Abstract
Interaction of cytokines with their cognate receptors leads to the activation of latent transcription factors – the signal transducers and activators of transcription (STAT) proteins – whose biological activities ultimately regulate many critical aspects of cell growth, survival and differentiation. Dysregulation of the JAK-STAT pathway is frequently observed in many primary human tumours, reflecting the importance of this pathway in the maintenance of cellular integrity. Here we review the current progress in STAT structure and function, and the contribution of STAT signalling to the pathogenesis of T-cell lymphomas.
Keywords: negative regulation, phophatases, STATs, T-cell lymphoma, truncated STATs
Introduction
It is now over a decade since the first signal transducer and activator of transcription (STAT) protein and its activating JAK kinases were identified as being a vital signalling pathway that mediates interferon signalling.1 Subsequently, the JAK-STAT pathway was shown to be involved in signalling by a plethora of cytokine and growth factor receptors.2 The vital contributions of the individual JAK and STAT proteins to numerous biological processes have been gleaned in vitro from cell culture studies and in vivo from mice harbouring targeted deletions of these genes.3 These crucial insights into the biological roles of STAT proteins and their function as essential regulators of cell proliferation, differentiation and survival in different cellular contexts paved the way for studies that revealed their critical role in malignant transformation and oncogenesis.4–6 Dysregulated STAT activation leads to increased angiogenesis and enhanced survival of tumours.5 Knockout studies have also highlighted the function of STAT proteins in the development and function of the immune system and of their roles in maintaining peripheral immune tolerance and tumour surveillance.7–10 In this review, we concentrate on the role of STAT signalling in T-cell lymphomas and consider the potential use of targeting STATs in the treatment of these diseases.
STAT structure and activation
STAT proteins were originally identified as being latent cytoplasmic transcription factors, which were only translocated to the nucleus upon Jak-mediated phosphorylation and dimerization following cytokine-induced activation of Jaks.1 Once in the nucleus the activated STAT dimers can bind to consensus DNA-recognition motifs, called gamma-activated sites (GAS), in the promoters of cytokine-inducible genes, resulting in transcriptional activation. The understanding of the JAK-STAT pathway has undergone several conceptual modifications based on new evidence for STAT dimerization, phosphorylation and nuclear–cytoplasmic shuttling (Fig. 1). In contrast to earlier notions, recent studies have revealed that STATs can exist as stable, non-phosphorylated dimers in the absence of cytokine stimulation.11 Studies on STATs 1, 3 and 5 have elegantly demonstrated that unphosphorylated STATs shuttle between the cytoplasm and the nucleus in the absence of cytokine activation.12–17 While non-phosphorylated STAT proteins constantly shuttle between the cytoplasm and nucleus, the phosphorylated STAT protein is retained in the nucleus and is only released upon dephosphorylation by nuclear phosphatases.18, 19 Therefore, nuclear retention, not translocation, of the active phosphorylated STAT protein is the important feature of cytokine-induced STAT activation.
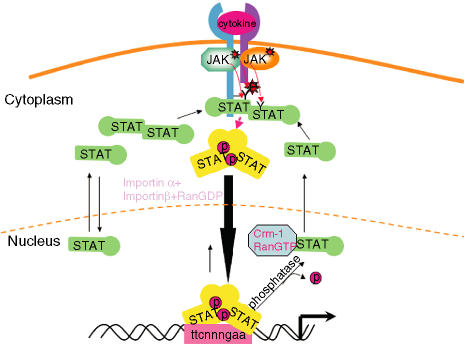
Figure 1.
The JAK-signal transducer and activator of transcription (JAK-STAT) signalling pathway. Ligand-induced activation of latent cytoplasmic JAK kinases leads to the docking and subsequent tyrosine phosphorylation of latent monomeric or N-domain-mediated dimeric STAT proteins, facilitating the formation of active SH2-domain-mediated homodimers of STAT proteins. The tyrosine-phosphorylated STAT dimers are actively transported to the nucleus using metabolic energy and the importin α/β and RanGDP complex. Once inside the nucleus, the active STAT dimers bind to the promoters of genes containing the consensus recognition motif (GAS motif-ttcnnngaa) and activate transcription of these genes. STATs can bind DNA as dimers or as N-domain-mediated tetramers. The active STAT protein is only released from DNA upon dephosphorylation by nuclear phosphatases, following cytokine withdrawal, and the inactivated protein is then actively exported out of the nucleus to the cytoplasm by the exportin Crm-1/RanGTP complex. Monomeric STAT proteins can also passively shuttle between the cytoplasm and the nucleus via carrier-free diffusion through nuclear pores facilitated by the interaction with nucleoporins.19
The mammalian STAT family comprises Stat1, Stat2, Stat3, Stat4, Stat5a, Stat5b and Stat6.20 The two Stat5 genes share > 90% homology but are encoded by separate genes.21 STAT proteins share an overall general structure that is organized into functional modular domains (Fig. 2). The three-dimensional crystal structures of core Stat1 and Stat3 proteins (amino acids 130–710) bound to DNA revealed a clamp-like structure for dimeric STAT proteins, including a coiled-coil domain comprising four β-sheets immediately preceding the DNA-binding domain, which facilitates multiple protein–protein interactions.20, 22, 23 Crystallization studies of isolated N-domains of Stat4 revealed that these form stable dimers in solution.24 Based on the physical properties of mutant STAT N domains, the favoured stable conformation appears to be a helical hook-like structure held together by molecular interactions involving a conserved hydrophobic residue (Phe 77 in Stat1).25 Mutagenesis studies of Stat1, Stat4 and Stat5a/b have shown that N-domain interactions between two activated STAT dimers facilitate the formation of stable tetramers on tandemly linked non-consensus GAS motifs present in naturally occurring promoters.26–29 All STAT proteins, except STAT2, have been shown to form higher-order tetramer complexes. N-domain-mediated tetramers of Stat1 can also form on single GAS motifs, and mutation of this domain in Stat1 affects the transcriptional activation of Stat1-responsive promoters with only a single discernable GAS motif.30 This raises the question as to whether the tetramer or the dimer is the physiologically relevant complex for cytokine-induced gene activation. The generation of mouse models of N-domain mutants of STAT proteins should resolve this issue. In addition to tetramerization, the N domain also regulates receptor recognition and phosphatase recruitment for some STATs.30, 31 Thus, for a relatively small domain, the N-domain is important in many functions for STAT signalling.
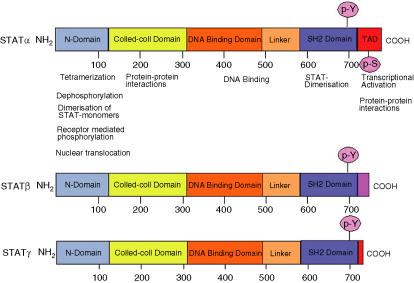
Figure 2.
Modular structure of signal transducer and activator of transcription (STAT) proteins. All STAT proteins share a common molecular topology and are organized into distinct functional domains. The NH2 terminus (N-domain) is involved in protein–protein interactions between adjacent STAT dimers on DNA, facilitating the formation of STAT tetramers. It is also involved in the formation of dimers between non-phosphorylated STAT monomers, which is important for receptor-mediated activation and nuclear translocation of certain STAT proteins. Interactions with STAT cofactors, which positively or negatively modulate their transcriptional activity, occurs via the N-domain, the adjacent coiled-coil domain and the carboxy-terminal transactivation domain (TAD). The conserved serine residue (p-S), which is phosphorylated upon cytokine stimulation and is important for maximal transcriptional activation, is located within the transactivation domain. In addition to the full-length form (STATα), STAT proteins can also be present as C-terminally truncated forms generated by alternative splicing (STATβ) or by proteolytic processing (STATγ). The C-termini of STATβ proteins differs from the corresponding STATα protein not only by being truncated but also by having additional amino acids inserted as a result of the splicing event.
Modulation of STAT activity by histone acetylases and deacetylases
STAT proteins synergize with other transcription factors to achieve transcriptional activation.20 Maximal transcriptional activation by STAT proteins additionally requires chromatin remodelling by histone acetylation/deacetylation. To achieve this, STAT proteins recruit the histone acetyl transferase (HAT) coactivator proteins, CBP/p300, via their transactivation domains.32 Stat1 can also functionally synergize with the HAT protein pCIP.33 Unlike the other STATs, Stat2 synergizes with another HAT protein, GCN5.34 Cytokine [interferon-α (IFN-α), interferon-γ (IFN-γ) and interleukin (IL)-2] induction of Stat1, Stat2 and Stat5 signalling requires chromatin recruitment of histone deacteylases (HDAC).35–37 Transcriptional activation by these STAT proteins can be inhibited by the treatment of cells with HDAC inhibitors, such as trichostatin A and SAHA. In contrast, HDACs may also negatively regulate the magnitude and duration of Stat5 transcriptional activity by recruitment to the STAT transcriptional complex via the nuclear co-repressor protein, SMRT, which directly associates with the coiled-coil domain of Stat5.38
Negative regulation of STAT activity
The duration and magnitude of STAT activation is critically regulated by a number of different mechanisms at the cytoplasmic and nuclear levels, which together ensure that these proteins function in a strictly controlled manner in normal cells. These include the actions of cytoplasmic and nuclear phosphatases, the interaction of inhibitory proteins, and covalent modifications that result in targeted degradation of the active STAT protein or inhibition of STAT transcription (Fig. 3). It is beyond the scope of this review to discuss this topic extensively and the reader is referred to other excellent reviews.39, 40
Figure 3.
Negative regulation of signal transducer and activator of transcription (STAT) signalling. Cytokine-induced STAT activation can be inhibited by SOCS proteins, the gene expression of which is regulated by STAT proteins, thus fulfilling a negative feedback loop. SOCS proteins inhibit STAT activation either by inhibition of the activating JAKs or by competition with STATs for receptor binding. Activated STAT proteins can be dephosphorylated by cytoplasmic and/or nuclear phosphatases. C-terminally truncated STAT proteins, STATβ and STATγ, behave as dominant-negative proteins to functionally compete with their full-length counterparts to alter or inhibit gene expression, respectively. Protein inhibitors of activated STAT proteins (PIAS) interact with STAT proteins to inhibit their DNA binding and/or potentially facilitate their covalent modification by sumoylation and subsequent degradation.
Phosphatases
STAT proteins are inactivated by the action of both cytoplasmic and/or nuclear phosphatases. The protein tyrosine phosphatase (PTP), SHP-2, directly interacts with Stat5 in the cytoplasm and induces its dephosphosphorylation, while Shp2–/– cells show hyper-phosphorylation of Stat5.41, 42 The cytosolic PTP, PTP1B, and a nuclear phosphatase, TC-PTP, have also been shown to inactivate Stat5a and Stat5b in in vitro experiments.43, 44 However, the physiological relevance of PTP-1B in Stat5 inactivation has not been confirmed. Analysis of cells from TC-PTP–/– mice have revealed that the nuclear isoform of this phosphatase, TC45, specifically dephosphorylates Stat1 and Stat3. but not Stat5 or Stat6.45
Protein inhibitors of activated STAT proteins
STAT protein activation can be inhibited by direct association with protein inhibitors of activated STAT proteins (PIAS). The mammalian PIAS family members include PIAS1, PIAS3, PIASx and PIASy and alternative splice variants of PIASx.46 PIAS1 and PIAS3 selectively inhibit DNA binding of Stat1 and Stat3, respectively.47, 48 In contrast, inhibition of IL-12-induced Stat4 activation by PIASx is mediated by the recruitment of HDAC1 and HDAC2 to the DNA-bound PIAS–STAT4 complex, and repression can be abolished by the HDAC inhibitor, trichostatin A.49 More recently, PIAS proteins were shown to induce sumoylation of STAT1, suggesting that this mechanism may be involved in the targeted proteasome-mediated degradation of activated STATs.50 PIAS1 knockout mice have confirmed its role as a negative regulator of STAT1 signalling, but not by protein sumoylation.51 Thus, the physiological importance of STAT sumoylation by PIAS proteins remains to be clarified.
Suppressors of cytokine-signalling proteins
Signalling by STAT proteins is crucially down-regulated by the cytokine-inducible suppressors of cytokine signalling (SOCS) proteins in an auto-regulatory negative feedback loop.52 There are eight members of the SOCS family, comprising CIS (cytokine-inducible SH2 domain protein) and SOCS1–7. Structurally they are all related, possessing a central SH2 domain and a conserved C-terminal motif, termed the SOCS box.52 Individual SOCS proteins adopt various strategies of inhibition to attenuate signal transduction from cytokines that act through the JAK/STAT pathway. These include direct binding to JAKs to inhibit their activity, binding to receptors to block the binding of JAKs and competing for STAT binding to activated receptors. Additionally, the SOCS box can interact with components of the E3–ubiquitin ligase complex, elongins B and C, which can target signalling proteins for proteasome-mediated degradation, although this has only been demonstrated with the oncogenic fusion protein, Tel.JAK2.53–55 Gene targeting studies have revealed numerous essential roles for these proteins in the immune system, and are reviewed extensively elsewhere.52 Recently, an unexpected role for SOCS proteins in maintaining the cytokine specificity of the initiating response was demonstrated by the altered IL-6-induced gene expression profile of SOCS3–/– macrophages.56, 57 Therefore, SOCS proteins are not only important in the attenuation of the cytokine-induced STAT signalling process, but also in maintaining the functional integrity of the cytokine-specific response.
Naturally occurring truncated STAT proteins
STAT proteins are naturally expressed not only as full-length proteins (STATα), but also as an alternatively spliced truncated form, which is missing the C-terminal transactivation domain (STATβ) (Fig. 2). STATβ forms have been identified for Stat1, Stat3, Stat4 and Stat5 and, like their full-length isoforms, they are activated in response to cytokine/interferon signalling to bind DNA, but differ in their ability to induce gene transcription.58–62 Although STATβ proteins behave as functional dominant-negative proteins when overexpressed in cells, physiologically, STATα and STATβ have specific, non-overlapping roles, as revealed by gene-deletion studies of Stat3 and Stat4 in mice. Mice expressing only Stat3α have a defective response to endotoxic shock, while exclusive expression of STAT3β rescues the embryonic lethality of a complete STAT3 deletion.63, 64 Exclusive expression of Stat4α or Stat4β leads to defective IL-12-induced proliferation and IFN-γ production in the respective transgenic mice.65 In each case there are significant changes in STAT3 or STAT4-induced transcription of target gene expression, indicating that physiologically STATβ is not a dominant-negative protein, but is independently capable of transcriptional activation. This suggests that cytokine-induced STAT signalling is a balance between the concerted action of full-length and naturally occurring variants of STAT proteins.
Proteolytic processing of STAT proteins
Initially, the generation of naturally occurring truncated STAT proteins was attributed to alternate splicing. Subsequent studies have shown that C-terminally truncated STAT proteins can also be generated by proteolytic processing. Processed forms of STAT3, STAT5a, STAT5b and STAT6 have all been identified in many different cell types and are generated by the action of cell-type proteases and of STAT-specific proteases.66 In myeloid cells a developmentally regulated serine protease generates exclusive expression of C-terminally truncated STAT5 proteins, the expression of which is essential for the maintenance of these cells in an immature, undifferentiated state.67–70 Similarly, in acute myeloid leukaemia (AML) patient-derived cell lines, a Stat3 and Stat5-specific serine protease activity has been identified, but the protease in these cell lines appears to differ from that detected in a murine haematopoietic cell line.71 Interestingly, the myeloid-specific Stat5 protease cleaves Stat5a and Stat5b to generate proteins that terminate at amino acids 720 and 724, respectively, while the truncated Stat5a generated by alternative splicing creates an altered C terminus beyond amino acid 740 and so it is possible that these proteins are functionally different.61, 69 Proteolytic cleavage of STATs has also been demonstrated for STAT6 in mast cells, for Stat3 and Stat5 in neutrophils and for Stat5 in T cells.72–75 The Stat6 serine protease, which is also sensitive to elastase inhibitors, does not cleave Stat5, but the neutrophil-derived Stat5 proteases will also cleave Stat3.76 The cellular distribution of the proteases varies for the different serine proteases, with the Stat5 proteases being cytoplasmic, nuclear or ubiquitously expressed, depending on the cellular context, while the Stat6 protease in mast cells is exclusively nuclear. While the exact identities of these proteases have not been established, it is possible that the STAT serine proteases represent a family of closely related proteins with different cell-type and STAT-specificities. One other protease implicated in STAT proteolytic processing is calpain, which can cleave Stat5 and Stat3 in platelets.77 Thus, STAT processing by proteases is an important negative regulatory mechanism in STAT biology.
Aberrant STAT signalling in T-cell lymphomas
The role of constitutively activated STATs, particularly Stat3 and Stat5, in cellular transformation and oncogenesis has been clearly established by in vitro and in vivo studies and is extensively reviewed elsewhere.4, 5, 78, 79
T-lymphoblastic lymphomas in STAT5 transgenic mice
Recent evidence from Stat5 transgenic mice indicates that over-expression of the wild-type protein may contribute to tumour formation. Studies in transgenic mice have shown that Stat5 proteins critically regulate CD8+ T-cell homeostasis.80 Additionally, mice exclusively expressing either wild-type Stat5a or Stat5b in the lymphoid compartment developed thymic CD8+ T-lymphoblastic lymphomas, with age.81 Moreover, co-expression of a T-cell receptor transgene in Stat5 transgenic mice profoundly increased the rate of lymphoma formation. The molecular mechanisms by which elevated expression of wild-type Stat5a or Stat5b proteins induces T-cell lymphomas is unclear at present. Gene expression profiling of wild-type and malignant splenic T cells from these transgenic models did not provide any major clues to the origins of tumour formation. Increased expression of a few selective genes, including protein tyrosine phosphatase non-receptor type 13 (PTPn13), which is a negative regulator of Fas-mediated apoptosis, was noted. However, these genes are more likely to be involved in maintenance rather than induction of the lymphomas. Nevertheless, these studies suggest that unlike Stat3, which behaves as an oncogene only when persistently activated or artificially dimerized, inappropriate expression of wild-type Stat5 can induce tumorigenesis.6, 78
Primary cutaneous T-cell lymphomas
The skin is the second most frequent extranodal sight, after the gastrointestinal tract, for lymphoma. Primary cutaneous T-cell lymphomas (CTCL) are a heterogeneous group of non-Hodgkin's lymphoma and are classified, depending on their clinical behaviour, into indolent or aggressive subtypes.82 Mycosis fungoides (MF) is the most common form of CTCL and is usually of an indolent nature. A less common variant of indolent CTCL is the primary cutaneous entity of CD30-positive anaplastic large cell lymphoma. Sezary Syndrome (SS) is a leukaemic form of CTCL with an aggressive clinical course.83 Tumour cells in CTCL are typically CD4+ T cells that preferentially migrate to the skin and which usually display an activated memory phenotype, and express T helper 2 (Th2)-like cytokines (IL-4, IL-5 and IL-10).84
Constitutive activation and dysregulation of STATs in MF and SS
Gene array studies used to examine gene-expression patterns in peripheral blood mononuclear cells from patients with SS have indicated that there is significant down-regulation of Stat4 expression, which is consistent with the loss of IL-12-mediated T helper 1 (Th1) responses and the characteristic Th2 skewing of T cells in this disease.85, 86 The high-affinity IL-2 receptor-α chain subunit (CD25), which is normally only expressed on activated T cells, is constitutively expressed in many patients with CTCL, suggesting dysregulated IL-2 receptor signalling in these lymphomas.87, 88 Analysis of peripheral blood lymphocytes (PBL) and tumour cell lines derived from patients with MF and SS revealed that Stat3, but not Stat5, is constitutively activated in these tumours in a cytokine-independent manner.89, 90 Another study revealed a cytokine-dependent constitutive activation of Stat3 and Stat5 in patients with SS.87 The reasons for these apparent differences between the two studies are unclear, but as with human T-cell lymphotrophic virus-1 (HTLV-1)-induced leukaemias, the activation of STATs in CTCL may proceed through an early cytokine-dependent and independent phases.91 Consistent with this, a recent study has shown that there is constitutive activation of Stat3 in tumour cells from skin biopsies of patients with MF, which was not seen in intradermal lymphocytes or non-malignant T-cell lines from patients with pretumour early stage (patch/plaque) disease.92 Furthermore, enforced expression of dominant-negative Stat3 in tumour cells increased apoptosis by 30–50%, indicating that persistently activated Stat3 strongly contributed to the survival of tumour cells.92
Cytokine-independent activation of Stat3 in a tumour cell line derived from a patient with SS was shown to be mediated by constitutive activation of Jak3. Inhibition of JAK phosphorylation by the JAK-specific inhibitor, tyrphostin AG490, also inhibited constitutive activation of Stat3 and induced the down-regulation of CD25 expression in SS and MF cell lines.89, 90 Treatment of SS cell lines with AG490 also inhibited growth and induced apoptosis in these cells. The constitutive activation of JAK3 in these tumour cells may be linked to the status of the tyrosine phosphatase, SHP-1, which negatively regulates IL-2R/JAK3 signalling. Interestingly, SHP-1 protein levels were shown to be decreased in T-cell lymphoma lines owing to methylation of the SHP-1 promoter.93 Furthermore, the decrease in SHP-1 protein levels in SS patient PBL and cell lines corresponded to the level of JAK3 phosphorylation.94 In contrast, constitutive activation of Stat3 and IL-2-induced activation of Stat5 has been shown to be independent of SHP-2 activity in a CTCL cell line.95 Inhibition of constitutive Stat3 activation in MF cell lines by expressing a dominant-negative C-terminally truncated Stat3 also blocked the spontaneous production of IL-5, and to a lesser extent IL-13, but not IL-6 production by these cells.96 Recent studies using murine models have shed new light on a new role of persistent Stat3 signalling in tumour biology. These studies have shown that constitutive activation of Stat3 leads to immune evasion by tumour cells through production of factors that inhibit the innate immune response mediated by dendritic cell differentiation/maturation and the promotion of T-cell tolerance.7 This is particularly interesting in view of recent in vitro findings, which demonstrate that the growth of malignant cells in CTCL is greatly enhanced by contact with immature dendritic cells.97 Thus, constitutive activation of Stat3 in MF and SS is a vital event in the growth and differentiation of malignant T cells in these diseases.
Constitutive SOCS3 expression
Constitutive activation of SOCS3 has also been reported in MF and SS tumour cell lines, associated with the persistent signalling of Stat3. Overexpression of dominant-negative C-terminally truncated Stat3 in these cells inhibits the constitutive expression of SOCS3, establishing a functional link between constitutive activation of Stat3 and SOCS3.92, 98 Inhibition of SOCS3 expression also increases the IFN-α sensitivity of the tumour cells, which show stronger Stat1 and Stat3 activation in response to IFN-α and IFN-γ treatment than the parental cells expressing high levels of SOCS3.98 Thus, examining the SOCS3 status of patients with MF and SS could predict the responsiveness of patients to IFN-α treatment. Constitutive expression of SOCS3 in MF and SS also has important physiological implications for the cytokine skewing observed in MF and SS. T-cell differentiation studies have shown that SOCS3 expression is greatly elevated in Th2 cells and inhibits IL-12-induced Stat4 activation.99–101 Moreover, studies using SOCS3 transgenic mice reveal exaggerated Th2 responses, resulting in the development of Th2-mediated allergic immune diseases.102 Thus, sustained expression of SOCS3 in malignant cells in MF and SS inhibits their Th1 response and promotes cytokine skewing to a Th2 phenotype.
Dominant-negative STAT5 expression
While the Jak3-Stat3 pathway is constitutively activated in SS, Stat5 activation is inducible. However, we have recently shown, in SS patients, a different form of dysregulation of Stat5.103 Characterization of Stat5 activation in freshly isolated PBL from healthy donors revealed that Stat5 is present only as a C-terminally truncated form in the nucleus of resting PBL, generated by the action of a protease.75, 103 Full-length Stat5 is only activated following mitogenic or antigenic stimulation of T cells, when the proteolytic activity is down-regulated.75, 103 In contrast, patients with SS showed elevated or exclusive expression of the C-terminally truncated Stat5 protein, even in potently activated T cells, which resulted in the loss of IL-2-induced Stat5-dependent gene expression of target genes such as PIM-1, CIS and BCL-2.103 Notably, CD25 expression was still inducible by IL-2, a finding that is consistent with the regulation of this gene by STAT3 in SS.89 Nevertheless, activation of either the full-length or C-terminally truncated Stat5 proteins was inducible, rather than constitutive, in SS patients. The loss of expression of known Stat5-dependent genes, even in patients that showed activation of some wild-type Stat5 protein, demonstrates the capacity of the C-terminally truncated Stat5 protein to behave as a physiological dominant-negative. Given the critical role of IL-2-induced Stat5 signalling in normal immune homeostasis and the maintenance of peripheral tolerance, the loss of this pathway has important implications for the pathogenesis of SS.9, 104, 105 Thus, sustained expression of C-terminally truncated Stat5 proteins may be one mechanism adopted by activated malignant T cells in SS to escape IL-2-induced apoptosis. Also consistent with dysregulation of the IL-2/Stat5 signalling pathway in SS, malignant cells typically show a marked loss of FAS expression and little or no IFN-γ production, both of which are required for the deletion of activated T cells during normal homeostatic regulation of the immune system.106, 107 As C-terminally truncated Stat5 proteins can be generated by post-translational processing through the action of proteases or by alternate splicing, it is possible that one or both of these mechanisms are progressively dysregulated in malignant T cells in SS. Future work on the characterization of the mechanisms underlying the regulation of Stat5 expression should provide important clues to the basis of the dysregulated expression of truncated Stat5 proteins in SS.
CD30+ anaplastic large T-cell lymphoma
Anaplastic large T-cell lymphoma (ALCL) represents a heterogeneous group of large cell lymphomas that are most frequently derived from cytotoxic T cells. The defining features of this disease consist of a proliferation of predominantly large lymphoid cells with strong expression of the cytokine receptor, CD30, and a characteristic growth pattern. There are three distinct entities: primary systemic anaplastic lymphoma kinase positive (ALK+) ALCL, ALK-negative (ALK–) ALCL, and primary cutaneous (PC) ALCL. ALK+ALCL predominantly affects young males and has a good prognosis when treated with chemotherapy. ALK–ALCL occurs in older patients (affecting both genders equally) and has an unfavourable prognosis. PC-ALCL appears to have different pathogenesis and better prognosis than systemic ALCL, and is usually ALK–.108
In 80% of ALCL there is a t(2:5) chromosomal translocation, which generates a fusion protein between the gene encoding nucleophosphomin (NPM), which is normally only expressed in neuronal tissues, and ALK.109 NPM/ALK is a constitutively activated tyrosine kinase, which can induce T-cell lymphomas in mice.110 Investigation of the mode of action of NPM/ALK in T-cell lymphomas from patients and in transfection experiments exogenously expressing the fusion protein, reveals that NPM/ALK-mediated lymphomagenesis involves STAT activation, in addition to the activation of pp60src, PI3K/Akt and phospholipase C-γ pathways.87, 111–113 ALK+ALCL cells contain constitutively activated Stat3 and Stat5, which physically associate with NPM/ALK in vivo.114–116 Phosphorylation of Stat5 and Stat3 is mediated by the direct action of ALK, although the role of Jak3 cannot be fully excluded because a JAK inhibitor can reduce, but not abrogate, STAT phosphorylation in ALCL cell lines.114 Expression of a dominant-negative Stat5b protein in NPM/ALK-transformed cells inhibited the cytokine-independent growth of these cells and induced apoptotic cell death upon factor withdrawal.116 Furthermore, transfer of NPM/ALK-transfected or NPM/ALK+ Stat5b dominant-negative transfected cells into severe combined immunodeficient (SCID) mice significantly reduces the rate of aggressive tumour formation and enhances the survival of affected animals. Thus, Stat5 plays a critical role in the rate of malignant transformation by NPM/ALK in CD30+ ALCL. However, as the NPM/ALK+ mice all eventually succumb to neoplastic death, irrespective of the presence of dominant-negative Stat5b expression, persistent Stat5 signalling is probably more important during the early stages of malignant transformation and can eventually become redundant because of signalling from other pathways that are also aberrantly activated.116
The constitutive activation of Stat3 is more striking than Stat5 in ALK+ALCL. In addition to direct tyrosine phosphorylation by NPM/ALK, tumour cells have acquired additional mechanisms to ensure that Stat3 is kept persistently active, indicating the importance of Stat3 signalling in the pathogenesis of ALK+ALCL. Tumour cells express the serine-threonine phosphatase, PP2A, a positive regulator of Stat3, which physically associates with Stat3 in these cells.115, 117 The mode of action of PP2A in the activation of Stat3 is not clear. However, treatment of cells with a PP2A inhibitor abrogates Stat3 tyrosine phosphorylation, suggesting that PP2A removes inhibitory serine or threonine phosphorylations on Stat3, which enhances its tyrosine phosphorylation.115 The functional relationship between ALK and PP2A in the activation of Stat3 is unclear because PP2A is also activated in ALK– cells. A second strategy employed by these tumour cells to activate Stat3 is to inactivate PIAS3, which, as mentioned earlier, specifically inhibits Stat3 DNA binding.115 Although Stat3 activation is associated with ALK expression, a recent study has shown that Stat3 expression is not strictly dependent on ALK, as a proportion of ALK–ALCL showed constitutive Stat3 activation.118 Interestingly in PC-ALCL, which has a much more favourable prognosis than systemic ALCL, Stat3 is not constitutively activated.
Stat3 signalling therefore appears to play a vital central role in malignant transformation of T cells in ALCL. The exact contributions of Stat3-signalling to the establishment and maintenance of tumours remains to be identified by future murine studies using cell-transfer experiments expressing dominant-negative Stat3 proteins. In vitro experiments expressing dominant-negative Stat3 proteins in ALK+/ALCL cell lines resulted in the induction of apoptosis and inhibition of cell cycle progression by activation of caspase 3 and decreased expression of Bcl-2, Bcl-XL, Mcl-1, cyclin D3 and c-myc.119 Survivin, a Stat3 target gene, which is frequently expressed at elevated levels in both ALK– and ALK+ALCL tumours and is associated with poor prognosis, was also down-regulated by inhibiting Stat3 signalling.119, 120 Thus, targeting Stat3 and its functional regulators offers multiple potential therapeutic strategies in this disease.
STAT targeting
As evidence for aberrant STAT signalling in various forms of human cancers has accumulated, the targeting of STAT signalling in cancer therapy has gained momentum. The success of STAT targeting has been proven by many in vitro experiments in cell culture experiments, but the in vivo proof of principle was shown in a gene therapy study by using a dominant-negative Stat3 to inhibit growth of the tumour in a murine model of melanoma.121 As the use of dominant-negative Stat5 and/or Stat3 has been successfully used in in vitro studies of ALCL and MF/SS tumour cell lines, this approach holds significant potential for the therapeutic intervention of these diseases.92, 115, 116, 119
An alternative approach to inhibiting STAT signalling is to inhibit the activity of the upstream activating kinase, such as ALK in ALK+ALCL. Small-molecule inhibitors could be designed to inhibit specifically the function of the tyrosine kinase, so that the tumour cells, which display constitutive activation of this protein, will be targeted. However, this approach has the drawback that it does not target the STATs specifically, but also affects expression of other proteins downstream of the aberrantly activated kinase. Nevertheless, this approach is being developed with the production of many inhibitors of known STAT activating kinases, such as AG490 for JAKs, and others targeting SRC, the EGF receptor.
Similarly, other physiological regulators of STAT function can be targeted, such as the putative Stat5 protease in PBL, the negative regulatory proteins SOCS and PIAS, and phosphatases. Dominant-negative versions of these regulatory proteins, or small-molecule inhibitors based on these proteins, could be effective either alone or in combination with direct STAT inhibitors to block constitutive or defective STAT signalling in tumours. On the other hand, strategies designed to activate the negative regulators of STATs, such as the SHP-1 phosphatase in SS, or PIAS3 in CD30+ALCL, could specifically inhibit STAT activity in tumour cells.
Finally, the use of histone deacetylase inhibitors (HDI), such as trichostatin A, depsipeptide and SAHA, which inhibit the activities of HDACs, may have beneficial effects at a more general level on STAT-dependent lymphomas because of the requirement for HDACs in STAT-mediated transcriptional activation of target gene expression. Of these HDIs, depsipeptide has emerged as a potentially effective therapy for patients with T-cell lymphoma and, like other HDIs, induce apoptosis of tumour cells.122, 123 Initial results from a phase-1 trial of depsipeptide (FR901228) on CTCL and unspecified peripheral T-cell lymphoma patients showed partial or complete responses, respectively.124 Future studies directed at understanding the exact molecular targets of these drugs in different T-cell lymphomas will help the design and use of more specifically targeted versions of these drugs.
Conclusions
STATs are modular signalling proteins, which efficiently and rapidly execute cytokine-mediated signalling, resulting in cell proliferation, differentiation and survival. The magnitude and duration of their activities are stringently regulated by a variety of different physiological mechanisms. The importance of STAT proteins to the normal homeostatic function of cells is displayed by their frequent dysregulation in human cancers. In T-cell lymphomas such as CTCL and ALCL, Stat3 and Stat5 are dysregulated, and the in vitro targeting of these proteins can inhibit tumour progression in these diseases. Much progress has been made in recent years in understanding the molecular mechanisms underlying the dysregulation of STATs in these T-cell lymphomas. Future studies aimed at elucidating the biochemical regulation of STAT proteins and their targets in peripheral T cells should greatly enhance our understanding of the mechanism of tumorigenesis in T-cell lymphomas and other cancers. In vitro experiments with dominant-negative Stat3 and Stat5 proteins indicate that future therapeutic strategies based on these proteins hold great potential for the treatment of CTCL and ALCL.
Acknowledgments
The authors would like to thank Dr Sean Whittaker for critical comments. The authors' research on this subject was funded by generous support from Professor Adrian Hayday; S.J. is funded by the Wellcome Trust and by the Medical Research Council, UK.
References
- 1.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Gadina M, Hilton D, Johnston JA, Morinobu A, Lighvani A, Zhou YJ, Visconti R, O'Shea JJ. Signaling by type I and II cytokine receptors: ten years after. Curr Opin Immunol. 2001;13:363. doi: 10.1016/s0952-7915(00)00228-4. [DOI] [PubMed] [Google Scholar]
- 3.Levy DE. Physiological significance of STAT proteins: investigations through gene disruption in vivo. Cell Mol Life Sci. 1999;55:1559. doi: 10.1007/s000180050395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 5.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang T, Niu G, Kortylewski M, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 8.Cheng F, Wang HW, Cuenca A, et al. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 9.Antov A, Yang L, Vig M, Baltimore D, Van Parijs L. Essential role for STAT5 signaling in CD25+ CD4+ regulatory T cell homeostasis and the maintenance of self-tolerance. J Immunol. 2003;171:3435. doi: 10.4049/jimmunol.171.7.3435. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. 1998;95:7556. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 12.Pranada AL, Metz S, Herrmann A, Heinrich PC, Muller-Newen G. Real time analysis of STAT3 nucleocytoplasmic shuttling. J Biol Chem. 2004;279:15114. doi: 10.1074/jbc.M312530200. [DOI] [PubMed] [Google Scholar]
- 13.Zeng R, Aoki Y, Yoshida M, Arai K, Watanabe S. Stat5B shuttles between cytoplasm and nucleus in a cytokine-dependent and -independent manner. J Immunol. 2002;168:4567. doi: 10.4049/jimmunol.168.9.4567. [DOI] [PubMed] [Google Scholar]
- 14.Meyer T, Begitt A, Lodige I, van Rossum M, Vinkemeier U. Constitutive and IFN-gamma-induced nuclear import of STAT1 proceed through independent pathways. EMBO J. 2002;21:344. doi: 10.1093/emboj/21.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begitt A, Meyer T, van Rossum M, Vinkemeier U. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc Natl Acad Sci USA. 2000;97:10418. doi: 10.1073/pnas.190318397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer T, Gavenis K, Vinkemeier U. Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp Cell Res. 2002;272:45. doi: 10.1006/excr.2001.5405. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharya S, Schindler C. Regulation of Stat3 nuclear export. J Clin Invest. 2003;111:553. doi: 10.1172/JCI15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer T, Marg A, Lemke P, Wiesner B, Vinkemeier U. DNA binding controls inactivation and nuclear accumulation of the transcription factor Stat1. Genes Dev. 2003:17. doi: 10.1101/gad.268003. 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marg A, Shan Y, Meyer T, Meissner T, Brandenburg M, Vinkemeier U. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J Cell Biol. 2004;165:823. doi: 10.1083/jcb.200403057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 21.Lin JX, Mietz J, Modi WS, John S, Leonard WJ. Cloning of human Stat5B. Reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. J Biol Chem. 1996;271:10738. [PubMed] [Google Scholar]
- 22.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 24.Vinkemeier U, Moarefi I, Darnell JE, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Bhandari R, Vinkemeier U, Van Den Akker F, Darnell JE, Jr, Kuriyan J. A reinterpretation of the dimerization interface of the N-terminal domains of STATs. Protein Sci. 2003;12:361. doi: 10.1110/ps.0218903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer WK, Reichenbach P, Schindler U, Soldaini E, Nabholz M. Interaction of STAT5 dimers on two low affinity binding sites mediates interleukin 2 (IL-2) stimulation of IL-2 receptor alpha gene transcription. J Biol Chem. 1997;272:31821. doi: 10.1074/jbc.272.50.31821. [DOI] [PubMed] [Google Scholar]
- 27.John S, Vinkemeier U, Soldaini E, Darnell JE, Jr, Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol. 1999;19:1910. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath CM, Wen Z, Darnell JE., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 29.Vinkemeier U, Cohen SL, Moarefi I, Chait BT, Kuriyan J, Darnell JE., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616. [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer T, Hendry L, Begitt A, John S, Vinkemeier U. A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of stat transcription factors. J Biol Chem. 2004;279:18998. doi: 10.1074/jbc.M400766200. [DOI] [PubMed] [Google Scholar]
- 31.Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol. 2004;5:208. doi: 10.1038/ni1032. [DOI] [PubMed] [Google Scholar]
- 32.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274:25343. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 33.Korzus E, Torchia J, Rose DW, et al. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 34.Paulson M, Press C, Smith E, Tanese N, Levy DE. IFN-Stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat Cell Biol. 2002;4:140. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- 35.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci USA. 2003;100:14742. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deactylase activity for signaling by STAT1. J Biol Chem. 2004;279:30358–68. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 37.Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol Cell Biol. 2003;23:4162. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima H, Brindle PK, Handa M, Ihle JN. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J. 2001;20:6836. doi: 10.1093/emboj/20.23.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenhalgh CJ, Hilton DJ. Negative regulation of cytokine signaling. J Leukoc Biol. 2001;70:348. [PubMed] [Google Scholar]
- 40.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 41.Chughtai N, Schimchowitsch S, Lebrun JJ, Ali S. Prolactin induces SHP-2 association with Stat5, nuclear translocation, and binding to the beta-casein gene promoter in mammary cells. J Biol Chem. 2002;277:31107. doi: 10.1074/jbc.M200156200. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Wen R, Yang S, Schuman J, Zhang EE, Yi T, Feng GS, Wang D. Identification of Shp-2 as a Stat5A phosphatase. J Biol Chem. 2003;278:16520. doi: 10.1074/jbc.M210572200. [DOI] [PubMed] [Google Scholar]
- 43.Aoki N, Matsuda T. A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: dephosphorylation and deactivation of signal transducer and activator of transcription 5a and 5b by TC-PTP in nucleus. Mol Endocrinol. 2002;16:58. doi: 10.1210/mend.16.1.0761. [DOI] [PubMed] [Google Scholar]
- 44.Aoki N, Matsuda T. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J Biol Chem. 2000;275:39718. doi: 10.1074/jbc.M005615200. [DOI] [PubMed] [Google Scholar]
- 45.ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu Y, Zhu W, Tremblay M, David M, Shuai K. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci. 2003;60:2561. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997:278. doi: 10.1126/science.278.5344.1803. 1803. [DOI] [PubMed] [Google Scholar]
- 48.Rogers RS, Horvath CM, Matunis MJ. SUMO modification of STAT1 and its role in PIAS-mediated inhibition of gene activation. J Biol Chem. 2003;278:30091. doi: 10.1074/jbc.M301344200. [DOI] [PubMed] [Google Scholar]
- 49.Arora T, Liu B, He H, Kim J, Murphy TL, Murphy KM, Modlin RL, Shuai K. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J Biol Chem. 2003;278:21327. doi: 10.1074/jbc.C300119200. [DOI] [PubMed] [Google Scholar]
- 50.Ungureanu D, Vanhatupa S, Kotaja N, Yang J, Aittomaki S, Janne OA, Palvimo JJ, Silvennoinen O. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood. 2003;102:3311. doi: 10.1182/blood-2002-12-3816. [DOI] [PubMed] [Google Scholar]
- 51.Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, Wu H, Shuai K. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5:891. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- 52.Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- 53.Johnston JA. Are SOCS suppressors, regulators, and degraders? J Leukoc Biol. 2004;75:743. doi: 10.1189/jlb.1003507. [DOI] [PubMed] [Google Scholar]
- 54.Zhang JG, Farley A, Nicholson SE, et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci USA. 1999;96:2071. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamizono S, Hanada T, Yasukawa H, et al. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem. 2001;276:12530. doi: 10.1074/jbc.M010074200. [DOI] [PubMed] [Google Scholar]
- 56.Croker BA, Krebs DL, Zhang JG, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4:540. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 57.Lang R, Pauleau AL, Parganas E, Takahashi Y, Mages J, Ihle JN, Rutschman R, Murray PJ. SOCS3 regulates the plasticity of gp130 signaling. Nat Immunol. 2003;4:546. doi: 10.1038/ni932. [DOI] [PubMed] [Google Scholar]
- 58.Zakharova N, Lymar ES, Yang E, Malik S, Zhang JJ, Roeder RG, Darnell JE., Jr Distinct transcriptional activation functions of STAT1alpha and STAT1beta on DNA and chromatin templates. J Biol Chem. 2003;278:43067. doi: 10.1074/jbc.M308166200. [DOI] [PubMed] [Google Scholar]
- 59.Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA. 1992;89:7836. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle JN. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Azam M, Erdjument-Bromage H, Kreider BL, et al. Interleukin-3 signals through multiple isoforms of Stat5. EMBO J. 1995;14:1402. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caldenhoven E, van Dijk TB, Solari R, Armstrong J, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J Biol Chem. 1996;271:13221. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 63.Yoo JY, Huso DL, Nathans D, Desiderio S. Specific ablation of Stat3beta distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell. 2002;108:331. doi: 10.1016/s0092-8674(02)00636-0. [DOI] [PubMed] [Google Scholar]
- 64.Maritano D, Sugrue ML, Tininini S, et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat Immunol. 2004;5:401. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]
- 65.Hoey T, Zhang S, Schmidt N, et al. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. 2003;22:4237. doi: 10.1093/emboj/cdg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakajima H, Suzuki K, Iwamoto I. Lineage-specific negative regulation of STAT-mediated signaling by proteolytic processing. Cytokine Growth Factor Rev. 2003;14:375. doi: 10.1016/s1359-6101(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 67.Meyer J, Jucker M, Ostertag W, Stocking C. Carboxyl-truncated STAT5beta is generated by a nucleus-associated serine protease in early hematopoietic progenitors. Blood. 1998;91:1901. [PubMed] [Google Scholar]
- 68.Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- 69.Lee C, Piazza F, Brutsaert S, Valens J, Strehlow I, Jarosinski M, Saris C, Schindler C. Characterization of the Stat5 protease. J Biol Chem. 1999;274:26767. doi: 10.1074/jbc.274.38.26767. [DOI] [PubMed] [Google Scholar]
- 70.Piazza F, Valens J, Lagasse E, Schindler C. Myeloid differentiation of FdCP1 cells is dependent on Stat5 processing. Blood. 2000;96:1358. [PubMed] [Google Scholar]
- 71.Xia Z, Salzler RR, Kunz DP, Baer MR, Kazim L, Baumann H, Wetzler M. A novel serine-dependent proteolytic activity is responsible for truncated signal transducer and activator of transcription proteins in acute myeloid leukemia blasts. Cancer Res. 2001;61:1747. [PubMed] [Google Scholar]
- 72.Chakraborty A, Tweardy DJ. Granulocyte colony-stimulating factor activates a 72-kDa isoform of STAT3 in human neutrophils. J Leukoc Biol. 1998;64:675. doi: 10.1002/jlb.64.5.675. [DOI] [PubMed] [Google Scholar]
- 73.Epling-Burnette PK, Garcia R, Bai F, Ismail S, Loughran TP, Djeu JY, Jove R, Wei S. Carboxy-terminal truncated STAT5 is induced by interleukin-2 and GM-CSF in human neutrophils. Cell Immunol. 2002;217:1. doi: 10.1016/s0008-8749(02)00522-1. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki K, Nakajima H, Kagami S, et al. Proteolytic processing of Stat6 signaling in mast cells as a negative regulatory mechanism. J Exp Med. 2002;196:27. doi: 10.1084/jem.20011682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hendry L, John S. Regulation of STAT signalling by proteolytic processing. Eur J Biochem. 2004;271:4613–20. doi: 10.1111/j.1432-1033.2004.04424.x. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki K, Nakajima H, Ikeda K, Tamachi T, Hiwasa T, Saito Y, Iwamoto I. Stat6-protease but not Stat5-protease is inhibited by an elastase inhibitor ONO-5046. Biochem Biophys Res Commun. 2003;309:768. doi: 10.1016/j.bbrc.2003.08.067. [DOI] [PubMed] [Google Scholar]
- 77.Oda A, Wakao H, Fujita H. Calpain is a signal transducer and activator of transcription (STAT)3 and STAT5 protease. Blood. 2002;99:1850. doi: 10.1182/blood.v99.5.1850. [DOI] [PubMed] [Google Scholar]
- 78.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945. [PubMed] [Google Scholar]
- 80.Kelly J, Spolski R, Imada K, Bollenbacher J, Lee S, Leonard WJ. A role for Stat5 in CD8+ T cell homeostasis. J Immunol. 2003;170:210. doi: 10.4049/jimmunol.170.1.210. [DOI] [PubMed] [Google Scholar]
- 81.Kelly JA, Spolski R, Kovanen PE, et al. Stat5 synergizes with T cell receptor/antigen stimulation in the development of lymphoblastic lymphoma. J Exp Med. 2003;198:79. doi: 10.1084/jem.20021548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willemze R, Meijer CJ. EORTC classification for primary cutaneous lymphomas: the best guide to good clinical management. European Organization for Research and Treatment of Cancer. Am J Dermatopathol. 1999;21:265. doi: 10.1097/00000372-199906000-00011. [DOI] [PubMed] [Google Scholar]
- 83.Foss F. Mycosis fungoides and the Sezary syndrome. Curr Opin Oncol. 2004;16:421. doi: 10.1097/00001622-200409000-00002. [DOI] [PubMed] [Google Scholar]
- 84.Edelson RL. Cutaneous T cell lymphoma: the helping hand of dendritic cells. Ann N Y Acad Sci. 2001;941:1. [PubMed] [Google Scholar]
- 85.Kari L, Loboda A, Nebozhyn M, et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med. 2003;197:1477. doi: 10.1084/jem.20021726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Showe LC, Fox FE, Williams D, Au K, Niu Z, Rook AH. Depressed IL-12-mediated signal transduction in T cells from patients with Sezary syndrome is associated with the absence of IL-12 receptor beta 2 mRNA and highly reduced levels of STAT4. J Immunol. 1999;163:4073. [PubMed] [Google Scholar]
- 87.Zhang Q, Nowak I, Vonderheid EC, Rook AH, Kadin ME, Nowell PC, Shaw LM, Wasik MA. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci USA. 1996;93:9148. doi: 10.1073/pnas.93.17.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Waldmann TA. T-cell receptors for cytokines: targets for immunotherapy of leukemia/lymphoma. Ann Oncol. 2000;11(Suppl. 1):101. [PubMed] [Google Scholar]
- 89.Eriksen KW, Kaltoft K, Mikkelsen G, et al. Constitutive STAT3-activation in Sezary syndrome. tyrphostin AG490 inhibits STAT3-activation, interleukin-2 receptor expression and growth of leukemic Sezary cells. Leukemia. 2001;15:787. doi: 10.1038/sj.leu.2402093. [DOI] [PubMed] [Google Scholar]
- 90.Nielsen M, Kaltoft K, Nordahl M, et al. Constitutive activation of a slowly migrating isoform of Stat3 in mycosis fungoides: tyrphostin AG490 inhibits Stat3 activation and growth of mycosis fungoides tumor cell lines. Proc Natl Acad Sci USA. 1997;94:6764. doi: 10.1073/pnas.94.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Migone TS, Lin JX, Cereseto A, Mulloy JC, O'Shea JJ, Franchini G, Leonard WJ. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 92.Sommer VH, Clemmensen OJ, Nielsen O, et al. In vivo activation of STAT3 in cutaneous T-cell lymphoma. Evidence for an antiapoptotic function of STAT3. Leukemia. 2004;18:1288. doi: 10.1038/sj.leu.2403385. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Q, Raghunath PN, Vonderheid E, Odum N, Wasik MA. Lack of phosphotyrosine phosphatase SHP-1 expression in malignant T-cell lymphoma cells results from methylation of the SHP-1 promoter. Am J Pathol. 2000;157:1137. doi: 10.1016/S0002-9440(10)64629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leon F, Cespon C, Franco A, et al. SHP-1 expression in peripheral T cells from patients with Sezary syndrome and in the T cell line HUT-78: implications in JAK3-mediated signaling. Leukemia. 2002;16:1470. doi: 10.1038/sj.leu.2402546. [DOI] [PubMed] [Google Scholar]
- 95.Lundin Brockdorff J, Woetmann A, Mustelin T, Kaltoft K, Zhang Q, Wasik MA, Ropke C, Odum N. SHP2 regulates IL-2 induced MAPK activation, but not Stat3 or Stat5 tyrosine phosphorylation, in cutaneous T cell lymphoma cells. Cytokine. 2002;20:141. doi: 10.1006/cyto.2002.1986. [DOI] [PubMed] [Google Scholar]
- 96.Nielsen M, Nissen MH, Gerwien J, et al. Spontaneous interleukin-5 production in cutaneous T-cell lymphoma lines is mediated by constitutively activated Stat3. Blood. 2002;99:973. doi: 10.1182/blood.v99.3.973. [DOI] [PubMed] [Google Scholar]
- 97.Berger CL, Hanlon D, Kanada D, et al. The growth of cutaneous T-cell lymphoma is stimulated by immature dendritic cells. Blood. 2002;99:2929. [PubMed] [Google Scholar]
- 98.Brender C, Nielsen M, Kaltoft K, Mikkelsen G, Zhang Q, Wasik M, Billestrup N, Odum N. STAT3-mediated constitutive expression of SOCS-3 in cutaneous T-cell lymphoma. Blood. 2001;97:1056. doi: 10.1182/blood.v97.4.1056. [DOI] [PubMed] [Google Scholar]
- 99.Egwuagu CE, Yu CR, Zhang M, Mahdi RM, Kim SJ, Gery I. Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: implications for Th cell lineage commitment and maintenance. J Immunol. 2002;168:3181. doi: 10.4049/jimmunol.168.7.3181. [DOI] [PubMed] [Google Scholar]
- 100.Yamamoto K, Yamaguchi M, Miyasaka N, Miura O. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor beta2 subunit. Biochem Biophys Res Commun. 2003;310:1188. doi: 10.1016/j.bbrc.2003.09.140. [DOI] [PubMed] [Google Scholar]
- 101.Matsumoto A, Seki Y, Watanabe R, et al. A role of suppressor of cytokine signaling 3 (SOCS3/CIS3/SSI3) in CD28-mediated interleukin 2 production. J Exp Med. 2003;197:425. doi: 10.1084/jem.20020939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seki Y, Inoue H, Nagata N, et al. SOCS-3 regulates onset and maintenance of T (H) 2-mediated allergic responses. Nat Med. 2003;9:1047. doi: 10.1038/nm896. [DOI] [PubMed] [Google Scholar]
- 103.Mitchell TJ, Whittaker SJ, John S. Dysregulated expression of COOH-terminally truncated Stat5 and loss of IL2-inducible Stat5-dependent gene expression in Sezary Syndrome. Cancer Res. 2003;63:9048. [PubMed] [Google Scholar]
- 104.Van Parijs L, Refaeli Y, Lord JD, Nelson BH, Abbas AK, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 105.Nelson BH. IL-2, regulatory T cells, and tolerance. J Immunol. 2004;172:3983. doi: 10.4049/jimmunol.172.7.3983. [DOI] [PubMed] [Google Scholar]
- 106.Meech SJ, Edelson R, Walsh P, Norris DA, Duke RC. Reversible resistance to apoptosis in cutaneous T cell lymphoma. Ann N Y Acad Sci. 2001;941:46. doi: 10.1111/j.1749-6632.2001.tb03710.x. [DOI] [PubMed] [Google Scholar]
- 107.Rook AH, Gottlieb SL, Wolfe JT, Vowels BR, Sood SS, Niu Z, Lessin SR, Fox FE. Pathogenesis of cutaneous T-cell lymphoma. implications for the use of recombinant cytokines and photopheresis. Clin Exp Immunol. 1997;107(Suppl. 1):16. [PubMed] [Google Scholar]
- 108.Kadin ME, Carpenter C. Systemic and primary cutaneous anaplastic large cell lymphomas. Semin Hematol. 2003;40:244. doi: 10.1016/s0037-1963(03)00138-0. [DOI] [PubMed] [Google Scholar]
- 109.Wasik MA. Expression of anaplastic lymphoma kinase in non-Hodgkin's lymphomas and other malignant neoplasms. Biological, diagnostic, and clinical implications. Am J Clin Pathol. 2002;118(Suppl. S81) doi: 10.1309/YCRV-7H6U-6E8B-95GU. [DOI] [PubMed] [Google Scholar]
- 110.Miething C, Grundler R, Fend F, et al. The oncogenic fusion protein nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) induces two distinct malignant phenotypes in a murine retroviral transplantation model. Oncogene. 2003;22:4642. doi: 10.1038/sj.onc.1206575. [DOI] [PubMed] [Google Scholar]
- 111.Cussac D, Greenland C, Roche S, et al. Nucleophosmin-anaplastic lymphoma kinase of anaplastic large-cell lymphoma recruits, activates, and uses pp60c-src to mediate its mitogenicity. Blood. 2004;103:1464. doi: 10.1182/blood-2003-04-1038. [DOI] [PubMed] [Google Scholar]
- 112.Bai RY, Ouyang T, Miething C, Morris SW, Peschel C, Duyster J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood. 2000;96:4319. [PubMed] [Google Scholar]
- 113.Bai RY, Dieter P, Peschel C, Morris SW, Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-gamma to mediate its mitogenicity. Mol Cell Biol. 1998;18:6951. doi: 10.1128/mcb.18.12.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zamo A, Chiarle R, Piva R, Howes J, Fan Y, Chilosi M, Levy DE, Inghirami G. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene. 2002;21:1038. doi: 10.1038/sj.onc.1205152. [DOI] [PubMed] [Google Scholar]
- 115.Zhang Q, Raghunath PN, Xue L, et al. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive T/null-cell lymphoma. J Immunol. 2002;168:466. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- 116.Nieborowska-Skorska M, Slupianek A, Xue L, et al. Role of signal transducer and activator of transcription 5 in nucleophosmin/anaplastic lymphoma kinase-mediated malignant transformation of lymphoid cells. Cancer Res. 2001;61:6517. [PubMed] [Google Scholar]
- 117.Woetmann A, Nielsen M, Christensen ST, et al. Inhibition of protein phosphatase 2A induces serine/threonine phosphorylation, subcellular redistribution, and functional inhibition of STAT3. Proc Natl Acad Sci USA. 1999;96:10620. doi: 10.1073/pnas.96.19.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Khoury JD, Medeiros LJ, Rassidakis GZ, et al. Differential expression and clinical significance of tyrosine-phosphorylated STAT3 in ALK+ and ALK- anaplastic large cell lymphoma. Clin Cancer Res. 2003;9:3692. [PubMed] [Google Scholar]
- 119.Amin HM, McDonnell TJ, Ma Y, et al. Selective inhibition of STAT3 induces apoptosis and G (1) cell cycle arrest in ALK-positive anaplastic large cell lymphoma. Oncogene. 2004;23:5426. doi: 10.1038/sj.onc.1207703. [DOI] [PubMed] [Google Scholar]
- 120.Schlette EJ, Medeiros LJ, Goy A, Lai R, Rassidakis GZ. Survivin expression predicts poorer prognosis in anaplastic large-cell lymphoma. J Clin Oncol. 2004;22:1682. doi: 10.1200/JCO.2004.10.172. [DOI] [PubMed] [Google Scholar]
- 121.Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, Jove R, Yu H. Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res. 1999;59:5059. [PubMed] [Google Scholar]
- 122.Pichardo DA, Querfeld C, Guitart J, Kuzel TM, Rosen ST. Cutaneous T-cell lymphoma: a paradigm for biological therapies. Leuk Lymphoma. 2004;45:1755. doi: 10.1080/10428190410001693560. [DOI] [PubMed] [Google Scholar]
- 123.Piekarz RL, Robey RW, Zhan Z, Kayastha G, Sayah A, Abdeldaim AH, Torrico S, Bates SE. T-cell lymphoma as a model for the use of histone deacetylase inhibitors in cancer therapy: impact of depsipeptide on molecular markers, therapeutic targets, and mechanisms of resistance. Blood. 2004;103:4636. doi: 10.1182/blood-2003-09-3068. [DOI] [PubMed] [Google Scholar]
- 124.Piekarz RL, Robey R, Sandor V, et al. Inhibitor of histone deacetylation, depsipeptide ( FR901228), in the treatment of peripheral and cutaneous T-cell lymphoma: a case report. Blood. 2001;98:2865. doi: 10.1182/blood.v98.9.2865. [DOI] [PubMed] [Google Scholar]