Abstract
Microbial structures activate Toll-like receptors (TLRs) and TLR-mediated cell signalling elicits and regulates host immunity. Most TLRs are poorly expressed but the underlying expression mechanism is not clear. Examination TLR sequences revealed that most human TLR genes deviated from using major human codons. CD14 resembles TLRs in sequence but its gene preferentially uses major codons. Indeed, CD14 expression on monocytes was higher than expression of TLR1 and TLR2. The TLR9 gene is abundant in major codons and it also showed higher expression than TLR1, TLR2 and TLR7 in transfected 293T cells. Change of the 5′-end 302 base pairs of the TLR2 sequence into major human codons markedly increased TLR2 expression, which led to increased TLR2-mediated constitutive nuclear factor-κB activation. Change of the 5′-end 381 base pairs of the CD14 sequence into prevalent TLR codons markedly reduced CD14 expression. These results collectively show that the deviation of TLR sequences from using major codons dictates the low TLR expression and this may protect the host against excessive inflammation and tissue damages.
Keywords: CD14, codon usage, expression, NF-κB activation, Toll-like receptor
Introduction
To the host immune system, microbial pathogens provide two major classes of signals, i.e. immunogenic epitopes and adjuvant stimuli. Antigenic epitopes are processed on antigen-presenting cells (APCs) before presentation to antigen-specific T cells.1 Adjuvant signals promote antigen processing and enhance T-cell activation through increasing major histocompatibility complex and costimulatory molecule expression on APCs and stimulating production of inflammatory cytokines.2 Host APCs and other cells recognize microbial adjuvant structures predominantly through Toll-like receptors (TLRs).3–5 TLRs are type I receptors with large extracellular domains characterized by leucine-rich repeats and cytoplasmic domains resembling that of the Drosophilla Toll protein and the interleukin-1 receptor (IL-1R), called the Toll/IL-1R homology (TIR) domain.6–8
The benefits of adjuvant in vaccination have long been described9 but the host receptors that recognize adjuvant stimuli are only now being characterized. The first mammalian TLR (i.e. TLR4) was identified based on its sequence similarity to Toll and IL-1R; functionally it was shown to activate nuclear factor-κB (NF-κB) upon dimerization.8 TLR4 was independently identified as a receptor for the bacterial wall component lipopolysaccharide (LPS), a well-characterized adjuvant, because a mutation in the TLR4 gene rendered C3H/HeJ mice hyporesponsive to LPS-induced systemic inflammation or septic shock.10 Nine additional TLRs, i.e. TLR2–TLR10, were subsequently identified based on their sequence features.11–15 All TLRs contain extracellular leucine-rich repeats and cytoplasmic TIR domains and some of these TLRs have been shown to recognize distinct microbial adjuvant structures, e.g. CpG DNA, double-stranded RNA, bacterial lipopeptides, etc.3–5 CD14 is also characterized by leucine-rich repeats but it is distinct from TLRs in that it lacks typical transmembrane/cytoplasmic domains. CD14 is a glycosylphosphatidylinositol-linked receptor.16 Being highly expressed on monocytes and macrophages, CD14 binds to LPS and mediates LPS-elicited cell signalling through TLR4.16,17
The fact that none of the TLRs were identified at the protein level based on their ability to recognize microbial structures is, to certain extent, the result of their poor TLR expression as pointed out previously.18 The mechanism underlying low TLR expression is not clear. In the present study, we observed that most human TLR genes deviated from using major codons and showed that partial optimization of the TLR2 gene using major codons could markedly improve TLR2 expression. We also showed that introduction of prevalent TLR codons into the CD14 sequence significantly reduced CD14 expression. Our results suggest that TLR expression is maintained at low levels through a robust mechanism acquired during evolution. The ability to increase TLR expression through codon optimization will also assist structural characterization of TLRs and investigation of TLR interaction with adjuvant structures.
Materials and methods
Cell culture, antibodies and expression vectors
The 293T cells were cultured in Dulbecco's modified Eagle's minimal essential medium supplemented with 10% (v/v) bovine calf serum (Hyclone, Logan, UT). Monoclonal antibodies against human TLR1, TLR2 and TLR9 were obtained from Imgenex Co. (San Diego, CA). The anti-CD14 antibodies [purified immunoglobulin G (IgG) and fluorescein isothiocyanate (FITC) -labelled] were purchased from Ancell Co. (Bayport, MN). The anti-myc monoclonal antibody was obtained from Roche Diagnostics (Mannheim, Germany). Monocytes were isolated from peripheral blood as previously described.19
Expression constructs
The pCD14, pTLR4 and pMD2 expression vectors were as previously described.20 Vectors for the expression of human TLR1, TLR2, TLR7 and TLR9 with C-terminal myc-His tags were constructed as follows. RNA was isolated from monocytes using the Trizol reagent (Life Technologies, Grand, Island, NY). The cDNA encoding these TLRs were amplified by reverse transcription polymerase chain reaction (RT-PCR) from the monocyte RNA using the following primers (5′ to 3′): TLR1: cggggtaccaccatgactagcatcttccattttg/gaagggccctagactatttctttgcttgct; TLR2: cggggtaccaccatgccacatactttgtggatg/gaagggccctaggactttatcgcagctc; TLR7: cggggtaccaccatggtgtttccaatgtggaca/gaagggcccctagaccgtttccttgaacac; TLR9: aggggtaccaccatgagactcatcagaaacattt/agaagggcccttatagacaatctgttctcatca.
The PCR products, all flanked with the KpnI and ApaI restriction sites, were cloned into the pcDNA3·1/myc-His vector to generate the pTLR1, pTLR2, pTLR7 and pTLR9 vectors. To express these TLRs with C-terminal myc-His tags, site-directed mutations were introduced to inactivate the 3′ stop codons. All these mutations were introduced at the first base of the stop codon, i.e. ‘t’ (the complementary nucleotides ‘a’ are underlined), which were deleted in the pTLR1 vector and mutated to ‘a’ in the other three vectors to generate the pTLR1-MH, pTLR2-MH, pTLR7-MH and pTLR9-MH vectors. All expression vectors or mutations were verified by sequencing.
Transfection and flow cytometry
The 293T cells were transfected in six-well plates using the GenePORTER 2 reagent (Gene Therapy Systems, La Jolla, CA) essentially as previously described.21 The cells were transfected with the TLR expression vectors each at 2 μg/well unless otherwise stated. As controls, 293T cells were transfected with the pcDNA3.1 vector. The cells were cultured for 36 hr and then analysed by flow cytometry. Briefly, transfected 293T cells were fixed in 1% (w/v) paraformaldehyde and then permeabilized in 0·2% (w/v) saponin for 10 min The cells were blocked for 30 min in 20% goat serum in phosphate-buffered saline (PBS) and then stained for 45 min with the anti-myc antibody (20 μg/ml). The cells were washed in PBS containing 0·1% (w/v) saponin and then stained with FITC-conjugated goat anti-mouse IgG. After washing, these cells were analysed on the FACScalibur using the cellquest software (BD Biosciences, San Jose, CA). Intracellular TLR9 in isolated monocytes was detected using a similar method. To detect CD14 and TLR expression on monocytes, the cells were washed in PBS and then incubated with antibodies against human TLR1, TLR2, TLR9 and CD14 or, as controls, isotype mouse IgG. Monocytes that were incubated with the primary antibodies, were then stained with FITC-conjugated goat anti-mouse IgG. The cells were washed in FACSwash (PBS containing 2·5% (v/v) bovine calf serum), fixed in 1% (w/v) paraformaldehyde in PBS (pH 7·6), and then analysed by flow cytometry.
Databases and sequence analyses
The average codon usage frequencies in the human genome were extracted from http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=Homo+sapiens+[gbpri]. Codon usage in human CD14 and TLR genes was analysed using the codonw software (http://bioweb).pasteur.fr/seqanal/interfaces/codonw.html#defaults) and codon frequencies generated from the CD14 and TLR genes were expressed as percentages, taking the overall frequencies of all codons encoding an amino acid as 100%, and aligned with the average human codon frequencies.
Codon optimization of the TLR2 coding sequence
Eleven sense (F1–F11) and 11 antisense (R1–R11) oligonucleotide primers were synthesized, as listed below, that overlap to span the 5′ 302 base pairs (bp) of the TLR2-coding sequence in the pTLR2-MH vector.
F1:CGGGGTACCACCATGCCCCACACCCTGTGGATG
F2:GGATGGTGTGGGTGCTGGGCGTGATCATCAGCCTG
F3:GCCTGAGCAAGGAGGAGAGCAGCAACCAGGCCAGC
F4:CCAGCCTGAGCTGCGACCGCAACGGCATCTGCAAG
F5:GCAAGGGCAGCAGCGGCAGCCTGAACAGCATCCCC
F6:TCCCCAGCGGCCTGACCGAGGCCGTGAAGAGCCTG
F7:GAGCCTGGACCTGAGCAACAACCGCATCACCTACATC
F8:ACATCAGCAACAGCGACCTGCAGCGCTGCGTGAAC
F9:TGAACCTGCAGGCCCTGGTGCTGACCAGCAACGGC
F10:ACGGCATCAACACCATCGAGGAGGACAGCTTCAGC
F11:TCAGCAGCCTGGGCAGCCTCGAGCACCTGGACCTG
R1:GCACCCACACCATCCACAGGGTGTG
R2:CCTCCTTGCTCAGGCTGATGATCACG
R3:CGCAGCTCAGGCTGGCCTGGTTGCT
R4:CGCTGCTGCCCTTGCAGATGCCGTTG
R5:TCAGGCCGCTGGGGATGCTGTTCAG
R6:TGCTCAGGTCCAGGCTCTTCACGGCC
R7:CGCTGTTGCTGATGTAGGTGATGCG
R8:GGGCCTGCAGGTTCACGCAGCGCTGC
R9:TGGTGTTGATGCCGTTGCTGGTCAG
R10:TGCCCAGGCTGCTGAAGCTGTCCTCC
R11:AGTTGTAGCTCAGGTCCAGGTGCTC
These primers were synthesized using major human codons. The 5′-GGTACC KpnI site and the 3′ CTCGAG XbaI sites are in bold letters and are incorporated in primers F1 and F11, respectively. A corresponding XbaI site was created in the pTLR2-MH vector through nonsense mutations (as indicated in Fig. 2b). The 22 primers were used, each at 0·5 μm, in PCR using pfu DNA polymerase (Promega, Madison, WI). The longest PCR product was purified and re-amplified using the F1 and R11 primers. The PCR products were cloned between the KpnI and the newly created XbaI sites and clones were sequenced to identify a clone that is free of mutations (pTLR2opt-MH).
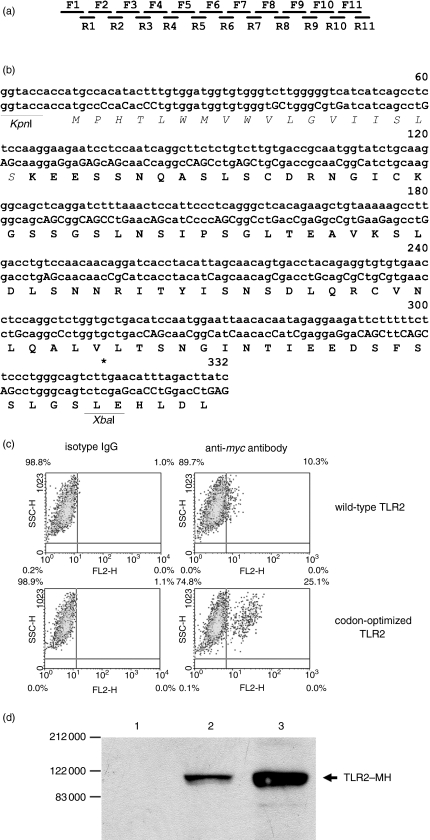
Figure 2.
Partial optimization of TLR2 codons increases TLR2 expression. (a) Schematic illustration of overlapping oligonucleotide primers used to synthesize the codon-optimized DNA fragment encoding the 5′ 302 bp of the TLR2 coding sequence. The sequences of the primers are described in the Materials and methods section. (b) The codon-optimized TLR2 sequence (lower sequence) is aligned with the corresponding wild-type sequence (upper sequence). The cloning sites (KpnI/XbaI) are indicated at each end of the sequence and the changed nucleotides in the optimized sequence are highlighted in upper case letters. The leader sequence of TLR2 was indicated using ‘italic’ letters. (c) The pTLR2-MH (wild-type TLR2) and pTLR2opt-MH (codon-optimized TLR4) vectors were transfected into 293T cells and the expression of TLR2 was detected, upon membrane permeabilization, with the anti-myc antibody (20 μg/ml) followed by FITC-conjugated goat anti-mouse IgG (anti-myc antibody). As controls, the transfected cells were stained with isotype IgG followed by the secondary goat antibody (isotype IgG). The percentages of TLR2-positive cells are indicated at the upper-right-hand corner of each panel. (d) Western blot analysis of TLR2-MH expression derived from the pTLR2-MH (lane 2) and pTLR2opt-MH (lane 3) vectors. As a control, 293T cells transfected with the pcDNA3·1 vector were similarly analysed (lane 1). The TLR2-MH bands are indicated.
Western blotting
Western blotting was carried out essentially as previously described.19 Cells were washed in PBS and then lysed on ice in a lysis buffer containing 20 mm Tris–HCl (pH 7·5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% (v/v) Nonidet P-40 and the complete protease inhibitor cocktail (Roche). Protein concentration was determined using the BCA reagent (Pierce Chemical Company, Rockford, IL). The lysates (50 μg proteins) were separated on 7·5% (w/v) sodium dodecyl sulphate–polyacrylamide gel electrophoresis gels and electro-blotted onto nitrocellulose membranes. Blots were blocked with 5% (w/v) non-fat milk in TBST buffer [50 mm Tris–HCl (pH 7·5), 150 mm NaCl, 0·1% (v/v) Tween-20] and were then probed with the mouse anti-myc antibody overnight at 4°. The blots were washed in TBST buffer and probed further with biotin-conjugated goat anti-mouse IgG and then horseradish peroxidase–streptavidin (Dako, Carpinteria, CA). Signals were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech, Buckinghamshire, UK).
Expression of CD14 using prevalent TLR codons
A 5′ 381-bp fragment of the CD14 sequence in the pCD14 vector, encoding the N-terminal 124 amino acids (including the leader sequence), was deleted with KpnI and XhoI. The corresponding DNA fragment was synthesized using prevalent TLR codons (GENEART GmbH, Regensburg, Germany) (Fig. 3a) and cloned into the pCD14 vector to generate the pCD14cod vector. Both pCD14 and pCD14cod were expressed in 293T cells in six-well plates at 0·25–2 μg/well and surface CD14 was detected by flow cytometry.
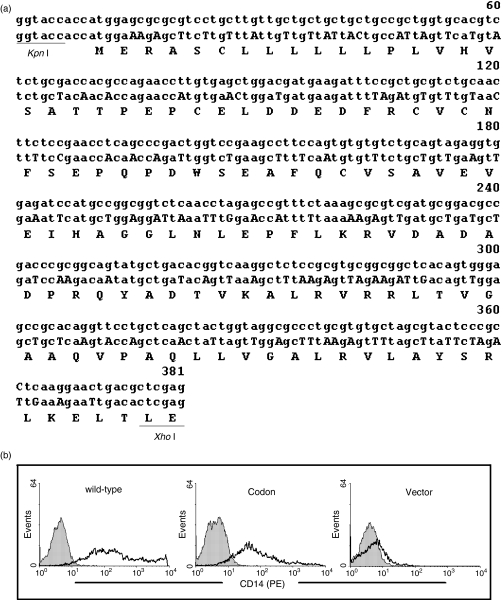
Figure 3.
Partial replacement of CD14 sequence with prevalent TLR codons reduces CD14 expression. (a) A 381-bp 5′ sequence of CD14 (middle line) was synthesized using prevalent TLR codons as described in Table 1. The changed nucleotides are highlighted with capital letters. The corresponding wild-type sequence (upper line) and the encoded amino acid sequence (lower line) were aligned. The synthesized CD14 sequence was cloned into the KpnI/XhoI site of the pCD14 vector to generate the pCD14cod vector. The single letter code is used to represent the amino acid sequence. (b) pCD14 (wild-type), pCD14cod (Codon) and, as a control, pcDNA3·1 (Vector) were used to transfect 293T cells at 0·25 μg/well in six-well plates. After 30 hr, the transfected 293T cells were stained with a mouse anti-CD14 monoclonal antibody (FITC-labelled) and analysed by flow cytometry (solid lines). The cells were also stained with mouse isotype IgG (FITC-labelled) as controls (filled histograms).
NF-κB luciferase assay
The assay was carried out as previously described.21 Briefly, 293T cells were transfected in 24-well plates with the pTLR2-MH or pTLR2opt-MH vectors or, as a control, the pcDNA3.1 vector. The expression vectors were used at 0, 1, 5, 25 and 125 ng/well and, in each experiment, were normalized to 125 ng/well with the pcDNA3.1 plasmid. In every experiment, the NF-κB-directed firefly luciferase (p5xNF-κB-Luc; Stratagene, La Jolla, CA) and the cytomegalovirus-driven Renilla luciferase (pRL-CMV; Promega) reporter vectors were cotransfected, each at 100 ng/well. Luciferase expression was determined 30 hr after transfection using the Dual Luciferase Assay (Promega). Where transfected cells were stimulated with proteoglycan (10 μg/ml), it was added to the transfected cells 24 hr after transfection and the cells were stimulated for 6 hr. NF-κB-directed firefly luciferase expression was determined and normalized to the CMV-driven Renilla luciferase expression and expressed as the percentage of the latter. The experiments were carried out in triplicate and data are presented as mean ± SD.
The 293T cells were also transfected with pTLR1, pTLR2, pTLR4, pTLR7, or pTLR9 in the presence of the luciferase reporter plasmids and the luciferase activities were measured in 30 hr. The 293T cells were also transfected with different doses of pTLR2 or pTLR4, i.e. 1000, 200, 40 and 8 ng/well, to examine constitutive NF-κB activation by these TLRs.
Results
Most human TLR genes exhibit low frequencies of major codons
Low TLR expression is frequently observed in published work and has been pointed out by Visintin et al.18 Protein expression is regulated at multiple key steps, e.g. transcription, translation and post-translational modifications. Besides these mechanisms, the codon usage adopted in a gene also affects the expression of a gene at the protein level. This has been clearly documented in some life forms.22,23 Although the impact of codon usage in the expression of vertebrate genes is less clear, genome-wide analyses have revealed preferential usage of certain codons for every amino acid in the proteins of humans and other mammals (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=Human+sapiens+[gbpro]). Modification of codon usage in the coding regions of expression vectors can indeed increase or decrease the expression of the encoded proteins.24
To examine whether the TLR genes have adopted common codon usage patterns, codon usage by all TLR genes and, as a comparison, by the CD14 gene was determined using the codonw program. CD14 was included in this study because it resembles the TLRs in sequence.16 The frequency of each codon in the CD14 and TLR genes was expressed as the percentage of the total number of codons in a gene that encode a specified amino acid. A major human codon was defined as the most frequently used codon for an amino acid genome-wide, which is available at (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=Human+sapiens+[gbpro]). Thus derived codon frequencies for the CD14 and TLR genes were compared with the human genome average to identify preferences in each gene for the set of major codons (Table 1).
Table 1.
Codon usage in the human genome and the CD14 and TLR genes
| Amino acids | Std | CD14 | TLR1 | TLR2 | TLR3 | TLR4 | TLR5 | TLR6 | TLR7 | TLR8 | TLR9 | TLR10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | TTT | 45·4 | 30·0 | 56·0 | 58·0 | 65·6 | 48·5 | 45·0 | 72·5 | 61·5 | 55·5 | 21·0 | 55·5 |
| TTC | 54·6 | 70·0 | 44·0 | 42·0 | 34·3 | 51·5 | 55·0 | 27·5 | 38·5 | 44·5 | 79·0 | 44·5 | |
| L | TTA | 7·5 | 0·0 | 21·3 | 19·8 | 14·3 | 14·7 | 8·8 | 19·0 | 13·0 | 18·5 | 0·0 | 19·2 |
| TTG | 12·5 | 4·8 | 21·3 | 20·7 | 21·8 | 14·7 | 16·8 | 9·5 | 11·2 | 19·8 | 4·7 | 18·3 | |
| CTT | 13·0 | 4·8 | 15·7 | 15·7 | 19·0 | 12·3 | 24·2 | 36·5 | 16·0 | 14·8 | 4·2 | 16·0 | |
| CTC | 20·0 | 25·8 | 12·0 | 11·5 | 13·7 | 17·7 | 20·5 | 8·0 | 19·8 | 10·5 | 25·5 | 12·0 | |
| CTA | 7·8 | 14·5 | 9·3 | 5·8 | 14·3 | 13·0 | 9·5 | 6·3 | 12·3 | 16·0 | 5·2 | 12·0 | |
| CTG | 39·6 | 50·0 | 20·3 | 26·5 | 17·0 | 27·7 | 20·5 | 20·7 | 27·9 | 20·3 | 60·3 | 22·3 | |
| I | ATT | 35·5 | 25·0 | 39·7 | 53·3 | 43·0 | 45·3 | 34·7 | 57·0 | 35·0 | 43·3 | 0·0 | 53·7 |
| ATC | 47·6 | 50·0 | 38·0 | 32·0 | 31·0 | 47·7 | 38·7 | 34·3 | 35·0 | 24·0 | 89·3 | 29·7 | |
| ATA | 17·0 | 25·0 | 22·3 | 12·3 | 26·0 | 6·7 | 13·3 | 8·7 | 30·0 | 33·0 | 10·7 | 8·3 | |
| V | GTT | 18·0 | 7·3 | 25·5 | 29·0 | 43·5 | 28·0 | 21·8 | 42·0 | 27·8 | 28·5 | 2·0 | 44·3 |
| GTC | 23·9 | 21·5 | 21·0 | 18·5 | 15·5 | 12·0 | 26·0 | 22·5 | 24·0 | 16·8 | 23·5 | 21·0 | |
| GTA | 11·8 | 14·3 | 16·3 | 18·5 | 23·0 | 18·0 | 21·8 | 9·8 | 9·3 | 19·0 | 2·0 | 18·5 | |
| GTG | 46·4 | 57·3 | 37·3 | 34·3 | 18·0 | 42·0 | 20·3 | 25·8 | 39·0 | 35·8 | 72·5 | 16·3 | |
| S | TCT | 18·4 | 13·8 | 29·3 | 26·2 | 29·5 | 22·2 | 12·5 | 25·3 | 21·7 | 24·7 | 8·0 | 28·0 |
| TCC | 22·0 | 31·0 | 11·8 | 25·0 | 19·2 | 14·8 | 32·0 | 15·5 | 20·7 | 19·3 | 24·2 | 7·0 | |
| TCA | 15·0 | 3·5 | 17·7 | 15·0 | 19·2 | 14·8 | 13·8 | 25·3 | 17·3 | 16·2 | 4·7 | 26·3 | |
| TCG | 5·5 | 24·2 | 2·3 | 3·8 | 1·3 | 2·5 | 2·8 | 2·8 | 1·2 | 3·2 | 8·0 | 1·8 | |
| AGT | 14·9 | 0·0 | 18·8 | 17·5 | 11·5 | 26·0 | 15·3 | 24·0 | 19·5 | 18·3 | 11·5 | 22·8 | |
| AGC | 24·2 | 27·7 | 20·0 | 12·5 | 19·2 | 29·0 | 23·7 | 7·0 | 19·5 | 18·3 | 43·7 | 14·0 | |
| P | CCT | 28·4 | 36·8 | 37·0 | 16·0 | 18·8 | 27·0 | 48·5 | 35·0 | 32·0 | 24·3 | 21·5 | 27·5 |
| CCC | 33·0 | 23·3 | 26·0 | 48·0 | 34·5 | 34·5 | 25·8 | 11·8 | 17·0 | 29·8 | 59·0 | 38·0 | |
| CCA | 27·4 | 13·3 | 22·3 | 32·0 | 47·0 | 38·5 | 22·5 | 53·5 | 40·5 | 35·3 | 3·5 | 31·0 | |
| CCG | 11·3 | 26·8 | 14·8 | 4·0 | 0·0 | 0·0 | 3·3 | 0·0 | 10·8 | 10·8 | 16·0 | 3·5 | |
| T | ACT | 24·3 | 5·5 | 42·8 | 35·5 | 28·3 | 33·3 | 37·5 | 42·0 | 34·8 | 31·0 | 12·8 | 26·8 |
| ACC | 36·2 | 39·0 | 23·8 | 20·0 | 26·0 | 30·8 | 22·5 | 23·8 | 24·5 | 23·8 | 61·5 | 23·3 | |
| ACA | 28·2 | 33·3 | 31·0 | 37·8 | 41·3 | 33·3 | 35·0 | 31·5 | 30·5 | 36·3 | 7·8 | 46·5 | |
| ACG | 11·4 | 22·3 | 2·5 | 6·8 | 4·3 | 2·5 | 1·0 | 2·8 | 10·3 | 9·0 | 18·0 | 3·5 | |
| A | GCT | 26·4 | 14·0 | 31·8 | 39·3 | 18·8 | 45·3 | 32·5 | 42·8 | 26·3 | 36·0 | 12·0 | 39·3 |
| GCC | 40·2 | 46·5 | 36·3 | 32·3 | 37·5 | 29·0 | 46·0 | 25·0 | 39·5 | 30·8 | 68·0 | 21·5 | |
| GCA | 22·8 | 16·3 | 27·3 | 25·0 | 37·5 | 25·8 | 19·0 | 25·0 | 26·3 | 30·8 | 14·8 | 35·8 | |
| GCG | 10·6 | 23·3 | 4·5 | 3·5 | 6·3 | 0·0 | 2·8 | 7·3 | 8·0 | 2·5 | 5·3 | 3·5 | |
| Y | TAT | 43·8 | 50·0 | 68·0 | 47·5 | 63·0 | 56·5 | 59·5 | 77·5 | 71·5 | 70·5 | 26·5 | 60·0 |
| TAC | 56·2 | 50·0 | 32·0 | 52·5 | 37·0 | 43·5 | 40·5 | 22·5 | 28·5 | 29·5 | 73·5 | 40·0 | |
| H | CAT | 41·2 | 14·5 | 40·5 | 75·0 | 53·0 | 52·0 | 60·0 | 77·5 | 64·5 | 54·0 | 17·0 | 56·5 |
| CAC | 58·8 | 85·5 | 59·5 | 25·0 | 47·0 | 48·0 | 40·0 | 22·5 | 35·5 | 46·0 | 83·0 | 43·5 | |
| Q | CAA | 26·0 | 14·5 | 50·0 | 31·0 | 39·0 | 28·5 | 37·5 | 72·0 | 46·5 | 52·5 | 14·0 | 65·5 |
| CAG | 74·0 | 85·5 | 50·0 | 69·0 | 61·0 | 71·5 | 62·5 | 28·0 | 53·5 | 47·5 | 86·0 | 34·5 | |
| N | AAT | 46·1 | 26·5 | 64·5 | 57·0 | 49·5 | 62·0 | 48·5 | 50·0 | 47·5 | 52·5 | 18·5 | 69·5 |
| AAC | 53·9 | 73·5 | 35·5 | 43·0 | 50·5 | 38·0 | 51·5 | 50·0 | 52·5 | 47·5 | 81·5 | 30·5 | |
| K | AAA | 42·8 | 11·0 | 54·0 | 51·0 | 65·0 | 57·0 | 45·0 | 62·5 | 56·0 | 62·5 | 26·0 | 72·0 |
| AAG | 57·2 | 89·0 | 46·0 | 49·0 | 35·0 | 43·0 | 55·0 | 37·5 | 44·0 | 37·5 | 74·0 | 28·0 | |
| D | GAT | 46·1 | 31·0 | 56·0 | 59·5 | 74·5 | 51·5 | 57·5 | 38·5 | 66·0 | 65·0 | 26·0 | 65·0 |
| GAC | 53·9 | 69·0 | 44·0 | 40·5 | 25·5 | 48·5 | 42·5 | 61·5 | 34·0 | 35·0 | 74·0 | 35·0 | |
| E | GAA | 42·1 | 45·0 | 72·5 | 62·5 | 59·5 | 62·0 | 64·0 | 69·0 | 59·0 | 68·0 | 17·0 | 72·5 |
| GAG | 57·9 | 55·0 | 27·5 | 37·5 | 40·5 | 38·0 | 36·0 | 31·0 | 41·0 | 32·0 | 83·0 | 27·5 | |
| C | TGT | 45·1 | 36·5 | 47·5 | 60·0 | 47·0 | 57·0 | 63·0 | 65·5 | 63·0 | 60·0 | 20·5 | 38·0 |
| TGC | 54·9 | 63·5 | 52·5 | 40·0 | 53·0 | 43·0 | 37·0 | 34·5 | 37·0 | 40·0 | 79·5 | 62·0 | |
| R | CGT | 8·2 | 13·7 | 4·5 | 9·3 | 6·7 | 6·5 | 6·0 | 10·2 | 10·8 | 15·2 | 13·7 | 5·7 |
| CGC | 19·1 | 50·0 | 9·2 | 9·3 | 3·3 | 9·7 | 9·2 | 6·2 | 0·0 | 10·8 | 50·0 | 0·0 | |
| CGA | 11·2 | 0·0 | 0·0 | 6·3 | 16·7 | 12·8 | 9·2 | 4·0 | 5·3 | 10·8 | 6·0 | 25·8 | |
| CGG | 20·7 | 18·2 | 4·5 | 15·7 | 13·3 | 6·5 | 6·0 | 4·0 | 10·8 | 15·2 | 21·2 | 2·8 | |
| AGA | 20·5 | 13·7 | 31·9 | 28·2 | 36·7 | 35·5 | 45·5 | 61·2 | 54·0 | 26·2 | 0·0 | 45·7 | |
| AGG | 20·3 | 4·5 | 50·0 | 31·3 | 23·3 | 29·0 | 24·2 | 14·3 | 19·0 | 21·7 | 9·2 | 28·5 | |
| G | GGT | 16·4 | 8·3 | 21·8 | 19·3 | 29·5 | 36·0 | 12·3 | 26·5 | 10·3 | 23·5 | 15·3 | 36·0 |
| GGC | 34·1 | 37·5 | 30·5 | 30·8 | 26·5 | 23·0 | 26·8 | 17·8 | 20·8 | 23·5 | 54·3 | 16·0 | |
| GGA | 24·9 | 33·3 | 26·0 | 33·8 | 23·5 | 25·8 | 39·0 | 50·0 | 38·0 | 29·5 | 11·8 | 24·0 | |
| GGG | 24·7 | 20·8 | 21·8 | 15·5 | 20·5 | 15·5 | 22·0 | 6·3 | 31·0 | 23·5 | 18·8 | 24·0 |
Codon frequencies in the human genome were obtained from http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=Homo+sapiens+[gbpri] and compared with that of the CD14 and TLR genes (accession numbers: NM_000591, U88540, U88878, U88879, NM_138554, AB060695, AB020807, AF245702, AF245703, AF245704, AF296673), which were generated using the codonw program. Codon frequency for each amino acid was expressed as a percentage, taking the number of codons in the human genome or the individual genes for each amino acid as 100%. The most frequently used codon, i.e. the major codon, for each amino acid residue is shown in bold type. One-letter codes were used for the amino acid residues.
Methionine and tryptophan, which are encoded by single codons, were not included for analysis. The three stop codons were also excluded. Examining the average codon frequencies in the human genome, as listed in Table 1, identified major codons for most amino acids except for arginine. Without exception, all these more frequently used codons ended with either ‘G’ or ‘C’. Examining the CD14 gene revealed that most amino acids in CD14, except for serine, proline and tyrosine residues, favoured major human codons. In the case of serine, the two most frequently used codons were AGC and TCC (31% and 27·7%, respectively) and both were also most frequently used in the human genome (24·2% and 22·0%, respectively). The overall usage of these two codons was increased in the CD14 gene compared with the human genome average. The same was true with the proline codons in the CD14 gene (Table 1). The deviation of tyrosine codon usage in the CD14 gene from the major tyrosine codon TAC was probably because the CD14 gene only contains two tyrosine residues. In summary, the CD14 gene clearly favoured the use of major human codons. Where major codons were preferentially adopted in the CD14 gene, the frequencies of these codons in the CD14 gene were mostly elevated compared with the genome average. Therefore, the CD14 gene appeared to have evolved for more effective expression at the protein level.
In contrast, most TLR genes showed reduced usage of major codons compared with the human genome average (Table 1). The most frequently used codons in the TLR genes mostly ended with ‘A’ or ‘T’. Where major human codons were adopted in TLR genes also as the most frequently used codons, the frequencies were mostly reduced compared with the genome average (Table 1). However, TLR9 was an exception. The TLR9 gene had adopted every major human codon as its most frequently used codon and the frequencies of these codons in the TLR9 gene were also substantially increased compared with the human genome average (Table 1). These data indicated that the usual low expression of TLRs was probably the result of deviation of the TLR genes from major human codons and they also implied that CD14 and TLR9 are probably more effectively expressed.
More CD14 expression is detected on monocytes than TLR1 and TLR2
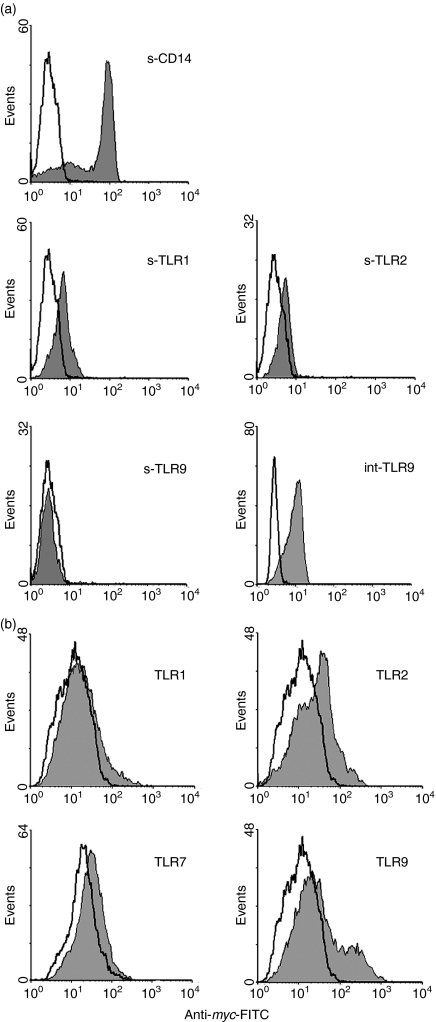
Monocytes express CD14 and these cells also express TLRs. Since the CD14 gene is distinct from the TLR1 and TLR2 genes in codon usage, the expression of these three receptors was compared on monocytes. As shown in Fig. 1(a), the level of CD14 on monocytes was much higher than that of TLR1 and TLR2, approximately 20–40-fold higher when mean fluorescence indices (MFI) were compared. The expression of TLR9 was similarly examined on monocytes, but it was not detectable on the cell surface. However, it was clearly detected upon permeabilization of the cell membrane, which is consistent with the report of its intracellular expression.25 TLR9 expression inside monocytes was found to be prominent although it could not be directly compared with that of surface CD14 and TLRs because of the different staining protocols used (Fig. 1a). The high expression of CD14 on monocytes, as compared with that of TLR1 and TLR2, agreed with the high frequencies of major human codons adopted in the CD14, but not TLR1 and TLR2, genes. However, these results are not sufficient to establish that the different CD14, TLR1 and TLR2 expression levels detected on monocytes resulted substantially from their differences in codon usage. CD14, TLR1 and TLR2 expression on monocytes was also regulated by other factors, e.g. intrinsic regulatory elements in the different genes. The use of different antibodies may also contribute to the apparent differences observed in CD14, TLR1 and TLR2 expression on monocytes.
Figure 1.
Expression of CD14, TLR1, TLR2 and TLR9. (a) Monocytes were isolated and stained with mouse antibodies specific for these receptors, or isotype mouse IgG, followed by FITC-labelled goat anti-mouse IgG. To detect intracellular TLR9 (int-TLR9), monocytes were fixed and permeabilized before staining. Open histograms represent signals obtained with isotype IgG and solid histograms represent signals obtained with the specific antibodies. ‘s-’denotes surface expression. (b) 293T cells were transfected with the TLR expression vectors and, after membrane permeabilization, were stained with the anti-myc antibody or isotype IgG. Open histograms represent signals obtained with isotype IgG and solid histograms represent signals obtained with the anti-myc antibody.
The TLR9-coding sequence is more effectively expressed than that of TLR1, TLR2 and TLR7
To assess more directly the effects of codon usage on TLR expression, the exact TLR coding sequences were cloned so that they could be expressed under the regulation of identical regulatory elements in the expression vector. This would eliminate the influences of specific endogenous regulatory elements associated with each TLR gene and would directly examine the effect of the coding sequences on TLR expression. The exact coding sequences of TLR1, TLR2, TLR7 and TLR9, to be compared, were cloned into the pcDNA3.1/myc-His vector. TLR7 was included in this experiment because it was most closely related to TLR9 in amino acid sequence and was also known to be intracellularly expressed (data not shown). Comparing the expression between these two TLRs is probably most meaningful. To standardize the detection protocol, all TLR sequences were cloned to express C-terminal myc-His tags. Thus all TLRs, irrespective of their subcellular localization, were detected using the same protocol by flow cytometry. As shown in Fig. 1(b), under these conditions, TLR9 was again more effectively expressed than the other three TLRs as judged by flow cytometry. These results are consistent with the higher usage of major codons in the TLR9 gene.
TLR2 expression is markedly increased upon partial codon optimization
The above evidence suggests that deviation from the usage of major codons in human TLR genes contributed to the usual low TLR expression. If this is true, optimization of codon usage in TLR genes could improve TLR expression. To test this hypothesis, the 5′ 302 bp of the TLR2 coding sequence were replaced with a synthetic DNA fragment in which all codons have been changed to major human codons (Fig. 2b). The encoded amino acid sequence was not affected by the optimized codons. This was achieved with 22 overlapping/complementary synthetic oligonucleotide primers and a complete DNA fragment was assembled from these primers by PCR (Fig. 2a). The optimized TLR2 sequence is presented in Fig. 2(b), in alignment with the corresponding wild-type TLR2 sequence. The resultant DNA fragment was cloned into the pTLR2-MH expression vector to replace the corresponding wild-type sequence. Both the pTLR2-MH and pTLR2opt-MH vectors were expressed in 293T cells and the expression of TLR2 from both vectors was detected by flow cytometry and Western blotting. As shown in Fig. 2(c), the percentage of TLR2-positive 293T cells after transfection with the pTLR2opt-MH vector (25·1%) was markedly increased compared with 293T cells transfected with the unmodified pTLR2-MH vector (10·3%) (Fig. 2c). It was also noted that the TLR2-positive cells that express the optimized TLR2 sequences also expressed much higher levels of TLR2 at the single-cell level. By Western blotting, the expression level of TLR2 detected in 293T cells that were transfected with the pTLR2opt-MH vector was also clearly higher than that in 293T cells transfected with the pTLR2-MH vector (Fig. 2d). Densitometry analysis, using the Image Scanner (Amersham Pharmacia Biotech), showed that the signal in lane 3 was approximately fourfold higher than that in lane 2 (data not shown). Collectively, these results show that the intrinsic low expression of most TLRs, except for TLR9, is directly affected by the low frequencies of major human codons in their genes. However, this can be experimentally corrected through codon optimization.
Partial modification of CD14 sequence using prevalent TLR codons markedly reduces CD14 expression
To examine whether the preferential adoption for major human codons by the CD14 gene contributes to its high expression observed on monocytes (Fig. 1a), the 5′ 381-bp CD14 sequence, encoding the N-terminal 124 residues (including the signal peptide), was replaced by prevalent TLR codons (Fig. 3a). The wild-type and codon-modified CD14 expression vectors were expressed in 293T cells and surface CD14 expression was examined by flow cytometry. As shown in Fig. 3(b), the 293(T) cells transfected with the wild-type CD14 expression vector exhibited a high level of CD14 expression, i.e. the mean fluorescence index (MFI) reached 198·65. This was consistent with a high level of CD14 being detected on monocytes (Fig. 1a). In contrast, the partially codon-modified CD14 vector conferred much less CD14 expression (MFI = 71·47) (Fig. 3b). As a control, 293T cells transfected with the pcDNA3.1 vector exhibited no detectable surface CD14. The above results were obtained when 293T cells were transfected with the vectors at 0·25 μg/well. At 2 μg/well, the modified CD14 vector showed less difference in CD14 expression compared with the wide-type CD14 vector (data not shown). These results show that the preferential usage of major human codons in wild-type CD14 contributes significantly to its high expression.
Over-expression of TLR2 constitutively activates NF-κB
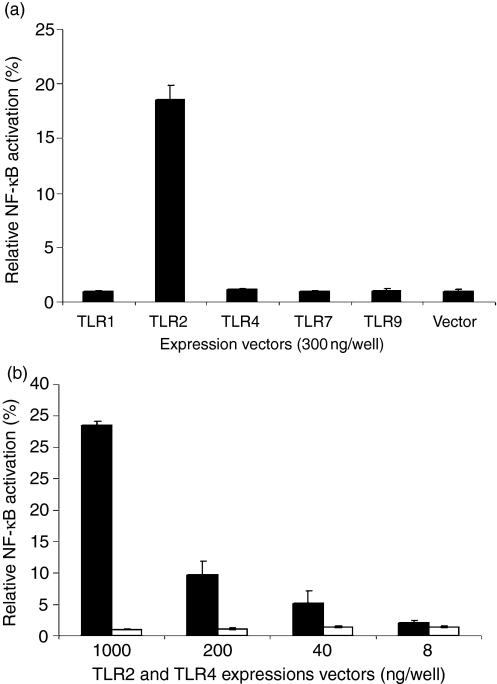
A common functional property of TLRs is the recognition of microbial adjuvant structures which is essential to trigger the production of inflammatory cytokines or host inflammation.3–5 Inflammation is tightly regulated so as to eradicate pathogens and, at the same time, prevent excessive tissue damage. Excessive or prolonged activation of TLRs may lead, or add, to pathological conditions. Constitutive NF-κB activation has been observed when TLR2 was over-expressed together with TLR1.26 In this study, when TLR1, TLR2, TLR4, TLR7 and TLR9 were over-expressed in 293T cells, only TLR2 was shown to cause constitutive NF-κB activation (Fig. 4a). Induction of constitutive NF-κB activation by TLR4 was previously reported when it was coexpressed with MD2 and CD14.27 However, TLR4 over-expression in the absence of CD14 and MD2, at up to 1 μg/well in 24-well plates, did not cause detectable constitutive NF-κB activation as shown in this study (Fig. 4b). In contrast, expression of pTLR2, at 40 ng/well in 24-well plates, was able to induce constitutive NF-κB activation which was increased in a dose-dependent manner with the increase of the pTLR2 dosage (Fig. 4b).
Figure 4.
Over-expression of TLR2, but not TLR1, TLR4, TLR7, or TLR9, causes constitutive NF-κB activation. (a) pTLR1, pTLR2, pTLR4, pTLR7 and pTLR9 were expressed in 293T cells in 24-well plates at 300 ng/well. pcDNA3·1 was used as a control (Vector). Also cotransfected were the two luciferase reporter vectors each at 100 ng/well. After 30 hr, the NF-κB-directed firefly luciferase expression and the CMV-driven Renilla luciferase activities were determined. NF-κB activation was presented as the firefly luciferase activities normalized to the activity of Renilla luciferase. (b) 293T cells were transfected with pTLR2 (black bars) or pTLR4 (open bars) in 24-well plates at different dosages, i.e. 1000, 200, 40 and 8 ng/well. The luciferase plasmids were similarly cotransfected. After 30 hr, luciferase expression was determined. All experiments were performed in triplicate. The results are representative of three similar experiments.
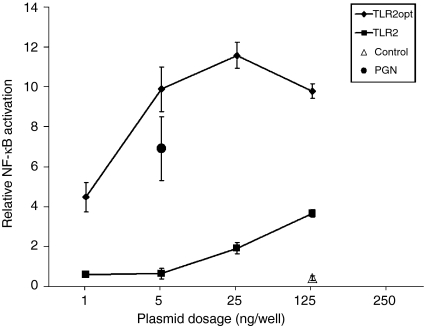
To assess the implications of elevated TLR2 expression, as a result of TLR2 codon optimization, we examined NF-κB activation in 293T cells that were transfected with the pTLR2-MH and pTLR2opt-MH expression vectors. When the doses of these vectors were serially increased, i.e. at 1, 5, 25 and 125 ng/well in 24-well plates, cells transfected with pTLR2-MH did not show detectable NF-κB activation at the doses of 1 and 5 ng/well, although stimulation of 293T cells, that have been transfected with pTLR2-MH at 5 ng/well, with peptidoglycan could effectively trigger NF-κB activation (Fig. 5). At the plasmid doses of 25 and 125 ng/well, pTLR2-MH conferred three- to sixfold increases in constitutive NF-κB activation (Fig. 5). In contrast, transfection of 293T cells with the codon-optimized pTLR2opt-MH vector led to more than seven-fold increase in NF-κB activation at 1 ng/well. At 5 ng/well, it caused a remarkable 18-fold increase. These results indicate the risk associated with TLR2 over-expression and stress, the essential requirement for mechanisms to restrict TLR2 expression. Although the over-expression of TLR1, TLR4, TLR7 and TLR9 did not cause NF-κB activation, it is possible that it confers higher sensitivity of the transfected cells to microbial structure stimulation.
Figure 5.
Constitutive NF-κB activation upon over-expression of TLR2. 293T cells were transfected with a serial dose of the pTLR2-MH or pTLR2opt-MH vectors, i.e. 1, 5, 25 and 125 ng/well in 24-well plates. Also cotransfected were the luciferase reporter vectors. After 30 hr, the NF-κB-directed firefly luciferase expression and the CMV-driven Renilla luciferase activities were determined. NF-κB activation was presented as the firefly luciferase activities normalized to the expression of Renilla luciferase. As a control, 293T cells were transfected with pcDNA3·1 (125 ng/well), in place of the TLR2 expression vectors. 293T cells transfected with pTLR2-MH at 5 ng/well were, after 24 hr, also stimulated with proteoglycan (for 6 hr) before luciferase measurement. Results are presented as means ± SD of triplicate experiments.
Discussion
The generally low TLR expression echoes the fact that, although the activities of TLRs as adjuvant receptors have long been described, none of these receptors were initially identified at the protein level based on their functional properties. In this study, we found that the sequences of human TLRs, except for TLR9, were highly deviated from using major human codons. The correlation between the low frequency of major codons in TLR genes and the low expression of these receptors was demonstrated through the expression of TLR1, TLR2, TLR7 and TLR9 in 293T cells. The expression of TLR9 was higher than expression of the other TLRs although it was not proportionally higher taking into account the high frequencies of major human codons in its gene. It is possible that the flow cytometry-based method for detecting TLR expression, based on the C-terminal myc tags, was not sufficiently sensitive to reflect the level of TLR9 expression. The demonstration that partial codon optimization of the TLR2 sequence was sufficient to increase TLR2 expression testifies to the important contribution of codon usage to the regulation of TLR expression. The demonstration that changing CD14 codons to prevalent TLR codons markedly reduced CD14 expression further stressed the role of codon choice in the control of gene expression. It is not clear why TLR expression should be regulated by such a rigid mechanism and in such a universal manner because temporally regulated protein expression, e.g. through regulatory elements in the individual TLR genes, should provide a greater degree of flexibility.
The presence of such a mechanism in the regulation of TLR expression stresses the importance of tight control of TLR expression. The benefits of systematically suppressed TLR expression may be deduced from the observations that over-expression of some TLRs may lead to constitutive TLR activation. Constitutive activation has been reported for TLR2 and TLR4 upon over-expression. Co-expression of TLR2 with TLR1 can result in TLR2/TLR1 heterodimers and in constitutive NF-κB activation.26 We also showed in this study that over-expression of TLR2, at 125 ng/well in 24-well plates, led to NF-κB activation in the absence of microbial structure stimulation (Fig. 5). Partial codon optimization of the TLR2 sequence resulted in increased TLR2 expression which led to effective NF-κB activation at low plasmid dosages (e.g. 1 ng/well). Over-expression of wild-type TLR4, in the presence of CD14 and MD2, causes constitutive NF-κB activation.27 Uncontrolled TLR4 activation can lead to excessive inflammation and severe tissue damage.28 Elevated TLR1, TLR2 and TLR4 expression has been reported in atherosclerotic plaques.29 Elevated TLR4 expression has been reported to contribute to the progression of intimal lesions.30 There is no published evidence showing that over-expression of other TLRs, other than TLR2, leads to constitutive NF-κB activation. We showed that over-expression of TLR4 alone, in the absence of CD14 and MD2, caused no detectable NF-κB activation. However, it is possible that over-expression of TLRs confers on host cells an increased sensitivity to microbial structure stimulation, leading to excessive inflammation and tissue damage.
Technically, our finding appears to remove a hurdle towards the understanding of TLR structures and TLR recognition of microbial structures. The specificity of TLRs for microbial structures is mostly determined through two approaches, i.e. loss of response to specific microbial structures in TLR-deficient mice or acquisition of responses to specific microbial structures by cells that have been transfected to express specific TLRs. Studies of direct interactions between TLRs and microbial structures are scarce.31,32 Investigations using purified TLRs can be particularly informative. Our demonstration that codon optimization could markedly increase TLR2 expression implied that TLRs could be more effectively expressed and purified, upon codon modification, for these studies.
Acknowledgments
This project is supported by Singapore National Medical Research Council grants: R-364-000-014-213 and R-364-000-019-213.
Abbreviations
- APC
antigen-presenting cell
- TIR
Toll/interleukin-1 receptor homology
- TLR
Toll-like receptor
References
- 1.Watts C. Capture and processing of exogenous antigens for presentation on MHC molecules. Annu Rev Immunol. 1997;15:821–50. doi: 10.1146/annurev.immunol.15.1.821. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–8. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 4.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 5.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–7. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 6.Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. 1991;351:355–6. doi: 10.1038/351355b0. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–15. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 8.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–7. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 9.Ramon G. Sur la toxine et sur l'anatoxine diphtheriques. Ann Inst Pasteur. 1924;38:1–10. [Google Scholar]
- 10.Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 11.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA. 1998;95:588–93. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi O, Kawai T, Sanjo H, Copeland NG, Gilbert DJ, Jenkins NA, Takeda K, Akira S. TLR6: a novel member of an expanding toll-like receptor family. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 13.Du X, Poltorak A, Wei Y, Beutler B. Three novel mammalian toll-like receptors. Gene structure, expression, and evolution. Eur Cytokine Netw. 2000;11:362–71. [PubMed] [Google Scholar]
- 14.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors, Htlr7, Htlr8, and Htlr9. Eur Cytokine Netw. 2000;11:372–8. [PubMed] [Google Scholar]
- 15.Chuang T, Ulevitch RJ. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta. 2001;1518:157–61. doi: 10.1016/s0167-4781(00)00289-x. [DOI] [PubMed] [Google Scholar]
- 16.Triantafilou M, Triantafilou K. Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 2002;23:301–4. doi: 10.1016/s1471-4906(02)02233-0. [DOI] [PubMed] [Google Scholar]
- 17.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA, Golenbock DT, Espevik T. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J Biol Chem. 2002;277:47834–43. doi: 10.1074/jbc.M207873200. [DOI] [PubMed] [Google Scholar]
- 18.Visintin A, Mazzoni A, Spitzer JH, Wyllie DH, Dower SK, Segal DM. Regulation of Toll-like receptors in human monocytes and dendritic cells. J Immunol. 2001;166:249–55. doi: 10.4049/jimmunol.166.1.249. [DOI] [PubMed] [Google Scholar]
- 19.Cao W, Bobryshev YV, Lord RS, Oakley RE, Lee SH, Lu J. Dendritic cells in the arterial wall express C1q: potential significance in atherogenesis. Cardiovasc Res. 2003;60:175–86. doi: 10.1016/s0008-6363(03)00345-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen K, Lu J, Wang L, Gan YH. Mycobacterial heat shock protein 65 enhances antigen cross-presentation in dendritic cells independent of Toll-like receptor 4 signaling. J Leukoc Biol. 2004;75:260–6. doi: 10.1189/jlb.0703341. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Tay PN, Cao W, Li W, Lu J. Integrin-nucleated Toll-like receptor (TLR) dimerization reveals subcellular targeting of TLRs and distinct mechanisms of TLR4 activation and signaling. FEBS Lett. 2002;532:171–6. doi: 10.1016/s0014-5793(02)03669-4. [DOI] [PubMed] [Google Scholar]
- 22.Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucl Acids Res. 1988;16:8207–11. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akashi H. Gene expression and molecular evolution. Curr Opin Genet Dev. 2001;11:660–6. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 24.Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–24. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur J Immunol. 2002;32:1958–68. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 26.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci USA. 2000;97:13766–71. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;4138:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 28.Beutler B. TLR4 as the mammalian endotoxin sensor. Curr Top Microbiol Immunol. 2002;270:109–20. doi: 10.1007/978-3-642-59430-4_7. [DOI] [PubMed] [Google Scholar]
- 29.Edfeldt K, Swedenborg J, Hansson GK, Yan ZQ. Expression of toll-like receptors in human atherosclerotic lesions: a possible pathway for plaque activation. Circulation. 2002;105:1158–61. [PubMed] [Google Scholar]
- 30.Vink A, Schoneveld AH, van der Meer JJ, et al. In vivo evidence for a role of toll-like receptor 4 in the development of intimal lesions. Circulation. 2002;106:1985–90. doi: 10.1161/01.cir.0000032146.75113.ee. [DOI] [PubMed] [Google Scholar]
- 31.Sato M, Sano H, Iwaki D, et al. Direct binding of Toll-like receptor 2 to zymosan, and zymosan-induced NF-κB activation and TNF-α secretion are down-regulated by lung collectin surfactant protein A. J Immunol. 2003;171:417–25. doi: 10.4049/jimmunol.171.1.417. [DOI] [PubMed] [Google Scholar]
- 32.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–53. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]