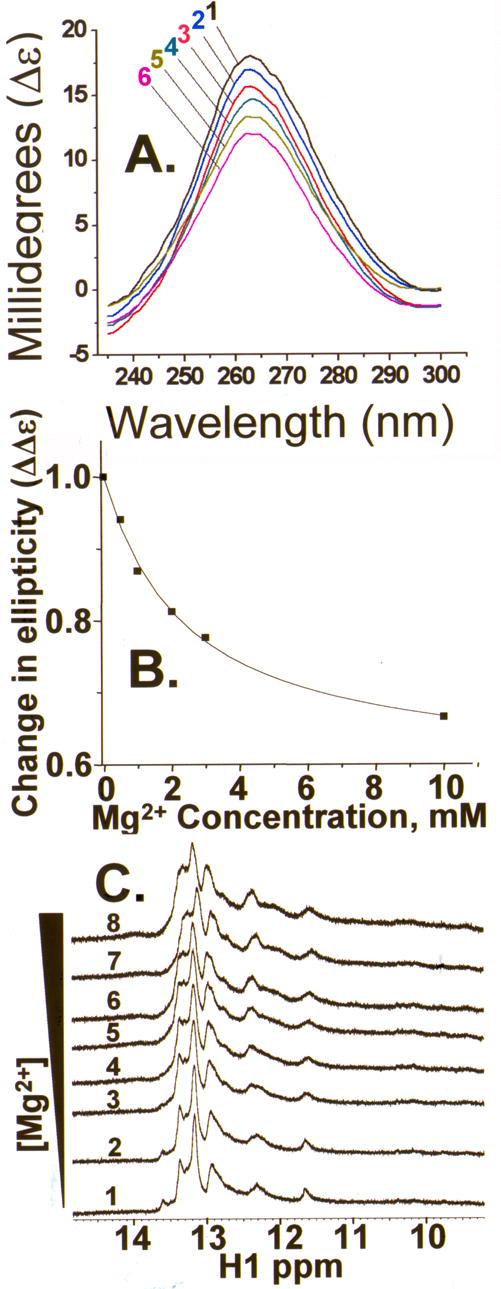
A. CD spectra (4 °C) of the bmtRNAMet D-domain half molecule in the absence and presence of various concentrations of Mg2+. The bmtRNAMet unmodified D-domain half molecule (144 μM) was titrated with Mg2+ from 0.0 to 10 mM and the CD spectrum collected at each concentration, spectrum numbered 1, 0.0 mM; 2, 0.5 mM; 3, 1.0 mM; 4, 3.0 mM; 5, 2.0 mM; and 6, 10 mM. Spectra shown are the average of six spectra for each concentration of Mg2+. B. Mg2+ binding by the bmtRNAMet D-domain half molecule. The change in ellipticity at 265 nm for each concentration of Mg2+ was plotted against the Mg2+ concentration. C. NMR spectra of bmtRNAMet D-half molecule in the absence and presence of various concentrations of Mg2+: spectrum 1, 0 mM; 2, 0.8 mM; 3, 2.3 mM; 4, 3.7 mM; 5, 4.9 mM; 6, 6.1 mM; 7, 7.1 mM; and 8, 8.1 mM. The imino proton base pairing region of the one dimensional spectrum is shown. Spectra were taken of the RNA in 2H-Tris-borate buffer at 4 °C (5% D2O).
If the unmodified bmtRNAMet D-half molecule formed a stable secondary structure composed of canonical, Watson-Crick base pairs, the base paired imino protons would be evident in the one-dimensional NMR spectrum of the RNA (in H2O). Destabilizing the secondary structure by addition of Mg2+ would result in broadening of the resonances of the base paired imino protons which would exchange more readily with H2O. In the absence of Mg2+ and at 4 °C, the NMR spectrum of the D-domain half molecule exhibited discrete, but broad, resonances in the spectral region attributed to the imino proton signals of A•U and G•C base pairs, at 11–14 ppm (Figure 6C). As the RNA was titrated with Mg2+, these resonances broadened further, but did not disappear even at 8.1 mM Mg2+ (Figure 6C). Interestingly, the appearance of the two distinct bands in gel electrophoresis of the D-half molecule corresponded to an RNA/Mg2+ molar ratio of ~1/25. This ratio could not be easily achieved under NMR conditions, and thus could account for the remaining broad imino proton resonances that are indicative of residual secondary structure. Thus, we conclude that the NMR results are consistent with those from UV-monitored thermal denaturations and CD. Mg2+ increases the dynamics and unfolding of the D-domain half molecule.
Conserved modifications affect D- and T-half thermal stability and tRNA reconstitution
Using the bimolecular system to reconstitute bmtRNAMet and ymtRNALeu1, we investigated the influence of conserved modifications on the individual half molecules and on the folding interactions required to form a functional, native tRNA. The natural modification pseudouridine (Ψ) is located at positions 27 (Ψ27) and 50 (Ψ50) in the D- and T-half molecules, respectively, of bmtRNAMet (Figure 1). Surprisingly, the D(Ψ27)-half molecule in the absence of Mg2+ exhibited a significantly reduced Tm (43.8 °C) and hyperchromicity (11%) as compared to its unmodified counterpart (54.3 °C and 18%; Table 1). As with the unmodified D-half molecule in the presence of Mg2+, the RNA began to melt at low temperature and no high temperature thermal transition was observed (data not shown). In the presence of Mg2+, introduction of Ψ27 reduced the hyperchromicity of the D-half (9.6 versus 11%). Thus, Ψ27 reduced the stacking, or ordering of bases in the D-half molecule whether in the presence or absence of Mg2+.
The introduction of Ψ50 into the T-half molecule lowered the Tm in the absence of Mg2+ by 7.5 °C (Table 1), and in the presence of Mg2+ by 2.5 °C. An increase in hyperchromicity was observed with or without Mg2+ (Table 1). Thus, Ψ50 in the T-half molecule appears to have contributed to ordering the structure (higher hyperchromicity). Ψ exhibits sequence-dependent effects on thermal stability and structural ordering in cytoplasmic tRNAs.3,24,25 In the anticodon stem, Ψ39 contributes thermal stability,3,24 whereas in the T-loop Ψ55 had little to no effect on thermal stability, but did contribute to base stacking.26,27
The thermal stabilities of the reconstituted bmtRNAMet formed from the Ψ-modified and unmodified D-half and T-half molecules were determined in the presence of 3.0 mM Mg2+ (Table 1). The Ψ modification stabilized the folding interaction that reconstituted the bmtRNAMet. The Tm of the reconstituted D(Ψ27):T(Ψ50) complex (41.4 °C) was 10.2 °C higher than that of the unmodified complex (31.2 °C; Table 1). The difference in stability also represented a free energy difference of 0.9 ± 0.2 kcal/mole (Table 1). The individual contributions of Ψ27 and Ψ50 to the reconstitution of the bmtRNAMet were approximately 10 °C and therefore, not additive.
We also studied the effect of the naturally-occurring modifications, Ψ27 and Ψ50, on the reconstitution of the bmtRNAMet using a gel mobility shift assay. The D-domain half molecule was titrated with the complementary T-half molecule in the presence of Mg2+ while complex formation was monitored. The bmtRNAMet D-domain half molecule exhibited a higher affinity for the T-half molecule when either Ψ27 or Ψ50 was present (Table 2). The presence of Ψ27 within the D-half increased its affinity for the T-half molecule with a reduction in the dissociation constant from approximately 44.5 to 25.3 μM (Table 2). The presence of Ψ50 in the T-half also reduced the Kd, but from 44.5 to 25.6 μM. The two Ψ modifications combined resulted in a Kd for complex formation did not differ from that of the individual Ψs, 26.8 μM (Table 2).
Table 2.
Mitochondrial tRNA folding interactions
| mtRNA constructs | D-half | T-half | Kda(μM) | ΔGb(kcal/mole) |
|---|---|---|---|---|
| bmtRNAMet | unmodified | unmodified | 44.5 | −5.9 |
| unmodified | Ψ50 | 25.6 | −6.3 | |
| Ψ27 | unmodified | 25.3 | −6.3 | |
| Ψ27 | Ψ50 | 26.8 | −6.2 | |
| ymtRNALeu | unmodified | unmodified | 16.0 | −6.6 |
| unmodified | Ψ55 | 16.8 | −6.6 | |
| D16 | unmodified | 13.3 | −6.7 | |
| D16 | Ψ55 | 12.9 | −6.7 | |
| D20 | unmodified | 9.3 | −6.9 | |
| D20 | Ψ55 | 10.4 | −6.8 | |
| D16 D20 | unmodified | 6.3 | −7.2 | |
| D16 D20 | Ψ55 | 5.9 | −7.2 |
Dissociation constant, Kd, error averaged was ± 4.3 μM for reconstitution of the bmtRNAMet from the variously modified half molecules and ±1.4 μM for reconstitution of ymtRNALeu.
Change in free energy calculated at 37 °, ΔG37, exhibited errors of ± 0.1 kcal/mole in reconstitution of both bmtRNAMet and ymtRNALeu.
Yeast mitochondrial tRNALeu1-ΔACVL and its E. coli counterpart have similar modifications (Figure 1). The tRNAs have two dihydrouridines (D16 and D20) in the D-domain and the Ψ55 modification common to cytoplasmic tRNAs. Again, the presence of modifications enhanced folding interactions of ymtRNALeu1-ΔACVL (Table 2). The dissociation constants were lowest for formation of the modified complexes and highest for the completely unmodified complexes. The introduction of D20 reduced the Kd from 16.0 to 9.3 μM, whereas D16 reduced the Kd from 16.0 to 13.3 μM. Together, the D16,20 modifications reduced the Kd to 6.3 μM. In contrast, Ψ55 of the T-half molecule, alone or in the presence of one or both of the Ds, had no affect on the affinity of the two half molecules for each other (Table 2). The contributions of modifications to the free energy of complex formation for both bmtRNAMet and ymtRNALeu1-ΔACVL were significant, and for bmtRNAMet comparable to the G° derived from thermal denaturations (Table 2). Thus, the conserved modifications Ψ and D increased the affinities of the D-half molecule for the T-half molecule for both mitochondrial tRNAs.
Conserved modifications affect aminoacylation of reconstituted tRNA
To assess whether the folding interactions of the D- and T-domain half molecules were biologically relevant, we determined if the resulting mtRNAMet and mtRNALeu complexes could be aminoacylated by their cognate tRNA synthetase. Though not as good of a substrate as the bovine mtRNAMet transcript, each of the bmtRNAMet complexes was successfully aminoacylated by human mitochondrial methionyl-tRNA synthetase (MetRSmt), indicating the formation of a functional tRNA complex. As would be expected, the half molecules alone were not aminoacylated, but acted as negative controls (Figure 7). Bacterial MetRS will aminoacylate a minihelix composed of the amino acid accepting stem,28 and both the D- and T-half molecules have significant recognition elements for the enzyme that could bind the half molecules.29,30 Interestingly, the presence of Ψ50 in the T-half of the molecule increased aminoacylation activity about 30%, while the presence of Ψ27 and Ψ50 increased the aminoacylation about 60%. The presence of Ψ27 alone had little effect on the aminoacylation of the complex of D-and T-domain half molecules. Collectively, the results indicate that these modifications promote the formation of the correctly folded structure in bmtRNAMet. The low degree of aminoacylation observed for the complexed half molecules compared to the transcript may be due in part to the break in the anticodon stem loop present in the half molecules. The anticodon is the primary determinant for aminoacylation by the bacterial MetRS and is likely to be of similar importance for human MetRSmt.31
Figure 7.
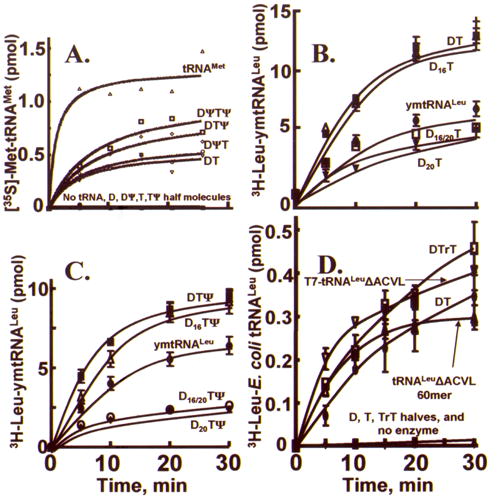
Aminoacylation of reconstituted bmtRNAMet, ymtRNALeu1-ΔACVL and E. coli tRNALeu-ΔACVL. A. The ability of human MetRSmt to aminoacylate the bmtRNAMet transcript and the reconstituted bmtRNAMet from D- and T-half molecules. Aminoacylation of unmodified and modified (Ψ) D-and T-half molecule complexes and transcript was measured as described in “Materials and Methods”. Assays contained 4.2 μM of the half molecule complexes, D:T (inverse triangles, ∇), D(Ψ):T (circles; ○), D:T(Ψ) (diamonds;⋄), D(Ψ):T(Ψ) (squares; □) or 0.06 μM bmtRNAMet transcript (triangles; Δ). A blank representing the amount of [35S]Met-tRNA present at time 0.0 min was subtracted from each point. The lines represent a best-fit of the data. These data represent the results of at least 3 independent measurements. B. and C. Aminoacylation activity of modified and unmodified, reconstituted ymtRNALeu1-ΔACVL and ymtRNALeu1 transcript. Aminoacylation of unmodified and modified (D16 and D20) D-domain half molecules interacting with the unmodified and modified (Ψ55) T-domain were aminoacylated and compared to that of the ymtRNALeu transcript. D16 and D20 represent the dihydrouridine modification at the position indicated within the tRNA D-arm. Ψ55 represents the pseudouridine modification in the TSL. Each reaction consisted of 4 μM RNA as individual D- and T-domains or complexes, or transcript and 50 nM LeuRSmt. D. Aminoacylation activity of reconstituted E. coli tRNALeu-ΔACVL, the T7-transcript and the unmodified, chemically synthesized 60mer of the same sequence. Each reaction consisted of 4 μM RNA as individual D- and T-domains or complexes (D:T), transcript (T7-tRNALeu-ΔACVL) or chemically synthesized 60mer (tRNALeu-ΔACVL 60mer) and 50 nM E. coli LeuRS. The T-arm with a ribothymidine modification at position 54 is represented by rT.
Modified and unmodified ymtRNALeu1-ΔACVL, reconstituted from the D- and T-half molecules, was aminoacylated by yeast mitochondrial LeuRS (LeuRSmt) and compared to the aminoacylation of the ymtRNALeu1 transcript. Half molecules, whether D- or T-domain, could not be aminoacylated. Eight different combinations of ymtRNALeu1-ΔACVL based D- and T-domain half molecules formed a tRNA that lacked the anticodon stem and loop and the variable loop. Four of these performed better than the transcript in aminoacylation assays (Figure 7B & C). The complexes formed from the unmodified D- and T-domains (D:T), and those formed from the D-domain modified with D16 or from the T-domain modified with Ψ55 and/or with D16 were aminoacylated at comparable levels and were also significantly better than the full length transcript. In contrast, the addition of D20 which aids in the folding interactions of the D- and T-domain half molecules, significantly decreased the aminoacylation activity of the complex relative to that of the full length ymtRNALeu1 transcript (Figure 7B and C). The structural contribution of D20, exhibited by the enhanced affinity of the modified D-half molecule for the T-half, may be advantageous for the tRNA’s interactions with other macromolecules (i.e. ribosome or EF-Tu), but it comes at the expense of aminoacylation efficiency by the LeuRS.
The E. coli and yeast mitochondrial LeuRSs are noted for their different aminoacylation efficiencies with their respective tRNAs.32 Therefore, the modified and unmodified E. coli tRNALeu bimolecular complexes and unimolecular constructs were aminoacylated with the cognate E. coli LeuRS for comparison to the aminoacylation results with mitochondrial tRNAs. In order to compare aminoacylation of a biomolecular interaction to a unimolecular species, an unmodified, and unimolecular 60-mer (tRNALeu ACVL 60-mer) was chemically synthesized with the AAS, DSL and TSL and was identical in the sequence of the E. coli tRNALeu-ΔACVL transcript (Figure 1B). As would be expected, the enzymatic and chemically synthesized E. coli tRNALeu–ΔACVL were aminoacylated at similar levels. The E. coli tRNALeu-ΔACVL composed of the unmodified D-domain half molecule annealed to the T54-modified, T-domain half molecule was aminoacylated similarly to the unimolecular E. coli tRNALeu-ΔACVL transcript (Figure 7D). In the absence of the T54 modification, the biomolecular complex of D- and T- half molecules and the equivalent chemically synthesized unimolecular tRNALeu analog were aminoacylated at significant levels, albeit slightly decreased compared to the complex formed from the T54-modified T-half molecule (Figure 7D). Individual D-domain and T-domain half molecules could not be aminoacylated.
DISCUSSON
A considerable number of mitochondrial tRNAs have truncated D- or T-domains. Many also have far fewer and less complex posttranscriptional modifications relative to their cytoplasmic counterparts. Yet, mitochondrial tRNAs are believed to fold into a global functional form similar to the L-shaped structure of cytoplasmic tRNAs (Figure 1).33 In contrast to proteins, the RNA polyanion places a heavy dependence on the type and concentration of counter-ions in the folding process.10,11,14,34,35 RNAs undergo entropy driven, counter-ion induced collapse to partially correct and partially misfolded, intermediate states.11,35–39 Therefore, electrostatic interactions probably dominate the free energy of folding interactions and result in stable tertiary structures being observed in the presence of divalent metal ions (Mg2+) or sometimes in high concentrations of monovalent ion.34,36,40,41
The susceptibility of truncated mtRNA to misfold into kinetic traps depends the local concentration of counter-ions and on the relative stabilities of the native versus many non-native interactions.42 Certain parameters diminish or eliminate kinetic traps of the RNA folding landscape,43 and Mg2+-induced folding from secondary structure-based, intermediate states to tertiary structure native states has been the parameter most often investigated.11,36,40,44 The counter-ion collapsed intermediate structures11,39,45 are considered somewhat similar to protein molten globule intermediates.45 The counter-ion dependent folding of native yeast tRNAPhe and the unmodified tRNAPhe transcript suggest that several folding pathways exist in which distinct, stable intermediate conformations can be observed.14,34 Thus, it is likely that Mg2+ serves as a counterion to RNA to facilitate folding interactions. In some cases, this creates the RNA counterpart to the protein molten globule, and will produce a fully folded or partially folded molecule.
Considering this understanding of the effect of Mg2+ on RNAs in general, the effect of Mg2+ on the D-half of bmtRNAMet is counterintuitive. In the absence of Mg2+, PAGE, UV-thermal denaturations, CD and NMR strongly indicated that the D-domain half molecule formed a single, folded conformation. However, at 1 mM Mg2+ two discrete D-half conformations exist. Only the slower migrating form in PAGE was capable of interacting with the T-half. Interestingly, this predominant conformer at 3 mM Mg2+ was largely unfolded based on thermal denaturations, CD and NMR. In comparison, the T-half molecule PAGE migration was not affected by Mg2+, and neither was its melting temperature. Thus, we conclude that, in contrast to conventional thought, Mg2+ unfolds the D-half molecule of bmtRNAMet. Most observations, including that of randomly generated RNA sequences,46 support the idea of magnesium-dependent folding events. We and others had found that the addition of Mg2+ to ASLs and TSLs did not enhance spectral properties; rather, low mM concentrations reduced the intensities and broadened the NMR line-widths of the imino base-paired protons.47,48 Also, the catalytic core of the bI5 group I intron does not appear to fold into its native structure, but is in equilibrium between expanded and multiple collapsed non-native states at physiological Mg2+ concentrations.49 At physiological concentrations, Mg2+ rescues the bmtRNAMet from a stable misfolded state, but does so by maintaining the D-half of the molecule in an open conformation until a functional folding interaction can be achieved with the T-half of the molecule. Since transcription occurs from the 5′ to the 3′ end, the D-domain half of all tRNAs is transcribed first. Stable co-transcriptional folding of the D-domain could hinder its interaction with the T-domain, which is transcribed later. The folding of RNA in vivo begins with the folding of the nascent, 5′-terminal polymer sequence, and the nucleoside sequence no doubt influences folding during transcription.50 Early and more recent studies have demonstrated that nascent RNAs could fold into and out of conformations before the correct fold was achieved.51,52 Studies of the sequential folding of tRNA indicated that the correct folding of helical stems occurs before the entire molecule is formed.53 The first completed stem during synthesis would be that of the D-domain, and the last completed stem would be that of the amino acid accepting domain (Figure 1). In the study of folding interactions of the 5′-domain of 16S rRNA, the rRNA guided the kinetic folding pathway yielding the expected tertiary interactions.54 However, nascent RNAs can form stable intermediates that differ in conformation from those found in renaturation of the complete molecule.55
Is it possible that site-specific coordination of Mg2+ to a specific oligonucleoside sequence and/or structure is common to mitochondrial tRNA folding interactions? An apparent Kd of 2.5 mM for Mg2+ binding with the D-half molecule suggests otherwise, but may reflect the non-specific coordination of Mg2+ to the phosphodiester backbone. Functional folding of the D-domain may require more than a single critical Mg2+ ion. The revised structure of yeast tRNAPhe, which is based upon the most recent crystallographic analyses,13 exhibits 11 metal ion coordination sites, only some of which are shown in Figure 1. Three Mg2+ ions are coordinated to the D-domain. Two, coordinated to G19OP and G20N7 and to G20OP and A21OP, appear at locations where moderate changes in backbone direction are found, and may not bind to the truncated D-loop of bmtRNAMet (Figure 1). However, the third Mg2+ is of particular interest for it is located at a tight turn between the 5′-side of the amino acid accepting stem and of the D-stem. This Mg2+ ion is fully hydrated with six waters which in turn appear coordinated to U8OP, A9OP, C11OP, U12O4 and U12OP. Thus, the Mg2+ coordination at this site could be the single most important ion interaction for the correct folding of the D-half.
RNAs are composed for the most part of just the four nucleoside chemistries of A, U, G and C, and mitochondrial RNAs are generally rich in A and U, in contrast to proteins with 20 different chemical side chains. Site specific modifications provide RNAs with chemical and structural alternatives, which are not otherwise possible.3 Mostly circumstantial evidence implicates the timing of modifications to their effects on RNA folding events, including folding and functional interactions that lead to native, biologically active RNA.4,8,56 Highly conserved and site specific posttranscriptional modifications occur early in the maturation of many RNAs. In particular, early events include the synthesis of pseudouridine (Ψ) and ribothymidine (T) in RNA processing.6,57 Conserved modified nucleosides are located predominantly in secondary structural motifs that are engaged in tertiary structure folding and functional interactions, i.e. in terminal loops at the ends of stems (hairpins), in the middle of stems (single and double strand bulges) or at the junctions of antiparallel duplex stems. Modified nucleosides have been implicated in the specific coordination of Mg2+ by native tRNAs, in lowering the concentration of Mg2+ required for tertiary structure interactions, and in thermal stabilization of RNA.3 Mg2+ induced low energy stages have been observed in native, modified tRNA separated by conformational transitions,14 leading to the possibility of multiple folding pathways to the native state both in vitro and in vivo.14,41,58 In contrast to native tRNA, tertiary structure interactions of unmodified transcripts have an absolute requirement for either high millimolar concentrations of Mg2+ or molar concentrations of monovalent ion.34,59,60
The incorporation of Ψ into the individual half molecules affected Tm and hyperchromicity differently from its effects on the reconstituted bmtRNAMet. Ψ’s contributions are sequence context dependent, particularly in regard to stems versus loops.3,24,25,61–65 With incorporation of Ψ, we observed significant decreases in Tm of both the D- and T-half molecules in the absence of Mg2+ (Table 1). In addition, there was a concomitant decrease in hyperchromicity for the D-half molecule, but the T-half molecule exhibited an increase in hyperchromicity with incorporation of Ψ50. We reported previously that the Tm of an ASL can be reduced by modification of the loop because the modification negates intraloop hydrogen bonding that extends the length of the stem.24,26,27 Yet, the very same modification increases order in the loop (hyperchromicity) because the modified nucleoside is a better base stacker than the unmodified nucleoside. In the absence of empirically determined structures for the D- and T-half molecules with and without Mg2+, we postulate that Ψ27 at the 5′-teminus of the ASL stem, and Ψ50 near the 5′-terminus of T-stem destabilize the transient half-molecule structures. Yet Ψ50 increases base stacking. The ubiquitous and highly conserved modification Ψ adds to the thermal stability of fully formed RNA duplexes. However, the increase in Tm with modification is usually nominal, between 1.5 and 4.5 °C (at pH 7),24,61–64 but sometimes higher, 6.7 °C.65 In contrast, Ψ contributed greatly to the thermal stability of the reconstituted bmtRNAMet. The complex formed between D(Ψ27) and T half molecules exhibited a Tm that was approximately 9 °C higher than that of the unmodified complex. A similar result was obtained for the complex of D with T(Ψ50). These contributions equate to an increase in stability of one additional kcal/mole. The enhanced stabilization by Ψ could be attributed to the relatively rich A-U sequence of bmtRNAMet compared to cytoplasmic tRNAs and rRNAs that have been studied. In the native and reconstituted bmtRNAMet, Ψ27 and Ψ50 are engaged in H-bonding at or near the 5′-termini of stems. However, their positive, individual contributions to complex stability were not additive. Their positions close to the tRNA “hinge” region, and at the ends of stems may have reduced their combined effects. There is redundancy built into the modification of cytoplasmic tRNAs,25 and the same may be true for Ψ27 and Ψ50 in mtRNA.
It appears that Mg2+ concentration does not substitute for a site-specific modification in the functional folding of at least one mitochondrial tRNA, human mtRNALys.8 This tRNA has a truncated D-domain incapable of D- to T-loop interactions conserved in cytoplasmic tRNAs. The truncated bmtRNAMet can not effectively achieve the functional conformation for aminoacylation in the absence of modification, whereas the unmodified ymtRNALeu was capable of acquiring the correct global fold, as was yeast cytoplasmic tRNAPhe.4 Conserved modifications that are introduced within the four helical junction of the mtRNA cloverleaf secondary structure may be critical to folding by ordering the structure, negating preferential Watson-Crick base pairs3 and channeling the folding pathway to the functional mtRNA structure. Thus, conserved modifications that are synthesized as early events in the processing of RNA may direct the folding pathway and maintain structural stability. This would be particularly important for mtRNAs with truncated domains that are more likely to enter kinetic traps while undergoing folding.
CONCLUSION
It is apparent that Mg2+ concentration is important for the functional folding interactions of at least one of the truncated mitochondrial tRNA, bmtRNAMet, and perhaps a number of other mtRNAs. Counter to general observations with RNA, the 5′-half of bmtRNAMet is relaxed, even disordered, by the presence of Mg2+, only to become re-ordered in a folding interaction with the 3′-half of the molecule to achieve a competent tRNA. Highly conserved modifications facilitate this folding interaction and function.
MATERIALS AND METHODS
RNA chemical synthesis, purification and analyses
The E. coli tRNALeu, bovine mitochondrial tRNAMet (bmtRNAMet) and yeast mitochondrial tRNALeu1(ymtRNALeu1) were chemically synthesized as D- and T-domain half molecules with and without modifications (Figure 1) (Dharmacon RNA Technologies, Inc., Lafayette, CO). Chemical synthesis permits site selected incorporation of modified nucleosides by using their respective phosphoramidites. The protected pseudouridine (Ψ) phosphoramidite was a product of Dharmacon RNA Technologies, and was incorporated at the native positions, 27 and 50 (Figure 1).16 The nucleoside D was purchased (TriLink Biotechnologies, San Diego, CA) and derivatized to the “ACE” chemistry (Dharmacon RNA Technologies). The modified and unmodified D- and T-half molecules of bmtRNAMet were synthesized to correspond to the native sequences of the tRNA. The modified and unmodified D- and T-domain half molecules of ymtRNALeu1-ΔACVL and E. coli tRNALeu-ΔACVL were synthesized to correspond to the native sequence of the tRNAs minus the anticodon and variable loop domains. The modified T-half molecule of E. coli tRNALeu-ΔACVL was synthesized with ribothymidine-54 or T54. Also, a unimolecular 60 nucleotide E. coli tRNALeu-ΔACVL was chemically synthesized. The RNAs were deprotected with TEMED-acetate (Dharmacon RNA Technologies), lyophilized and redissolved in H2O. Ionic exchange HPLC66 and polyacrylamide gel electrophoresis (PAGE) under denaturing conditions (89 mM Tris base, 89 mM boric acid, 7 M urea, 2.0 mM EDTA, pH 8.3) were used to confirm the purity of the RNAs.
RNA transcription products
RNAs that are chemically synthesized are sometimes not fully-deprotected and thus, are only partially active. RNAs produced by transcription in vitro do not have specifically sited modified nucleosides and may have added 3′-nucleosides. The aminoacylation activities of chemically synthesized and transcribed products were compared with each other and with tRNAs reconstituted from D- and T-half molecules. A full-length bovine mitochondrial tRNAMet (bmtRNAMet) transcript was prepared as previously described.31 The ymtRNALeu1-ΔACVL transcript was constructed without the anticodon stem and loop domain. Deletion of the anticodon stem and loop domain was accomplished using primers ymtDNALeu1-ΔAC-FWD [5′-GGAATTGGTAGA CACGATTACTTTACAGTATGAAGG-3′] and ymtDNALeu1-ΔAC-REV [5′-CCTTCATACTG TAAAGTAATCGTGTCTACCAATTCC-3′] by polymerase chain reaction (PCR) based mutagenesis. Each 50 μl PCR reaction contained 25 ng of the plasmid ymtDNALeu1,32 10 pmoles of each forward and reverse primer, 0.2 mM dNTPs, 1X Pfu DNA polymerase buffer and 2.5 U Pfu DNA polymerase (Stratagene, La Jolla, CA). The PCR reaction was digested with 20 U Dpn I prior to being used for transformation of E. coli DH5α (Stratagene, La Jolla, CA). The plasmid, pYMtJLH1, was isolated using a Qiaprep mini-prep spin kit (Qiagen, Valencia, CA) and the DNA was sequenced to confirm the deletion of the anticodon stem loop (SeqWright, Houston, TX). To delete both the anticodon stem loop and the variable loop, primers ymtDNALeu1-ΔACVL-FWD [5′-GGAATTGGTAGACACGATTATGAAGGTTCAAGTCC-3′] and ymtDNALeu1-ΔACVL-REV [5′-GGACTTGAACCTTCATAATCGTGTCTACCAATTCC-3′] were used in a PCR reaction as described above except that the plasmid pYMtJLH1 was used as template.
The plasmid pymtDNA Leu1 containing the gene of ymtRNALeu UAA and pymtJLH3 containing the gene of yeast mitochondrial tRNALeu1 with the anticodon and variable loop deletions were digested with BstN I and then used as template for in vitro transcription reactions.32,67,68 The ethanol precipitated RNA transcript was re-suspended and purified by a denaturing PAGE. The RNA, detected by UV shadowing, was excised, crushed, and then extracted twice by overnight incubation with shaking at 37 °C (0.5 M NH4-acetate, 1 mM EDTA, pH 8.0). The combined supernatants were filtered (0.2 μm syringe filter) and concentrated to 500 μl with butanol. Glycogen (0.5 mg/ml) was added and the RNA was precipitated overnight with ethanol at −80 °C.68 The tRNA pellet was collected by centrifugation, washed twice with 70% ethanol and vacuum dried. The concentration of the tRNA was determined by absorbance at 260 nm using the extinction coefficient of 8.785 × 105 L/mol·cm for ymtRNALeu UAA and 6.194 × 105 L/mol•cm for yeast mitochondrial tRNA Leu1-ΔACVL.69 Purified tRNALeu was denatured at 80 °C for 1 min, followed by the addition of 1 mM MgCl2 and quick-cooled on ice.
Polyacrylamide gel mobility-shift assay
The electrophoretic mobility of the various individual tRNA constructs, the half molecule D- and T-domains, and their interactions (mobility shift) was assessed with PAGE under the native conditions of a 15% polyacrylamide gel with Tris-borate, TB, buffer (89 mM Tris base, 89 mM boric acid, pH 8.3). The concentration of Mg2+ in the TB running buffer and the gel-formation reaction was 3 mM unless otherwise noted. PAGE was conducted at 4 ± 0.5 °C using a temperature controlled gel electrophoresis (Novex Mini-cell Thermoflow, Invitrogen, Carlsbad CA). Gel shift analyses of D-domain half molecules interacting with T-domain half molecules were conducted with several combinations composed of modified and unmodified D- and T-domains. Concentrations of the D-half molecule were kept constant and titrated with increasing amounts of T-half molecules. The reaction mixtures were heated at 50 °C for 5 min and cooled to room temperature before Mg2+ was added. Prior to applying the RNA samples to the gel, they were diluted with loading buffer (3/1; TB, 50% glycerol; 0.025% w/v Bromophenol Blue; 0.025% w/v Xylene Cyanol FF). After electrophoresis, gels were stained with ethidium bromide (0.5 μg/ml) for 10 min and digitally photographed (Bio Rad Imager, Quantity One software; Hercules, CA). The RNA bands were quantified by analyzing the density of the bands using ImageQuant software (Molecular Dynamics, Amersham Biosciences, Piscataway, NJ). Background correction for all the lanes was taken from a blank area on the gel. Percent complex was determined by the density of the bands and normalized. The most complex formed in a gel analyses represented 100%, whereas the lane of only the D-domain half molecule represented 0%. Dissociation constants, Kd, were determined from graphs of bound and free D-domain half molecule using a ‘one-site binding model’ (Prism, GraphPad Software, San Diego, CA).
RNA extraction from polyacrylamide gels and denaturating gel electrophoresis
The D-half molecule (30 μM) was subjected to PAGE in four adjacent lanes and in the presence of 3mM Mg2+. After running the gel, one lane was removed and stained with ethidium bromide to reveal the position of the slow migrating D-half molecule. Stained and unstained gel pieces were aligned by the bands of the D-half molecule revealed by UV irradiation. Using the stained gel piece as a guide, an area of the unstained gel corresponding to the migration of the D-half molecule was removed and placed on top of a 20 mM EDTA, 15% polyacrylamide gel in the absence of Mg2+. The D-half molecule was run from the gel slices into the gel resulting in a gel shift to the rapidly migrating D-half molecule. The opposite experiment was performed in which the rapidly migrating D-half molecule with a gel in the absence of Mg2+ was subjected to PAGE with 3 mM Mg2+ resulting in a slow moving D-half molecule.
Temperature controlled UV-spectroscopy
The thermal stability of the RNAs was investigated in the presence and absence of Mg2+ (Varian, Cary III, temperature controlled, UV-spectrometer).24,70 The D- and T-domain half molecules were diluted in TB buffer with and without Mg2+ and to a concentration of ~0.5–2.0 μM. The concentration of Mg2+ was 3 mM unless otherwise indicated. The temperature range for each study was 5 °C to 90 °C, with a ramp rate of 0.5 °C per minute. Two readings were acquired each minute. The Tm and thermodynamic parameters for each RNA were extracted from the denaturation/renaturation data using the line fitting and analysis program MeltWin.70 E. coli tRNAMet (Sigma, St. Louis, MO) was used for comparison of Tms and hyperchromicity.
Temperature controlled circular dichroism (CD) spectropolarimetry
Circular dichroism, CD, spectra were recorded using a Jasco J600 spectropolarimeter and an interfaced computer.71,72 Temperature was controlled at 4 °C with 1 cm path length, jacketed, cylindrical sample cell and RNA sample concentrations adjusted to ~0.2 absorbance units at 260 nm. RNA samples were titrated with Mg2+ to final concentrations of 0, 0.5, 1, 2, 3 and 10 mM. CD data are baseline corrected for signals due to the cell and buffer.
Nuclear magnetic resonance (NMR) spectrometry
The affect of Mg2+ on the bmtRNAMet D-half molecule was studied using proton NMR (600 MHz, Varian Instruments). The bmtRNAMet D-domain half molecule (0.8 mM) in 2H Tris (Cambridge Isotope Laboratories)-Borate buffer (5% D2O) was titrated with 33 mM Mg2+. Seven additions of Mg2+ were conducted and spectra taken at 0, 0.8, 2.3, 3.7, 4.9, 6.1, 7.1 and 8.1 mM Mg2+. One dimensional spectra with a sweep width of 12000 Hz were acquired at 4 °C using ‘watergate’73 for suppression of the H2O signal and a delay of 1.5 sec. In the presence of Mg2+, spectra exhibited line broadening. Thus, each spectrum was acquired with 1024 scans. In the absence of Mg2+, only 512 scans were sufficient to achieve a similar signal to noise ratio.
Isolation of aminoacyl-tRNA synthetases and aminoacylation of tRNAs, transcripts, and D-domain and T-domain half molecule complexes
The human MetRSmt was purified as described previously.31 The D-domain and T-domain half molecules were resuspended in diethylpyrocarbonate-treated H2O to a final concentration of 30 μM. The hybrids were formed by combining the various half molecules at a final concentration of 6.5 μM in a solution containing 3 mM MgCl2 and heating at 50 °C for 5 min. The samples were then allowed to cool at room temperature for 30 min prior to the aminoacylation assay. Aminoacylation of the bmtRNAMet transcript, individual half molecules, and half molecule complexes were conducted as described.31 Reaction mixtures (60 μL) contained 50 mM Tris-HCl pH 7.6, 10 mM MgCl2, 2.5 mM ATP, 1 mM spermine, 10 U SUPERase•In™ (Ambion, Austin, TX), 20 μM [35S]-Methionine (~ 7000 cpm/pmol), 4 μg of partially purified human MetRSmt, and either 4.2 μM final concentration of half molecules or ~ 0.05 μM bmtRNAMet transcript. The reaction was carried out at 25 °C. At the indicated time points, 10 μL was removed and precipitated with ice-cold 5 % trichloroacetic acid (TCA) with the addition of unfractionated yeast RNA (yeast ribonucleate, Sigma) at 0 °C for 10 min. The precipitated tRNA was collected on nitrocellulose filters (Millipore Corporation, Billerica, MA) and the radioactivity incorporated was determined by liquid scintillation counting. The zero time point has been subtracted from each value.
Yeast mitochondrial LeuRS (LeuRSmt) was expressed in E. coli, isolated and purified. Cultures were inoculated with a single BL21+ (codon plus) colony harboring the plasmid pYM3-17,74 which encodes the wildtype yeast LeuRSmt fused to an N-terminal six-histidine tag. A 3 ml culture (Luria Broth; 100 μg/ml ampicillin; 34 μg/ml chloramphenicol) incubated at 37 °C overnight was used to inoculate a 500 ml LB culture and then grown at 30 °C. At an OD600 between 0.6 and 0.8, the expression of the recombinant yeast mitochondrial protein was induced (1 mM isopropyl-β-D-thiogalactopyranoside; 2 hours; 30 °C). The cells were collected by centrifugation (6,000g; 15 min). The recombinant E. coli LeuRS proteins were expressed using the same procedures with the exception that E. coli BL21 was transformed with plasmid p15ec3-175 and cells were grown and induced at 37 °C. The cells were harvest and LeuRS was affinity purified using His-Select resin (Sigma, St. Louis, MO) as previously described.32 The purified protein was concentrated (Centricon-50; Amicon, Bedford, MA) and the final protein concentration determined (Bio-Rad Protein Assay).
For aminoacylation of ymtRNALeu1 half molecule complexes, complex formation was achieved at 100 μM of both the D- and the T-domain halves incubated in 60 mM Tris-HCl, pH 7.5 and 1 mM DTT, 60 °C, 2 min. RNA was cooled at room temperature for 30 min, MgCl2 added (3 mM) and incubated for an additional 30 min at room temperature. Each aminoacylation reaction contained 4 μM folded ymtRNALeu1 transcripts, individual half molecules or complexed D- and T-half molecules and 50 nM yeast mitochondrial LeuRS in 60 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 21 μM [3H]-L-leucine, 150 μCi/ml.76,77 The reactions were initiated by the addition of 4 mM ATP and then quenched at specific time points by transfer to filter pads (Whatman, Clifton, NJ) that had been pre-soaked with 5% trichloroacetic acid (TCA). The pads were washed and the radioactivity on each was quantified by scintillation counting as previously described.32 For aminoacylation of E. coli tRNALeu or complexed D- and T- half molecule complexes, 4 μM tRNALeu or 4 μM D-domain halves with 6 μM of T-domain halves were added to each aminoacylation reaction and then conducted as described above.
Acknowledgments
The authors wish to acknowledge the contributions of William Graham (NCSU) in the purification of the chemically synthesized RNAs, Ann Scheunemann (NCSU) for conducting the Ca2+ experiments, and Lise Barley-Maloney (NCSU) in the writing of manuscript. We thank Dr. Rob Kaiser (Dharmacon RNA Technologies) for aid and advice in the synthesis of the dihydrouridine phosphoramidite, Audrey Nelson (NCSU) for help in the initial thermal denaturations, and Keith Gagnon and Dr. Paul Wollenzien (NCSU) for conducting the photoactivated crosslinking of 32P-end labeled RNA. Christopher Jones was a National Science Foundation REU awardee (MCB9986011 to P.F.A.). The research was supported in part by the National Science Foundation (MCB9986011 to P.F.A.), and by the National Institutes of Health (2-RO1-GM23037 to P.F.A, 2-RO1-GM32734 to L.S., and 2-RO1-GM63107 to S.M.).
Footnotes
Angela C. Spencer: Greenville Technical College, Greenville, SC 29606
References
- 1.Wittenhagen LM, Kelley SO. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem Sci. 2003;28:605–11. doi: 10.1016/j.tibs.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Coulbault L, Herlicoviez D, Chapon F, Read MH, Penniello MJ, Reynier P, Fayet G, Lombes A, Jauzac P, Allouche S. A novel mutation in the mitochondrial tRNAAsn gene associated with a lethal disease. Biochem Biophys Res Commun. 2005;329:1152–1154. doi: 10.1016/j.bbrc.2005.02.083. [DOI] [PubMed] [Google Scholar]
- 3.Agris PF. The importance of being modified: Roles of modified nucleosides and Mg2+ in RNA structure and function. In: Cohn W, Moldave K, editors. Progress in Nucleic Acid Research and Molecular Biology. Vol. 53. Academic Press; NY: 1996. p. 79. [DOI] [PubMed] [Google Scholar]
- 4.Nobles KN, Yarian CS, Guenther RH, Agris PF. Highly conserved modified nucleosides influence Mg2+_dependent tRNA folding. Nucl Acids Res. 2001;30:4751–4760. doi: 10.1093/nar/gkf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucl Acids Res. 2005;33:D139–140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrell BG, Seidman JG, Guthrie C, McClain WH. Transfer RNA biosynthesis: the nucleotide sequence of a precursor to serine and proline transfer RNAs. Proc Natl Acad Sci U S A. 1974;71:413–416. doi: 10.1073/pnas.71.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helm M, Brule H, Degoul F, Cepanec C, Leroux JP, Giege R, Florentz C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucl Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helm M, Giege R, Florentz C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999;38:13338–13346. doi: 10.1021/bi991061g. [DOI] [PubMed] [Google Scholar]
- 9.Rainaldi G, Volpicella M, Licciulli F, Liuni S, Gallerani R, Ceci LR. PLMItRNA, a database on the heterogeneous genetic origin of mitochondrial tRNA genes and tRNAs in photosynthetic eukaryotes. Nucl Acids Res. 2003;31:436–438. doi: 10.1093/nar/gkg080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onoa B, Tinoco I., Jr RNA folding and unfolding. Curr Opin Struct Biol. 2004;14:374–379. doi: 10.1016/j.sbi.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Thirumalai D, Lee N, Woodson SA, Klimov DK. Early events in RNA Folding. Annu Rev Phys Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 12.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J Mol Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 13.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serebrov V, Clarke RJ, Gross HJ, Kisselev L. Mg2+ induced tRNA folding. Biochemistry. 2001;40:6688–6698. doi: 10.1021/bi002241p. [DOI] [PubMed] [Google Scholar]
- 15.Sakurai M, Ohtsuki T, Watanabe K. Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucl Acids Res. 2005;33:1653–1661. doi: 10.1093/nar/gki309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriya J, Yokogawa T, Wakita K, Ueda T, Nishikawa K, Crain PF, Hashizume T, Pomerantz SC, McCloskey JA, Kawai G, Hayashi N, Yokoyama S, Watanabe K. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry. 1994;33:2234–2239. doi: 10.1021/bi00174a033. [DOI] [PubMed] [Google Scholar]
- 17.Larkin DC, Williams AM, Martinis SA, Fox GE. Identification of essential domains for Escherichia coli tRNALeu aminoacylation and amino acid editing using minimalist RNA molecules. Nucl Acids Res. 2002;30:2103–2113. doi: 10.1093/nar/30.10.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittenhagen LM, Kelley SO. Dimerization of a pathogenic human mitochondrial tRNA. Nat Struct Biol. 2002;9:586–590. doi: 10.1038/nsb820. [DOI] [PubMed] [Google Scholar]
- 19.Behlen LS, Sampson JR, Uhlenbeck OC. An ultraviolet light-induced crosslink in yeast tRNAPhe. Nucl Acids Res. 1992;20:4055–4059. doi: 10.1093/nar/20.15.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montero M, Alonso MT, Carnicero E, Cuchillo-Ibáòez I, Albillos A, Garcia AG, Garcia-Sancho J, Alvarez J. Chromaffin-cell stimulation triggers fast millimolar mitochondrial Ca2+ transients that modulate secretion. Nature Cell Biology. 2000;2:57–61. doi: 10.1038/35000001. [DOI] [PubMed] [Google Scholar]
- 21.Clark CL, Cecil PK, Singh D, Gray DM. CD, absorption and thermodynamic analysis of repeating dinucleotide DNA, RNA and hybrid duplexes [d/r(AC)]12·[d/r(GT/U)]12 and the influence of phosphorothioate substitution. Nucl Acids Res. 1997;25:4098–4106. doi: 10.1093/nar/25.20.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agris PF, Dao V, Basti M, Guenther R. Ribosome-independent anticodon to codon binding assessed by circular dichroism: Roles of base modifications, Mg2+ and 2′OH. Biospectroscopy. 1996;2:205–217. [Google Scholar]
- 23.Sosnick TR, Fang X, Shelton VM. Application of circular dichroism to study RNA folding transitions. Methods Enzymol. 2000;317:393–409. doi: 10.1016/s0076-6879(00)17026-0. [DOI] [PubMed] [Google Scholar]
- 24.Yarian CS, Cain R, Basti MM, Ansari G, Guenther RH, Sochacka E, Malkiewicz A, Agris PF. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA’s position 39. Nucl Acids Res. 1999;27:3543–3549. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agris PF. Decoding the Genome, A Modified View. Nucl Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koshlap KM, Guenther RH, Sochacka E, Malkiewicz A, Agris PF. A distinctive RNA Fold: the solution structure of an analog of the yeast tRNAPhe TΨC domain. Biochemistry. 1999;38:8647–8656. doi: 10.1021/bi990118w. [DOI] [PubMed] [Google Scholar]
- 27.Sengupta R, Vainauskas S, Yarian C, Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM, Agris PF. Modified constructs of tRNA’s TΨC-domain to probe substrate conformational requirements of m1A58 and m5U54-tRNA methyltransferases. Nucl Acids Res. 2000;28:1374–1380. doi: 10.1093/nar/28.6.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gale AJ, Shi JP, Schimmel P. Evidence that specificity of microhelix charging by a class I tRNA synthetase occurs in the transition state of catalysis. Biochemistry. 1996;35:608–615. doi: 10.1021/bi9520904. [DOI] [PubMed] [Google Scholar]
- 29.Sugiura I, Nureki O, Ugaji-Yoshikawa Y, Kuwabara S, Shimada A, Tateno M, Lorber B, Giege R, Moras D, Yokoyama S, Konno M. The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules. Structure. 2000;8:197–208. doi: 10.1016/s0969-2126(00)00095-2. [DOI] [PubMed] [Google Scholar]
- 30.Senger B, Fasiolo F. Yeast tRNA(Met) recognition by methionyl-tRNA synthetase requires determinants from the primary, secondary and tertiary structure: a review. Biochimie. 1996;78:597–604. doi: 10.1016/s0300-9084(96)80006-x. [DOI] [PubMed] [Google Scholar]
- 31.Spencer AC, Heck A, Takeuchi N, Watanabe K, Spremulli LL. Characterization of the human mitochondrial methionyl-tRNA synthetase. Biochemistry. 2004;43:9743–9754. doi: 10.1021/bi049639w. [DOI] [PubMed] [Google Scholar]
- 32.Hsu JL, Rho SB, Vannella KM, Martinis SA. Functional divergence of a unique C-terminal domain of leucyl-tRNA synthetase to accommodate its splicing and aminoacylation roles. J Biol Chem. 2006 doi: 10.1074/jbc.M601606200. in press. [DOI] [PubMed] [Google Scholar]
- 33.Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C. Search for characteristic structural features of mammalian mito-tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shelton VM, Sosnick TR, Pan T. Altering the intermediate in the equilibrium folding of unmodified yeast tRNAPhe with monovalent and divalent cations. Biochemistry. 2001;40:3629–3638. doi: 10.1021/bi002646+. [DOI] [PubMed] [Google Scholar]
- 35.Heilman-Miller SL, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. I Equilibrium stabilization by cations. J Mol Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 36.Pyle AM. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 37.Russell R, Millett IS, Doniach S, Hershlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nature Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 38.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nature Struct Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 39.Murthy VL, Rose GD. Is counterion delocalization responsible for collapse in RNA folding? Biochemistry. 2000;39:14365–14370. doi: 10.1021/bi001820r. [DOI] [PubMed] [Google Scholar]
- 40.Woodson SA. Compact but disordered states of RNA. Nature Struct Biol. 2000;7:349–352. doi: 10.1038/75106. [DOI] [PubMed] [Google Scholar]
- 41.Wilson TJ, Zhao ZY, Maxwell K, Kontogiannis L, Lilley DM. Importance of specific nucleotides in the folding of the natural form of the hairpin ribozyme. Biochemistry. 2001;40:2291–2302. doi: 10.1021/bi002644p. [DOI] [PubMed] [Google Scholar]
- 42.Woodson SA. Folding mechanisms of group I ribozymes: role of stability and contact order. Biochem Soc Trans. 2001;30:1166–1169. doi: 10.1042/bst0301166. [DOI] [PubMed] [Google Scholar]
- 43.Trieber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 44.Tinoco I, Jr, Bustamante B. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 45.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci US A. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat Struct Mol Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- 47.Stuart JW, Koshlap KM, Guenther R, Agris PF. Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J Mol Biol. 2003;334:901–918. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 48.Cabello-Villegas J, Winkler ME, Nikonowicz EP. Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J Mol Biol. 2002;319:1015–1034. doi: 10.1016/S0022-2836(02)00382-0. [DOI] [PubMed] [Google Scholar]
- 49.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nat Struct Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 50.Schroeder R, Grossberger R, Pichler A, Waldsich C. RNA folding in vivo. Curr Opin Struct Biol. 2002;12:296–300. doi: 10.1016/s0959-440x(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 51.Kramer FR, Mills DR. Secondary structure formation during RNA synthesis. Nucl Acids Res. 1981;9:5109–5124. doi: 10.1093/nar/9.19.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan T, Sosnick T. RNA folding during transcription. Annual Review of Biophysics and Biomolecular Structure. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 53.Boyle J, Robillard GT, Kim SH. Sequential folding of transfer RNA. A nuclear magnetic resonance study of successively longer tRNA fragments with a common 5′ end. J Mol Biol. 1980;139:601–625. doi: 10.1016/0022-2836(80)90051-0. [DOI] [PubMed] [Google Scholar]
- 54.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J Mol Biol. 2005;351:508–519. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 55.Sosnick TR, Pan T. RNA folding: models and perspectives. Curr Opin Struct Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 56.Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA. 2001;7:833–845. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Björk GR. Biosynthesis and function of modified nucleosides. In: Söll D, RajBhandary UL, editors. tRNA: Structure, biosynthesis and function. ASM; Wash. D.C.: 1995. pp. 165–205. [Google Scholar]
- 58.Schroeder R, Grossberger R, Pichler A, Waldsich C. RNA folding in vivo. Curr Opin Struct Biol. 2002;12:296–300. doi: 10.1016/s0959-440x(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 59.Zheng M, Wu M, Tinoco I., Jr Formation of a GNRA tetraloop in P5abc can disrupt an interdomain interaction in the Tetrahymena group I ribozyme. Proc Natl Acad Sci USA. 2001;98:3695–3700. doi: 10.1073/pnas.051608598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silverman SK, Zheng M, Wu M, Tinoco I, Jr, Cech TR. Quantifying the energetic interplay of RNA tertiary and secondary structure interactions. RNA. 1999;5:1555–1674. doi: 10.1017/s1355838299991823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 62.Cabello-Villegas J, Nikonowicz EP. Solution structure of psi32-modified anticodon stem-loop of Escherichia coli tRNAPhe. Nucl Acids Res. 2005;33:6961–6971. doi: 10.1093/nar/gki1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meroueh M, Grohar PJ, Qiu J, SantaLucia J, Jr, Scaringe SA, Chow CS. Unique structural and stabilizing roles for the individual pseudouridine residues in the 1920 region of Escherichia coli 23S rRNA. Nucl Acids Res. 2000;28:2075–2083. doi: 10.1093/nar/28.10.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sumita M, Desaulniers JP, Chang YC, Chui HM, Clos L, 2nd, Chow CS. Effects of nucleotide substitution and modification on the stability and structure of helix 69 from 28S rRNA. RNA. 2005;11:1420–1429. doi: 10.1261/rna.2320605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, Agris PF. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry. 2000;39:13390–13395. doi: 10.1021/bi001302g. [DOI] [PubMed] [Google Scholar]
- 66.Guenther RH, Gopal DH, Agris PF. Purification of Transfer RNA Species by Single-Step Ion-Exchange HPLC. J Chromatogr. 1988;444:79–87. doi: 10.1016/s0021-9673(01)94010-5. [DOI] [PubMed] [Google Scholar]
- 67.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucl Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 70.McDowell JA, Turner DH. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 71.Guenther RH, Hardin CC, Sierzputowska-Gracz H, Agris PF. Magnesium-induced conformational transition in a DNA analog of the yeast tRNAPhe anticodon stem-loop. Biochemistry. 1992;31:11004–11011. doi: 10.1021/bi00160a009. [DOI] [PubMed] [Google Scholar]
- 72.Dao V, Guenther RH, Agris PF. The Role of 5-methylcytidine in the anticodon arm of yeast tRNAPhe: Site-specific Mg2+ binding and coupled conformational transition in DNA analogs. Biochemistry. 1992;31:11012–11019. doi: 10.1021/bi00160a010. [DOI] [PubMed] [Google Scholar]
- 73.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 74.Rho SB, Martinis SA. The bI4 group I intron binds directly to both its protein splicing partners, a tRNA synthetase and maturase, to facilitate RNA splicing activity. RNA. 2000;6:1882–1894. doi: 10.1017/s1355838200001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinis SA, Fox GE. Non-standard amino acid recognition by Escherichia coli leucyl-tRNA synthetase. Nucleic Acids Symp Ser. 1977;26:125–128. [PubMed] [Google Scholar]
- 76.Lincecum TL, Jr, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grotli M, Martinis SA, Cusack S. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 77.Mursinna RS, Lincecum TL, Jr, Martinis SA. A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry. 2001;40:5376–5381. doi: 10.1021/bi002915w. [DOI] [PubMed] [Google Scholar]


