Abstract
Background
The type III secretion system (TTSS) is an important virulence determinant of Gram-negative bacterial pathogens. It enables the injection of effector proteins into the cytosol of eukaryotic cells. These effectors ultimately manipulate the cellular functions of the infected organism. Salmonella enterica serovar Typhimurium encodes two virulence associated TTSSs encoded by the Salmonella Pathogenicity Islands (SPI) 1 and 2 that are required for the intestinal and systemic phases of the infection, respectively. However, recent studies suggest that the roles of these TTSSs are not restricted to these compartments. The regulation of TTSSs in Salmonella is very complex with several regulators operating to activate or to repress expression depending on the environmental conditions.
Results
We performed a systematic analysis of the regulation of type III effectors during growth in vitro. We have tested the ability of seven regulatory genes to regulate ten effector genes. Each regulator was expressed in the absence of the other six to avoid cascade effects. Our results confirm and extend the previously reported regulation of TTSS1 and TTSS2 effectors by InvF-SicA and SsrB respectively.
Conclusion
The set of strains constructed for this study can be used to quickly and systematically study the regulation of newly identified effector genes of Salmonella enterica. The approach we have used can also be applied to study complex regulatory cascades in other bacterial species.
Background
The type III secretion system (TTSS) is a complex molecular machine found in numerous Gram-negative bacterial pathogens of animals and plants [1]. This secretion system encodes a syringe-like organelle that injects effector proteins directly into the cytosol of eukaryotic cells. The effectors ultimately affect host cell physiology.
S. enterica serovar Typhimurium (hereafter referred to simply as Typhimurium) possesses two virulence associated TTSSs encoded by the Salmonella Pathogenicity Islands (SPI) 1 and 2 [2]. TTSS1 (encoded by SPI1) delivers effectors that mediate the invasion of epithelial cells and the intestinal events of Typhimurium infection [3-5]. TTSS2 is required during the systemic phase of Typhimurium infection [6]. It secretes effectors that allow the survival and multiplication of the pathogen within macrophages [7]. The regulation of the Typhimurium TTSSs is complex with several regulatory proteins involved (Figure 1). SPI1 encodes five regulators, four of which are known to be involved in regulatory cascades that lead to the expression of genes inside and outside of SPI1 [8]. The central regulator of SPI1 gene expression is HilA, a member of the OmpR/ToxR family [5,8]. HilA directly activates the expression of two SPI1 operons that encode TTSS1 apparatus components [5,9]. One of these operons begins with the invF gene, which encodes a transcriptional activator of the AraC family. InvF activates the expression of TTSS1 effectors encoded both inside and outside of SPI1 [10,11]. The activity of InvF requires the SicA co-regulator which is also encoded within SPI1 [12,13]. The third and fourth regulators are HilC and HilD, both of which belong to the AraC family. Both can directly activate the expression of hilA [14,15]. They can also activate the expression of the invF operon independently of HilA [16,17]. The fifth regulator encoded within SPI1, SprB, contains a LuxR/UhpA helix-turn-helix motif, however no target genes for SprB have been identified [18].
Figure 1.
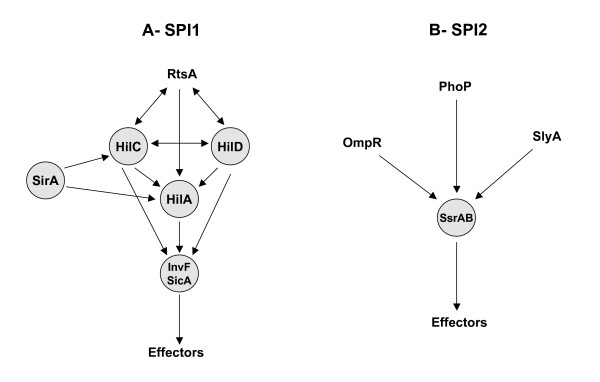
Regulatory cascades activating the expression of SPI1 (A) and SPI2 (B) related type III effectors. The regulators analyzed in this study are represented in circled boxes.
SPI1 gene expression is also regulated by factors encoded outside of SPI1 (Figure 1A). The SirA/BarA two-component system is an ancient regulatory system with orthologs found throughout the γ-proteobacteria that is involved in virulence gene expression, exoenzyme and antibiotic production, motility, and biofilm formation [19,20]. In Typhimurium, SirA has been shown to bind and activate the promoters of hilC and hilA [21], although an alternate model has also been proposed in which SirA acts through hilD rather than through hilA and hilC [22]. Additional regulators of SPI1 gene expression encoded outside of SPI1 include the two-component regulatory systems PhoPQ [23], PhoBR [24], and OmpR/EnvZ [24], and the AraC-like transcriptional activator RtsA [25]. How these regulatory inputs are integrated is not yet known.
The central regulator of SPI2, and some TTSS2 effector genes located outside of SPI2, is the SsrAB two-component system [26]. SsrB has been shown to bind and activate the promoter of the srfH/sseI gene that encodes a TTSS2 effector [27]. Similarly, SsrB is thought to directly activate the expression of SPI2 operons as well as other effector genes located outside of SPI2 [28,29]. The expression of ssrA and ssrB is activated by factors encoded outside of SPI2 (Figure 1B). These include the two-component systems OmpR/EnvZ [30], PhoPQ [31], and the slyA gene [32]. Recently the ydgT gene was identified as a negative regulator of SPI2 gene expression [33].
The effectors secreted by TTSS1 are required for the invasion of intestinal epithelial cells [4,34]. In contrast, the SPI2 genes are induced after Typhimurium has invaded or is phagocytized by eukaryotic cells [29,31,35-37]. These observations led to the hypothesis that TTSS1 is needed to invade intestinal cells, but is not required during the subsequent phases of Typhimurium pathogenesis, while TTSS2 is expressed only when the bacteria reside within eukaryotic cells. Several recent reports suggest a more complicated role of the Typhimurium TTSSs. For example, SPI1 mutants have a replication defect and are unable to synthesize a normal SCV inside epithelial cells suggesting that SPI1 genes are involved in these functions [38]. Conversely, SPI2 genes have been shown to be involved in the induction of the inflammatory response caused by S. enterica serovar Dublin in a bovine ligated-ileal-loop model [39] and to be essential for the full virulence of Typhimurium in murine infectious enterocolitis [40,41]. These results suggest that each TTSS might be expressed in different compartments within the host, and that at least some effectors might be involved in more than one phase of the infection.
Given the complexity of these regulatory hierarchies, we took a systematic in vitro approach to categorizing the regulatory inputs of effector genes in Typhimurium. Besides examining the effects of individual regulatory mutations on individual effectors, we wanted to test each regulatory gene in the absence of other regulators. For example, the regulatory hierarchy for SPI1 is complex in that SirA activates hilA and hilC, HilC also activates hilA, and HilA and HilC in turn activate invF. InvF combined with SicA then activates effector genes. Since mutations anywhere in the cascade can cause loss of effector gene expression, we chose to test individual members of the cascade for activation of each effector in the absence of all other members of the cascade. This type of approach could reveal if, for instance, sirA, hilA, or hilC can regulate any effectors independently of invF. This approach might also reveal novel regulatory patterns for individual effectors, for example, effector genes regulated by both SPI1 and SPI2 regulators. Therefore, in this report we have examined the effects of seven regulatory genes on ten effector genes. The expression of each effector gene was examined in a set of strains lacking all but one of the seven regulators. Our results confirm and extend the known regulation of TTSS1 and TTSS2 effectors by invF-sicA and ssrB respectively.
Results and discussion
Construction of Typhimurium multi-regulator mutant strains harboring lacZY fusions to type III effector genes
We constructed a Typhimurium strain that lacks seven regulators, six of which are known to affect the expression of the type III secretion systems of this bacterium. The TTSS1-encoding region of SPI1, which harbors five regulatory genes (hilC, hilD, hilA, invF and sprB) and a co-regulator (sicA), was deleted in a strain that harbors a mutation of the sirA gene (see Materials and Methods). Transduction of an ssrB mutation into this strain resulted in YD038 which lacks seven regulatory genes (hereafter known as the multi-mutant). Chromosomal lacZY transcriptional fusions to ten effector genes were then constructed in YD038. These included fusions to genes that encode effectors secreted by TTSS1 (sopA, sopB and sopE2), TTSS2 (sspH2, sifA, sifB, sseI, and sseG) or both (slrP and sspH1). These ten fusions were also transduced into the wild type strain, 14028, using phage P22HTint.
Effects of individual regulators on the expression of the type III secreted effectors
Low copy number plasmids individually encoding seven regulators of SPI1 or SPI2 (or the vector control) were transformed into each of the ten strains that carry a lacZY fusion in the multi-mutant background. Each plasmid was constructed in such a way as to have the regulatory gene expressed from the vector's lac promoter. Expression and regulatory activity of the resulting constructs was confirmed using complementation tests with known target genes (Figure 2). The only exception was sprB for which the target genes are not known. The resulting 80 strains (and the ten additional strains harboring the same fusions in the wild-type background) were then grown in triplicate to log phase under either SPI1-inducing conditions (LB broth standing at 37°C overnight [42]) or SPI2-inducing conditions (MgM broth shaking at 37°C until OD595 of 0.2 to 0.8). The β-galactosidase activity of the strains was then determined. The results of SPI1-inducing conditions are shown in Table 1 and the results of SPI2-inducing conditions are shown in Table 2. For all of the discussion below, any effect less than 3-fold is considered to be no effect.
Figure 2.
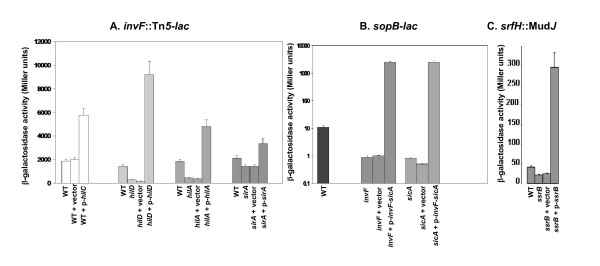
Functional verification of regulator-encoding plasmids. Each regulatory gene, cloned into the pWSK29 vector, was introduced into a strain containing a mutation of the corresponding regulator and a lacZY reporter to a gene known to be activated by the regulator (The hilC-encoding plasmid was introduced into the wild type strain and compared to the control vector). β-galactosidase assays were performed on log-phase cultures (using ONPG) and the results represent a mean of at least three independent assays. (A) An invF::Tn5-lacZY reporter was used to verify the plasmids encoding hilA, hilC, hilD and sirA. (B) A sopB-lacZY fusion was used for the plasmid encoding invF and sicA; and (C), a srfH::MudJ reporter for the plasmid encoding ssrB.
Table 1.
Regulatory dependence of type III secreted effectors under SPI-1 inducing conditions.
| A | B | C | D | E | F | G | H | I | ||
| wild type | MM+ pWSK29 | MM sprB+ | MM hilC+ | MM hilD+ | MM hilA + | MM invF+ sicA + | MM ssrB+ | MM sirA + | ||
| 1 | sopA-lacZ | 7685 ± 1026 | 23 ± 1 | 1 ± 2 | 0 ± 2 | 1 ± 2 | 9 ± 3 | 4982 ± 625 | 9 ± 3 | 1 ± 1 |
| 2 | sopB-lacZ | 10981 ± 922 | 148 ± 11 | 85 ± 2 | 75 ± 8 | 82 ± 4 | 199 ± 15 | 56160 ± 4842 | 3095 ± 1711 | 113 ± 8 |
| 3 | sopE2-lacZ | 14251 ± 4224 | 1872 ± 252 | 2425 ± 748 | 2477 ± 281 | 1093 ± 391 | 3735 ± 65 | 31415 ± 1109 | 3963 ± 90 | 2574 ± 69 |
| 4 | slrP-lacZ | 385 ± 44 | 305 ± 5 | 3511 ± 349 | 295 ± 164 | 448 ± 78 | 403 ± 87 | 1282 ± 138 | 308 ± 39 | 621 ± 23 |
| 5 | sspHl-lacZ | 796 ± 196 | 689 ± 26 | 697 ± 165 | 668 ± 98 | 949 ± 103 | 960 ± 146 | 2906 ± 585 | 827 ± 165 | 666 ± 67 |
| 6 | sspH2-lacZ | 360 ± 34 | 326 ± 38 | 412 ± 128 | 225 ± 16 | 389 ± 71 | 453 ± 90 | 359 ± 24 | 15685 ± 1481 | 296 ± 11 |
| 7 | sifA-lacZ | 21664 ± 538 | 10321 ± 719 | 9970 ± 528 | 9232 ± 665 | 9611 ± 887 | 10176 ± 817 | 10329 ± 1153 | 14085 ± 429 | 16738 ± 1612 |
| 8 | sifB-lacZ | 65 ± 2 | 939 ± 81 | 1393 ± 849 | 19 ± 5 | 1537 ± 470 | 809 ± 150 | 30 ± 6 | 14334 ± 2904 | 25 ± 6 |
| 9 | sseI-lacZ | 70 ± 4 | 60 ± 10 | 69 ± 22 | 32 ± 2 | 43 ± 5 | 61 ± 15 | 47 ± 10 | 5154 ± 1291 | 60 ± 15 |
| 10 | sseG-lacZ | 1931 ± 36 | 2046 ± 440 | 1153 ± 257 | 3001 ± 297 | 2083 ± 442 | 2872 ± 314 | 2444 ± 554 | 25454 ± 314 | 919 ± 73 |
The lac fusion being measured is listed in the left column. The strain background is listed along the top. Each strain harbors a lac fusion to an effector and a plasmid encoding a regulator or the vector control. The values in bold are those showing greater than 3-fold differences compared to the MM + pWSK29 strain that are discussed in the text. Each value represents the mean of at least three experiments ± standard deviation. MM, multi-regulator mutant [Δ(avrA-invH)1 ssrB::cat sirA4::hyg].
Table 2.
Regulatory dependence of type III secreted effectors under SPI-2 inducing conditions.
| A | B | C | D | E | F | G | H | I | ||
| wild type | MM + pWSK29 | MM sprB+ | MM hilC+ | MM hilD+ | MM hilA + | MM invF+sicA + | MM ssrB+ | MM sirA + | ||
| 1 | sopA-lacZ | 1221 ± 35 | 35 ± 13 | 24 ± 7 | 19 ± 7 | 26 ± 7 | 48 ± 17 | 6129 ± 1152 | 75 ± 36 | 33 ± 6 |
| 2 | sopB-lacZ | 3573 ± 34 | 99 ± 5 | 61 ± 7 | 61 ± 15 | 78 ± 10 | 44 ± 3 | 219791 ± 11811 | 33 ± 15 | 46 ± 5 |
| 3 | sopE2-lacZ | 3530 ± 270 | 5406 ± 547 | 4807 ± 538 | 7285 ± 1182 | 2347 ± 35 | 7141 ± 852 | 49681 ± 5581 | 7141 ± 2059 | 5222 ± 350 |
| 4 | slrP-lacZ | 1650 ± 24 | 1222 ± 92 | 3091 ± 43 | 1108 ± 132 | 1211 ± 163 | 1688 ± 523 | 1858 ± 238 | 1901 ± 311 | 2146 ± 34 |
| 5 | sspH1-lacZ | 3744 ± 140 | 1908 ± 254 | 1772 ± 471 | 1686 ± 161 | 1420 ± 116 | 1950 ± 128 | 3558 ± 232 | 3947 ± 978 | 1951 ± 154 |
| 6 | sspH2-lacZ | 6482 ± 1779 | 764 ± 123 | 1006 ± 48 | 655 ± 80 | 625 ± 84 | 981 ± 35 | 850 ± 102 | 62305 ± 14432 | 890 ± 111 |
| 7 | sifA-lacZ | 9995 ± 1326 | 13634 ± 1478 | 11691 ± 1186 | 10521 ± 1782 | 12603 ± 1877 | 13028 ± 2017 | 11010 ± 1742 | 8625 ± 784 | 21071 ± 1272 |
| 8 | sifB-lacZ | 2209 ± 503 | 1085 ± 77 | 711 ± 89 | 49 ± 3 | 1264 ± 139 | 1074 ± 207 | 43 ± 6 | 14677 ± 1092 | 43 ± 8 |
| 9 | sseI-lacZ | 3543 ± 314 | 119 ± 23 | 168 ± 40 | 130 ± 19 | 127 ± 32 | 128 ± 25 | 108 ± 16 | 11049 ± 2830 | 165 ± 10 |
| 10 | sseG-lacZ | 3017 ± 780 | 1444 ± 93 | 700 ± 49 | 1503 ± 109 | 1146 ± 118 | 690 ± 121 | 1083 ± 96 | 4229 ± 221 | 802 ± 99 |
The lac fusion being measured is listed in the left column. The strain background is listed along the top. Each strain harbors a lac fusion to an effector and a plasmid encoding a regulator or the vector control. The values in bold are those showing greater than 3-fold differences compared to the MM + pWSK29 strain that are discussed in the text. Each value represents the mean of at least three experiments ± standard deviation. MM, multi-regulator mutant [Δ (avrA-invH)1 ssrB::cat sirA4::hyg].
• TTSS1 effectors
Regardless of growth conditions, the fusions to sopA, sopB, and sopE2 produced more β-galactosidase in the wild type than in the multi-mutant vector control suggesting that a regulatory factor is missing in the multi-mutant (Tables 1 and 2). For sopA the invF-sicA plasmid was the only plasmid that restored expression. Expression of the sopB fusion was also restored by the invF-sicA plasmid. These results confirm previous findings in which invF-sicA activates sopA and sopB [10,25]. Interestingly, the ssrB plasmid also partially restored sopB expression, but only in SPI1-inducing conditions. This is a novel result but the effect of ssrB is quite small compared to the effect of invF so it is not clear if it is physiologically relevant.
The expression of the sopE2 fusion was decreased in the multi-mutant background and the invF-sicA plasmid restored expression (Tables 1 and 2). The activation of sopE2 by invF-sicA has not been previously reported. However, a homolog of sopE2, the sopE gene, has been shown to be regulated by invF-sicA [12]. The sopE2 gene is present in all Salmonella serovars examined to date, while the presence of sopE is sporadic [43]. The sopE gene was not included in this study because it is not present in our wild-type 14028 strain. Our results suggest that in addition to their similar function, sopE and sopE2 are both regulated by invF-sicA.
Additionally, all of the above results indicate that the regulators above invF in the SPI1 regulatory cascade (sirA, hilA, hilC, hilD) cannot regulate sopA, sopB, or sopE2 in the absence of invF.
• slrP and sspH1
SspHl and SlrP are known to be secreted by both TTSS1 and TTSS2 [28,44]. The sspH1 fusion was not decreased in the multi-mutant background compared to the wild-type in either SPI1 or SPI2-inducing conditions, suggesting that none of the seven regulators control sspH1. However, the invF-sicA plasmid activated the sspH1 fusion by 4-fold compared to the vector control in SPI1-inducing conditions (Table 1). Previously Miao and Miller found that the expression of sspH1 is constitutive [44], so it is not clear if the 4-fold effect of a plasmid-encoded regulator is physiologically significant.
Expression of the slrP fusion did not decrease in the multi-mutant background in either growth condition, but it was activated 12-fold and 4-fold in the presence of the sprB and invF-sicA plasmids, respectively. This effect was only seen in SPI1-inducing conditions. Both of these results are interesting since no target has been identified for sprB, and invF-sicA was not known to regulate slrP. It has been reported that while chromosomal hilC and hilD have no effect on slrP, plasmid-encoded hilC and hilD do moderately activate slrP expression [25]. In this study, the slrP fusion was induced neither by hilC nor by hilD in the multi-mutant background suggesting that the previously observed regulation of slrP by hilC and hilD is through invF.
To test the activation of the slrP fusion by chromosomal sprB and invF-sicA, mutations in these regulators were introduced into an otherwise wild-type background carrying a chromosomal slrP-lacZY fusion. None of the mutations affected the expression of the slrP fusion (Figure 3). This is consistent with the lack of a decrease in the multi-mutant background and a previous report in which plasmid-encoded rtsA, hilC, and hilD had effects on slrP, but little or no difference in expression was seen with the same regulators expressed from the chromosome [25].
Figure 3.
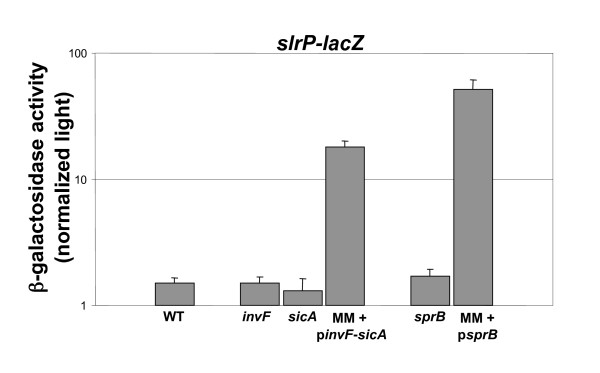
Effect of chromosomal regulators on the expression of slrP. β-galactosidase assays were performed on log-phase cultures (using Beta-glo reagent). The results represent a mean of at least three independent assays. WT, wild type; invF, invF mutant; sicA, sicA mutant; sprB, sprB mutant, MM + pinvF-sicA, multi-mutant containing the plasmid encoding invF and sicA; MM + psprB, multi-mutant containing the plasmid encoding sprB.
• TTSS2 effectors
Regardless of growth condition, four of the five fusions to TTSS2 effector genes (sspH2, sifB, sseI, and sseG) were activated by the ssrB-encoding plasmid (Tables 1 and 2). This confirms the known activation of these genes by the ssrAB two-component system [29,44,45]. Expression of the sifA fusion was not affected in the multi-mutant background compared to the wild-type, regardless of growth condition. Additionally, none of the plasmid-encoded regulators altered sifA expression. This result was unexpected since the expression of sifA has previously been shown to depend on ssrA, which encodes the sensor kinase component of the SsrAB two-component system [44].
To try to reconcile this discrepancy, we constructed a second chromosomal lac fusion to sifA using the method developed by Ellermeier et al. (see Methods). This fusion was moved into the wild type and the multi-mutant backgrounds. The multi-mutant strain was then transformed with the ssrB-encoding plasmid or the pWSK29 vector. This sifA fusion was not regulated by ssrB under SPI1-inducing conditions and was only regulated 2-fold under SPI2-inducing conditions (Table 3). SifA expression was almost twice as high in the wild-type strain as it was in either the ΔssrA, ΔssrB, or ΔssrAB strains. Furthermore, sifA expression was 2-fold higher in the multi-mutant background containing the ssrB plasmid than it was in the vector control strain (Table 3).
Table 3.
Regulation of sifA by ssrAB.
| LB | MgM | |
| WT sifA-lac | 5.5 ± 1.6 | 26.3 ± 4.8 |
| MM sifA-lac | 3.6 ± 0.5 | 16.8 ± 2.9 |
| MM sifA-lac + p-ssrB | 4.8 ± 0.8 | 29.6 ± 6.9 |
| MM sifA-lac + pWSK29 | 3.8 ± 0.5 | 15 ± 5.6 |
| Δ ssrA sifA-lac | 3.8 ± 1.1 | 14.4 ± 2.7 |
| Δ ssrB sifA-lac | 3.9 ± 1.4 | 14.2 ± 2.5 |
| Δ ssrAB sifA-lac | 2.7 ± 2.4 | 15.6 ± 2.6 |
The β-galactosidase activities of strains harbouring a sifA-lac fusion were measured using ONPG. Each value represents the mean Miller units of at least three experiments ± standard deviation. WT, wild type 14028; MM, multi-mutant [Δ(avrA-invH)1 ssrB::cat sirA4::hyg].
To test the regulation of sifA by ssrB in another way, we constructed a plasmid that harbours the luxCDABE operon from Photorhabdus luminescens under the control of the sifA promoter. This plasmid was placed into the wild type, the ssrA mutant, and the ssrB mutant containing either the pssrB plasmid or the vector control. These strains were grown in LB or MgM media and the light emitted by the cells captured using a Xenogen IVIS imaging system (Alameda, CA) (Figure 4A) and measured using a luminometer (Figure 4B). The light emitted by the wild type strain is higher in MgM compared to LB showing that the PsifA promoter is induced in the MgM medium (Figure 4). The activity of the PsifA also decreased strongly in the ssrA and the ssrB mutants after growth in both MgM and LB. The pssrB plasmid restored the production of light in the ssrB mutant (Figure 4). These results are consistent with previous findings that sifA is regulated by the ssrAB two-component regulatory system. It is not clear why the regulatory effects are seen so clearly with plasmid-encoded fusions and not observed with chromosomal fusions. It is possible that additional layers of regulation are imposed on the chromosomal fusions such as supercoiling or chromatin structure that are relieved on the plasmid.
Figure 4.
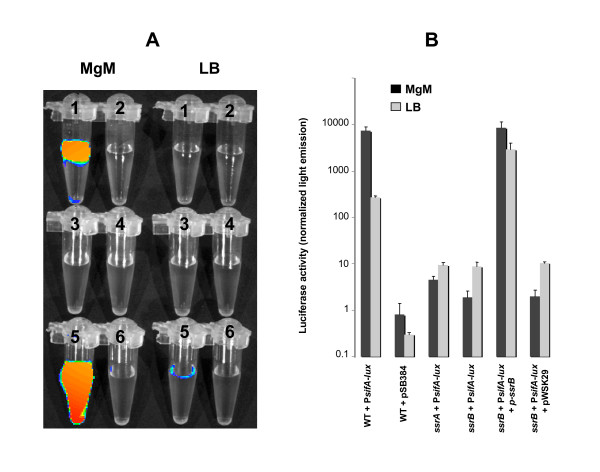
ssrB activates the promoter of sifA. The expression of the luxCDABE operon under the control of the PsifA promoter was analyzed in the wild type and the isogenic ssrA mutant and ssrB mutant with or without complementation. A, the strains were grown in MgM or in LB for six hours, and the light emitted by bacterial suspensions of similar cell density were captured using the Xenogen IVIS imaging system (Alameda, CA) and displayed as pseudocolor images with blue representing the lowest and red the highest light intensity. 1, wild type + PsifA-lux; 2, wild type + pSB384 vector control; 3, ssrA + PsifA-lux; 4, ssrB + PsifA-lux; 5, ssrB + PsifA-lux + pssrB; 6, ssrB + PsifA-lux + pWSK29 control. B, quantification of the activation of the PsifA promoter. The light emitted by 100 microliters of culture was measured using a luminometer. The values represent the mean ± SEM of three independent experiments.
Expression of the sifB fusion was activated by ssrB, as expected [29,44,45]. However, sifB was also repressed by sirA, hilC, and invF. This is an interesting and novel result because a TTSS2 effector is being repressed by the SPI1 regulatory cascade. No other effectors are activated by one system and repressed by the other. Even more unusual is that sirA and hilC had their effects in the multi-mutant background lacking invF. This indicates that sirA and hilC are acting on sifB independently of the standard SPI1 regulatory pathway.
Conclusion
We have taken a systematic approach to study the regulation of type III effector genes during growth in vitro. Our results confirm that sopA and sopB are activated by invF-sicA. In addition we have determined that sopE2 and sspH1 are regulated by invF-sicA. The regulation of sopE2 is very strong under either SPI1 or SPI2-inducing conditions while the regulation of sspH1 is weak and only observed under SPI1-inducing conditions. The sspH2, sifB, sseI and sseG fusions were shown to be strongly activated by ssrB. The most interesting regulation was observed with sifA and sifB. Chromosomal fusions to sifA were largely unaffected by any regulator. However, a plasmid-based sifA fusion was strongly regulated by ssrB. The sifB fusion was strongly regulated by ssrB, as expected. However, it was also strongly repressed by sirA, hilC, and invF. This is the first indication that sirA and hilC might regulate an effector gene independently of invF. The set of strains constructed for this study can now be used to quickly and systematically study the regulation of newly identified effector genes.
Methods
Growth conditions, enzymes, reagents, and transduction
The strains and plasmids used in this study are described in Additional file 1. The bacterial strains were grown in LB (Luria-Bertani) medium at 37°C. For induction in the Magnesium minimal medium (MgM) [46] cells from LB overnight cultures were washed with MgM prior to 1:50 dilution. The following antibiotics were obtained from Sigma and used at the following concentrations when required: kanamycin (kan, 50 μg/ml), ampicillin (amp, 100 μg/ml), chloramphenicol (cam, 20 μg/ml), tetracycline (tet, 20 μg/ml), and hygromycin (hyg, 200 μg/ml). Sucrose was supplemented at 219 mM final concentration to select against sacB. The β-galactosidase chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) was obtained from Molecular Probes (Eugene, OR) and used at a final concentration of 40 μg/ml.
General molecular biology techniques were performed essentially as described [47]. Restriction and modification enzymes were purchased from Invitrogen (Carlsbad, CA) or New England Biolabs (Beverly, MA), and used as recommended by the manufacturers. PCR primers were purchased from IDT Inc. (Coralville, IA). Plasmids were extracted using kits from Qiagen (Valencia, CA). P22 transduction was performed as described [48].
β-galactosidase assay
The β-galactosidase activities were measured using o-nitrophenyl β-D-galactopyranoside (ONPG, Sigma, St. Louis MO) using the method described by Miller [49] or the Beta-Glo™ assay system (Promega, WI) according to the manufacturer's recommendations. Briefly the assay consisted of mixing an equal volume of a log phase growing culture (OD590 0.2–0.8) and a detection reagent. The latter contains (i) a luciferin-galactoside substrate (6-O-β-galactopyranosyl-luciferin) that is cleaved by the β-galactosidase to form luciferin and galactose, and (ii) a firefly luciferase that catalyzes the luciferin to generate light. After at least 1 hour of incubation at room temperature the light produced is measured in a Turner Designs TD-20/20 luminometer or a Wallac 1420 VICTOR3 plate reader and the results expressed as light per OD595 per μl of culture. The results were expressed as the mean ± SD of at least three different experiments.
Mutation of regulatory genes
The invF, sicA, sprB, ssrA, and ssrB genes were replaced with the cat cassette from pKD3 using the λ-red recombination system as described [50].
Construction of a multi-regulator mutant strain
The TTSS genes of SPI1 (~35 kb), but not the manganese transport genes [51], were deleted using the pRE112 suicide vector [52]. This deleted avrA through invH. To do this, PCR was performed using primers BA1093 and BA1094 (see Additional file 2) with 14028 genomic DNA as template. The 1366 bp product contains the 3' end of the invH gene, stm2901, and the 5' end of stm2902. A second PCR was performed using primers BA1095 and BA1096 (see Additional file 2). The 1255 bp product contains the 3' end of sitC, the sitD gene, and the 5' part of the avrA gene. Both products were cloned into pCR2.1 Topo (Invitrogen). The sitD-containing fragment was removed with SacI and cloned into the SacI site of the stm2901-containing clone. This placed the two fragments adjacent to each other and created the Δ(avrA-invH)1 deletion. This 2894 bp fragment was removed with KpnI and PvuII and cloned into the suicide vector pRE112 that had been digested with KpnI and SmaI to yield the plasmid pYD25. This plasmid contains (i) a cat gene that confers a resistance to chloramphenicol, (ii) the R6K replication origin that requires the pir gene, and (iii) the sacB gene of Bacillus subtilis that is toxic in Gram-negative bacteria grown in the presence of high concentrations of sucrose [53]. Allelic exchange was performed by mobilizing pYD25 from BW20767 into JVR140 (sirA4::hyg). The chloramphenicol resistant transconjugants harbor the pYD25 plasmid integrated into the chromosome, resulting in the duplication of the region upstream and downstream of TTSS1. Isolated colonies were grown in LB without chloramphenicol selection until the exponential phase and plated onto LB lacking NaCl and containing 219 mM sucrose to select for loss of the integrated plasmid by homologous recombination. The colonies obtained were screened for Δ(avrA-invH)1 using PCR with primers hybridizing upstream and downstream of the TTSS1 region. Finally, the resulting Δ(avrA-invH)1 sirA4::hyg strain was used as a recipient for the P22HTint transduction of an ssrB::cat mutation from MJW129 [29] to make the multi-mutant YD038 strain. YD038 lacks the co-regulator sicA, and seven regulators, sirA, ssrB, hilA, hilC, hilD, invF, and sprB.
Construction of lacZY fusions to effector genes
To make lacZY transcriptional fusions to effector genes, the 3' portion of each gene (including the stop codon but without the transcription terminator) was amplified using PCR and cloned into pCR2.1 TOPO (Invitrogen, Carlsbad, CA). These gene portions were then removed as EcoRI fragments and cloned into the EcoRI site upstream of the promoterless lacZY gene of the pVIK112 pir-dependent suicide vector [54]. The resulting plasmids were mobilized from BW20767 into YD038. The correct integration of the plasmids into the chromosome of the resulting kanamycin-resistant transconjugants was verified by PCR. Fusions were transduced using phage P22HTint into other backgrounds as needed. A second fusion to sifA was constructed as described previously [55]. First the lambda red recombination system was used to integrate a kanamycin resistance gene flanked by FRT (Flp recombinase target) sites from the pKD4 plasmid [50]. The FRT-kan-FRT cassette was placed after the stop codon, but before the transcription terminator, of sifA. Second, the temperature sensitive pCP20 plasmid [56] that encodes Flp recombinase was introduced into the strain. Flp-mediated excision of the kan cassette left a single FRT site that serves as an integration point for the fusion. The third step was to integrate the pCE36 suicide plasmid that has a FRT site directly upstream of the promoterless lacZY [55]. After the integration of pCE36, the pCP20 plasmid was cured by growth at 37°C.
Construction of plasmids encoding regulators of the Salmonella type III secretion systems
Each of the regulatory genes missing in the multi-mutant was amplified using PCR (the primers used are described in Additional file 2), cloned into the pCR2.1 TOPO vector, released by enzymatic digestion and cloned under the control of the lac promoter of pWSK29. The hilD, sirA, and hilA genes were released from the TOPO vector as 1353 bp SacI-XbaI, 936 bp KpnI-XhoI, and 1920 bp EcoRI fragments respectively and cloned into the corresponding sites of pWSK29 to yield the plasmids pYD17, pYD28 and pYD29 respectively. The 971 bp SpeI-XhoI fragment containing the sprB gene was cloned into the XbaI-XhoI sites of pWSK29 to give the pYD15 plasmid. The hilC gene was released as a 1541 bp PvuII-XbaI fragment and ligated to the EcoRV-XbaI sites of pWSK29 to produce the plasmid pYD16. A 852 bp EcoRI fragment and a 622 bp KpnI-XhoI fragment containing the invF and the sicA genes were cloned into the corresponding sites of pWSK29 to yield the plasmids pYD23 and pYD38 respectively. The 5353 bp ApaLI-XhoI fragment from pYD23 was ligated to the 1527 ApaLI-XhoI fragment from pYD38 to give the pYD40 plasmid that harbours the invF and sicA genes as a bi-cistronic operon under the control of the lac promoter. Each construction was verified by the complementation of a corresponding regulatory mutant (Figure 2), except for sprB for which a complementable function has not yet been discovered [18]. We sequenced both strands of the Plac-sprB region of the sprB-encoding plasmid to ensure that the sequence was correct.
Construction of the PsifA-lux reporter plasmid
The promoter region and the 5'part of the sifA gene was PCR-amplified from 14028 genomic DNA using the primers YD1 and YD2 (see Additional file 2). The product was cloned into the pCR2.1 TOPO vector (Invitrogen) and then released as an EcoRI fragment that was subsequently cloned upstream of the luxCDABE operon of the pSB384 vector [57]. The correct orientation of the PsifA promoter in the resulting pYD56 plasmid was verified by PCR.
Cell imaging and luminometry
Liquid cultures were grown in LB or MgM media for six hours. The cultures were brought to similar OD600 and 200 μl of cell suspensions were placed in tubes and imaged using the Xenogen IVIS imaging system (Alameda, CA). The images are displayed as pseudocolors with blue representing the lowest and red the highest light intensity. For luminometry the light emitted by 100 μl of culture was measured using a Turner Designs TD-20/20 luminometer.
Authors' contributions
BMMA provided the original idea for this study. YD designed and conducted the experiments. RK and JLD participated in the molecular biology and enzymatic assays. YD and BMMA wrote the article. All authors read and approved the final manuscript.
Supplementary Material
Bacterial strains and plasmids used in this study.
Primers used in this study.
Acknowledgments
Acknowledgements
We would like to thank Dr Roy Curtiss III, Arizona State University, for his generous support that enabled us to carry out a substantial part of this study. We are grateful to Dr Lionello Bossi, CNRS, Gif-sur-Yvette, France for providing us with bacterial strains. We thank Jeroen van Reeuwijk for constructing JVR140. This publication was made possible by grant nos. AI50002 (to B. M. M. A.) and AI24533 (to Roy Curtiss III) from the National Institute of Allergy and Infectious Diseases.
Contributor Information
Yakhya Dieye, Email: Yakhya.Dieye@asu.edu.
Jessica L Dyszel, Email: dyszel.1@osu.edu.
Rebin Kader, Email: rebin.kader@asu.edu.
Brian MM Ahmer, Email: ahmer.1@osu.edu.
References
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen-Wester I, Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/S1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 2001;3:1335–1344. doi: 10.1016/S1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- Wallis TS, Galyov EE. Molecular basis of Salmonella-induced enteritis. Mol Microbiol. 2000;36:997–1005. doi: 10.1046/j.1365-2958.2000.01892.x. [DOI] [PubMed] [Google Scholar]
- Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3:1281–1291. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- Hensel M. Salmonella pathogenicity island 2. Mol Microbiol. 2000;36:1015–1023. doi: 10.1046/j.1365-2958.2000.01935.x. [DOI] [PubMed] [Google Scholar]
- Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003;5:501–511. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- Jones BD. Salmonella invasion gene regulation: a story of environmental awareness. Journal of microbiology (Seoul, Korea) 2005;43:l10–117. [PubMed] [Google Scholar]
- Lostroh CP, Bajaj V, Lee CA. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol Microbiol. 2000;37:300–315. doi: 10.1046/j.1365-2958.2000.01991.x. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Miller VL. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. Journal of bacteriology. 1999;181:4949–4954. doi: 10.1128/jb.181.16.4949-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberg K, Galan JE. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-l)-encoded transcriptional activators InvF and hilA. Infection and immunity. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Miller VL. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. Embo J. 2001;20:1850–1862. doi: 10.1093/emboj/20.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Miller VL. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol Microbiol. 2000;35:949–960. doi: 10.1046/j.1365-2958.2000.01772.x. [DOI] [PubMed] [Google Scholar]
- Boddicker JD, Knosp BM, Jones BD. Transcription of the Salmonella Invasion Gene Activator, hilA, Requires HilD Activation in the Absence of Negative Regulators. Journal of bacteriology. 2003;185:525–533. doi: 10.1128/JB.185.2.525-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Lee CA. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol Microbiol. 2001;40:1289–1299. doi: 10.1046/j.1365-2958.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- Baxter MA, Fahlen TF, Wilson RL, Jones BD. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infection and immunity. 2003;71:1295–1305. doi: 10.1128/IAI.71.3.1295-1305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar S, Schechter LM, Lostroh CP, Lee CA. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol Microbiol. 2003;47:715–728. doi: 10.1046/j.1365-2958.2003.03322.x. [DOI] [PubMed] [Google Scholar]
- Eichelberg K, Hardt WD, Galan JE. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol Microbiol. 1999;33:139–152. doi: 10.1046/j.1365-2958.1999.01458.x. [DOI] [PubMed] [Google Scholar]
- Heeb S, Haas D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol Plant Microbe Interact. 2001;14:1351–1363. doi: 10.1094/MPMI.2001.14.12.1351. [DOI] [PubMed] [Google Scholar]
- Teplitsi M, Ahmer BMM. The control of secondary metabolism, motility, and virulence by the two-component regulatory system BarA/SirA of Salmonella and other g-proteobacteria. In: Prüb BM, editor. Global regulatory networks in enteric bacteria. Trivandrum, Kerala, India: Research Signpost; 2005. pp. 107–132. [Google Scholar]
- Teplitski M, Goodier RI, Ahmer BM. Pathways Leading from BarA/SirA to Motility and Virulence Gene Expression in Salmonella. Journal of bacteriology. 2003;185:7257–7265. doi: 10.1128/JB.185.24.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005;57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- Pegues DA, Hantman MJ, Behlau I, Miller SI. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar typhimurium. Journal of bacteriology. 2000;182:1872–1882. doi: 10.1128/JB.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Slauch JM. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. Journal of bacteriology. 2003;185:5096–5108. doi: 10.1128/JB.185.17.5096-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Walthers D, Oropeza R, Kenney LJ. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol. 2004;54:823–835. doi: 10.1111/j.1365-2958.2004.04317.x. [DOI] [PubMed] [Google Scholar]
- Miao EA, Scherer CA, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ, Miller SI. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- Worley MJ, Ching KH, Heffron F. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- Lee AK, Detweiler CS, Falkow S. OmpR regulates the two-component system SsrA-ssrB in Salmonella pathogenicity island 2. Journal of bacteriology. 2000;182:771–781. doi: 10.1128/JB.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma JJ, Groisman EA. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol Microbiol. 2005;57:85–96. doi: 10.1111/j.1365-2958.2005.04668.x. [DOI] [PubMed] [Google Scholar]
- Linehan SA, Rytkonen A, Yu XJ, Liu M, Holden DW. SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infection and immunity. 2005;73:4354–4362. doi: 10.1128/IAI.73.7.4354-4362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17460–17465. doi: 10.1073/pnas.0505401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Baumler AJ. Molecular Pathogenesis of Salmonella enterica Serotype Typhimurium-Induced Diarrhea. Infection and immunity. 2003;71:1–12. doi: 10.1128/IAI.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infection and immunity. 1999;67:5690–5698. doi: 10.1128/iai.67.11.5690-5698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, Finlay BB. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002;4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- Bispham J, Tripathi BN, Watson PR, Wallis TS. Salmonella pathogenicity island 2 influences both systemic salmonellosis and Salmonella-induced enteritis in calves. Infection and immunity. 2001;69:367–377. doi: 10.1128/IAI.69.1.367-377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B, Li Y, Owen D, Vallance BA, Finlay BB. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infection and immunity. 2005;73:3219–3227. doi: 10.1128/IAI.73.6.3219-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- Mirold S, Rabsch W, Rohde M, Stender S, Tschape H, Russmann H, Igwe E, Hardt WD. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9845–9850. doi: 10.1073/pnas.96.17.9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Miller SI. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen-Wester I, Stecher B, Hensel M. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infection and immunity. 2002;70:1403–1409. doi: 10.1128/IAI.70.3.1403-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes BK, Brown NF, Valdez Y, Brumell JH, Finlay BB. Expression and secretion of salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J Biol Chem. 2004;279:49804–49815. doi: 10.1074/jbc.M404299200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Maloy SR, Stewart VJ, Taylor RK. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Janakiraman A, Slauch JM, Maguire ME. SitABCD is the alkaline Mn(2+) transporter of Salmonella enterica serovar Typhimurium. Journal of bacteriology. 2002;184:3159–3166. doi: 10.1128/JB.184.12.3159-3166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 1998;207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. Journal of bacteriology. 1985;164:18–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogeraki VS, Winans SC. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene. 1997;188:69–75. doi: 10.1016/S0378-1119(96)00778-0. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Janakiraman A, Slauch JM. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290:153–161. doi: 10.1016/S0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Fip-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- Winson MK, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GS. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS microbiology letters. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Goodier RI, Ahmer BM. Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int J Med Microbiol. 2006;296:449–466. doi: 10.1016/j.ijmm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bacterial strains and plasmids used in this study.
Primers used in this study.


