Abstract
The assembly of major histocompatibility complex (MHC) class I molecules with their peptide ligands in the endoplasmic reticulum (ER) requires the assistance of many proteins that form a multimolecular assemblage termed the ‘peptide-loading complex’. Tapasin is the central stabilizer of this complex, which also includes the transporter associated with antigen processing (TAP), MHC class I molecules, the ER chaperone, calreticulin, and the thiol-oxidoreductase ERp57. In the present report, we investigated the requirements of these interactions for tapasin protein stability and MHC class I dissociation from the peptide-loading complex. We established that tapasin is stable in the absence of either TAP or MHC class I interaction. In the absence of TAP, tapasin interaction with MHC class I molecules is long-lived and results in the sequestration of existing tapasin molecules. In contrast, in TAP-sufficient cells, tapasin is re-utilized to interact with and facilitate the assembly of many MHC class I molecules sequentially. Furthermore, chemical cross-linking has been utilized to characterize the interactions within this complex. We demonstrate that tapasin and MHC class I molecules exist in a 1: 1 complex without evidence of higher-order tapasin multimers. Together these studies shed light on the tapasin protein life cycle and how it functions in MHC class I assembly with peptide for presentation to CD8+ T cells.
Keywords: MHC, protein stability, stoichiometry, TAP, tapasin
Introduction
Tapasin is a 48 000 molecular weight (MW), type I transmembrane glycoprotein that is essential for the proper assembly of major histocompatibility complex (MHC) class I molecules with their peptide ligands in the endoplasmic reticulum (ER).1 This process of assembling MHC class I heavy chains, light chains [β2-microglobulin (β2m)] and peptide results in an appropriately folded molecule that is competent for transport from the ER to the cell surface and recognition by CD8+ T cells.
Peptide ligands are generated in the cytosol by the action of proteasomes and trimming peptidases.2, 3 Peptides are then translocated into the ER by a heterodimeric member of the ABC family of transporters, termed the transporter associated with antigen processing (TAP).4, 5 In the ER, MHC class I heavy chains are synthesized and interact rapidly with the chaperone calnexin.6, 7 MHC class I heavy chains then dissociate from calnexin and interact with β2m, calreticulin, ERp57 and a preformed complex of TAP and tapasin.8, 9 This peptide-loading complex comprising MHC class I heavy chains, β2m, calreticulin, ERp57, TAP and tapasin is essential for the efficient assembly of MHC class I molecules.1, 10 Biochemical purification has revealed that four MHC class I molecules, calreticulin molecules and tapasin molecules bind to each TAP transporter.1
Each component in the peptide-loading complex has specific functions in assisting MHC class I assembly with peptide. TAP translocates peptides into the ER and thus represents the portal of entry of peptides.11, 12 Tapasin recruits MHC class I molecules to the peptide entry portal by interactions with both TAP and MHC class I molecules1, 13, 14 The soluble ER chaperones – calreticulin and ERp57 – assist in the folding of many proteins destined for the cell surface or for secretion.15, 16 Calreticulin specifically interacts with monoglucosylated N-linked glycans of nascent ER glycoproteins and assists in the folding of MHC class I molecules.15, 17 ERp57 is a thioredoxin family member containing two CxxC motifs. Thioredoxin family members use these motifs to assist in the formation and dissolution of disulfide bonds within nascent proteins to achieve a properly folded final molecule.18 ERp57 is known to form disulfide intermediates with target proteins when assisting in their folding.19 Within the peptide-loading complex we have previously demonstrated that ERp57 and tapasin form a disulfide-linked intermediate.20 However, when the tapasin–ERp57 disulfide intermediate is abolished, the redox status of MHC class I molecules is altered, indicating that tapasin and ERp57 may work together to achieve appropriately folded and oxidized MHC class I molecules.20 Tapasin also has multiple additional roles in facilitating MHC class I assembly.
Tapasin stabilizes the peptide-loading complex by interacting with all proteins in the complex.21 The N-terminal domain of tapasin is necessary for interaction with MHC class I molecules, calreticulin and ERp57, while the C-terminal domain of tapasin is sufficient for interaction with TAP.21 By having discrete binding sites at opposite ends of the molecule, tapasin bridges TAP and MHC class I molecules and stabilizes the entire peptide-loading complex. In addition to its bridging function, tapasin interaction with MHC class I molecules results in the formation of appropriate MHC class I–peptide complexes that are transported to the cell surface.1, 22 Thus, in the presence of tapasin, high levels of MHC class I–peptide complexes are observed at the cell surface and these complexes are stable, persisting at the cell surface for several hours.23 As tapasin aids in class I assembly and folding, it is considered a ‘chaperone’ for MHC class I molecules. Overall, cells containing stable and functional tapasin exhibit enhanced MHC class I levels owing to enhanced MHC class I–peptide complex formation and stability.24
Tapasin also affects TAP stability. Tapasin enhances TAP1 levels, resulting in enhanced TAP1/2-mediated peptide translocation into the ER.21, 22, 25 Therefore, by promoting heterodimer assembly and/or stabilizing the peptide-binding site on TAP, tapasin is thought to act as a ‘chaperone’ for TAP.26 The presence of tapasin therefore leads to enhanced overall peptide levels in the ER for MHC class I assembly.
Biochemical purification of the peptide-loading complex suggests that each TAP1/2 heterodimer associates with four tapasin molecules, four MHC class I molecules and four calreticulin molecules.1 This 4: 1 stoichiometry may result from the formation of: a tapasin tetramer that interacts with a single binding site on TAP; tapasin dimers that associate with TAP at two independent sites; or tapasin monomers that each associate with the TAP heterodimer at four sites. In the present study we tested for the existence of tapasin dimers or tetramers by using chemical cross-linking.
Tapasin interaction with both TAP and MHC class I molecules results in stabilization and promotion of assembly. One of the objectives of the current study was to determine how these interactions affect tapasin stability. We undertook these tests by examining cells deficient for either MHC class I interaction or TAP interaction. In our analysis of the kinetics of tapasin interactions in these cells, we present data arguing that, in wild-type cells, each tapasin molecule is reused to assist the folding of multiple MHC class I molecules sequentially. However, in the absence of a peptide supply, tapasin is effectively permanently bound and sequestered by MHC class I molecules. As previous purification of the TAP complex revealed a stoichiometry of four tapasin–MHC class I–calreticulin molecules for every TAP transporter, we have also taken advantage of deficient cells and chemical cross-linking to examine the question of whether tapasin can form multimers to achieve the 4: 1 tapasin to TAP stoichiometry.
Materials and methods
Cell lines, antibodies and reagents
The β2m-deficient Burkitt's lymphoma Daudi and its restored counterpart, Daudi.β2m, have been described previously.8 The human B-lymphoblastoid cell line, 45.1, is the parent of the TAP-deficient mutant line, .174.27
Antisera recognizing the N- and C-terminal peptides of tapasin (R.gp48N and R.gp48C, respectively) have been described previously.21 The TAP1-specific antiserum, R.RING4C, recognizes the C-terminal peptide of TAP1.13 Rabbit antiserum specific for calnexin was obtained from Dr Ari Helenius (Swiss Feberal Institute of Technology, Zurich, Switzerland). The calreticulin-specific rabbit antiserum was obtained from Affinity Bioreagents (Golden, CO).21 Rabbit antiserum, R.ERp57, generated against recombinant ERp57, has been described previously.20 The cross-linker, ethylene glycol bis[succinimidyl succinate] (EGS) was purchased from Pierce Biotechnology (Rockford, IL).
Western blot analysis
Cells were extracted in lysis buffer containing Triton-X-100. Postnuclear supernatants were subject to twofold serial dilutions and denatured by using Laemmli sample buffer. After heating, the samples were applied to sodium dodecyl sulphate (SDS) polyacrylamide gels for separation. Proteins were transferred to Immobilon-P membranes. Non-specific binding sites on the membranes were blocked by using 3% (w/v) skim milk in phosphate-buffered saline (PBS) containing 0·2% (v/v) Tween 20. Primary antibodies were incubated in blocking buffer followed by washing and incubation with alkaline phosphatase secondary antibody conjugate. VistraECF substrate (Amersham, Piscataway, NJ) was incubated with the blot before scanning by using a Fluorimager instrument (Molecular Dynamics, Palo Alto, CA). The fluorescence intensity was analysed by using imagequant software (Molecular Dynamics).
Radiolabelling, immunoprecipitation and cross-linking
Prior to labelling, cells were washed in PBS and methionine-starved in Dulbecco's modified Eagle's minimal essential medium (DMEM), lacking methionine, for 30–60 min at 37°. [35S]Methionine was added for the desired incubation time. For pulse-chase studies, after 30 min of labelling, complete media containing excess methionine was added to the cells and aliquots were removed at various time-points for analysis. Following labelling (and/or chase), cells were washed once in PBS and extracted in 0·01 m Tris, pH 7·4, containing 1% (v/v) Triton-X-100, 0·15 m NaCl, and the protease inhibitors, phenylmethylsulphonyl fluoride (PMSF) and N-ethylmaleimide, as previously described.21 Postnuclear supernatants were precleared by using Protein G–Sepharose. Precleared supernatants were then subjected to immunoprecipitation with antibody and Protein G–Sepharose. Bound proteins were eluted and reimmunoprecipitated as indicated in the figure legends.
Proteins were cross-linked by the addition of EGS (0·4 mm final concentration) to lysis buffer [1% (v/v) Triton-X-100, 0·13 m NaCl, 0·02 m bicine, pH 8·0] immediately prior to solubilization of the cells. After incubation on ice for 30 min to allow cross-linking and extraction, the cross-linker was quenched by using glycine (10 mm final concentration), and postnuclear supernatants were precleared and immunoprecipitated as described above.
Results
Tapasin steady-state levels are unaffected by interactions with MHC class I or with TAP
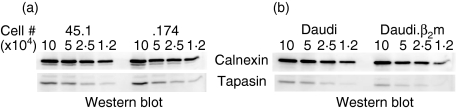
Tapasin enhances the protein levels of both TAP and MHC class I; however, the effect of these interactions on tapasin protein levels is unclear. To determine the effects of TAP interaction or of MHC class I interaction on tapasin levels, we examined the tapasin steady-state protein levels in the absence of each of these interactions. By using fluorescence-based quantitative Western blotting, we found that the amount of total tapasin protein is similar in the TAP-deficient .174 cell line and its wild-type counterpart, 45.1 (Fig. 1a). Identical levels of calnexin were also found, demonstrating that equivalent cell lysates were applied to the gel. These data indicate that the absence of TAP interaction does not affect overall tapasin expression.
Figure 1.
Steady-state levels of tapasin in β2-microglobulin (β2m)-deficient Daudi cells and transporter associated with antigen processing (TAP)-deficient .174 cells. Extracts of 45.1 cells and of TAP-deficient .174 cells. (a), or of β2m-deficient Daudi and of Daudi.β2m (β2m-restored) cells (b), were serially diluted, corresponding to the number of cell equivalents indicated. Proteins were transferred to membranes and probed for calnexin or TAP1.
In Daudi cells (which are deficient for β2m) tapasin does not interact appreciably with MHC class I molecules or calreticulin.8 Therefore, to examine the effect of MHC class I interaction on tapasin stability, we used β2m-deficient Daudi cells and their wild-type counterpart, Daudi cells transfected with β2m (Daudi.β2m). The tapasin protein levels in Daudi and Daudi.β2m were found to be identical, indicating that the MHC class I (and calreticulin) interaction that occurs in Daudi.β2m does not enhance tapasin levels (Fig. 1b). Therefore, the absence of either TAP or MHC class I interaction does not alter the steady-state protein levels of tapasin.
Tapasin remains associated with class I molecules in .174 cells, resulting in fewer tapasin molecules being available for association with newly synthesized class I molecules
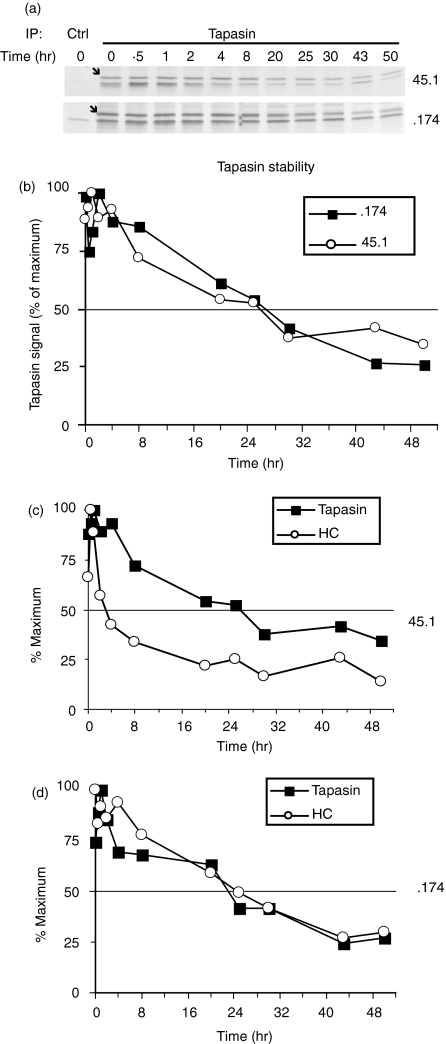
To examine the influence of TAP on the interaction between tapasin and MHC class I molecules, pulse–chase analysis of tapasin immunoprecipitates was performed in 45.1 and .174 cells (Fig. 2). Consistent with the identical overall levels of tapasin observed in these two cell types, tapasin stability is also identical in 45.1 and .174 cells (Fig. 2a,2b). Tapasin-associated MHC class I heavy chains are observed as the band migrating slightly faster than tapasin (Fig. 2a, lower band). MHC class I heavy chains dissociate from tapasin with a half-time of 4 hr in 45.1 cells, but show prolonged association in .174 cells (half-time 24 hr, compare the open circles of Fig. 2c with those of Fig. 2d). Therefore, tapasin and MHC class I heavy chains appear to be permanently associated and degraded with identical kinetics in TAP-deficient .174 cells.
Figure 2.
Kinetics of tapasin–major histocompatibility complex (MHC) class I interactions in the presence or absence of transporter associated with antigen processing (TAP). Pulse–chase analysis of 45.1 and of TAP-deficient .174 cells was performed by using the tapasin-specific antibody R.gp48C. Eluted proteins were electrophoresed on 12% polyacrylamide gels (a). Quantification of the tapasin (bands marked with an arrow) and MHC class I bands from (a) are shown in (b), (c) and (d). (b) Comparison of tapasin stability in 45.1 (open circles) and .174 cells (closed squares). (c) and (d) Analyses of 45.1 and .174 cells, respectively, for tapasin (closed squares) and associated heavy chains (HC, open circles).
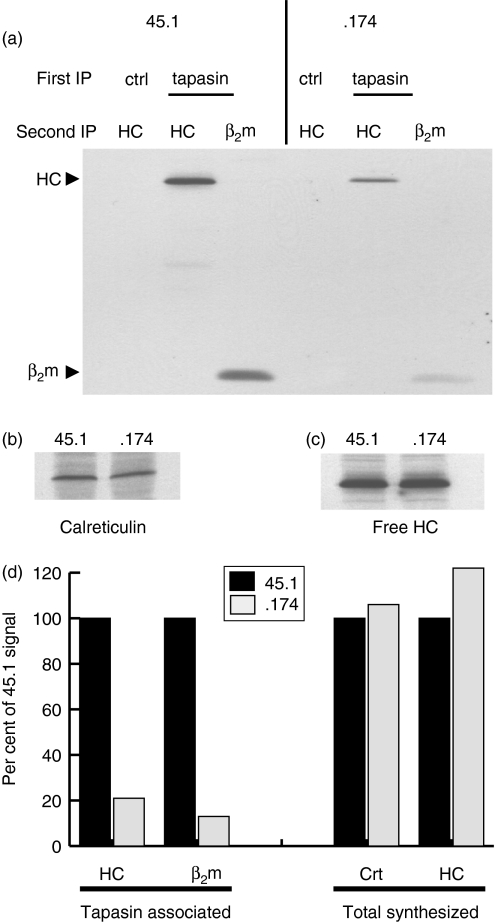
This prolonged interaction in .174 cells allowed us to test whether, in wild-type cells, each tapasin molecule is reutilized to assist in the folding of many MHC class I molecules sequentially. We predicted that, if each tapasin molecule is reused to assist multiple MHC class I molecules sequentially, we would detect a higher level of newly synthesized MHC class I molecules in association with tapasin in 45.1 cells compared to tapasin in .174 cells (Fig. 3). This is because, in .174 cells, all pre-existing tapasin molecules are essentially ‘permanently bound’ to previously synthesized (non-radioactive) MHC class I molecules and are not available for binding newly synthesized (radioactively labelled) MHC class I molecules. Therefore, in .174 cells, the only tapasin molecules available for MHC class I interaction are the newly synthesized tapasin molecules. 45.1 cells contain this newly synthesized pool of tapasin for MHC class I binding, but these cells also contain pre-existing tapasin molecules that have assisted in the folding of a previous MHC class I molecule and permitted its export from the ER. Therefore, these two pools of tapasin molecules in 45.1 cells provide a larger overall pool with which newly synthesized MHC class I molecules can associate. Indeed we detected four- to fivefold more newly synthesized MHC class I molecules associated with tapasin in 45.1 cells than in .174 cells (Fig. 3a,3d).
Figure 3.
Reduced available tapasin for class I interaction in transporter associated with antigen processing (TAP)-deficient .174 cells. Cells were metabolically labelled for 90–120 min, then lysed in digitonin-containing buffer to preserve the major histocompatibility complex (MHC) class I interaction with TAP and tapasin. (a) Bound proteins after control (ctrl) or tapasin immunoprecipitations (IP) were eluted and reimmunoprecipitated by using 3B10.7 [anti-class I heavy chain (HC)] or rabbit anti-β2-microglobulin (β2m) serum. Sodium dodecyl sulphate–polyacrylamide gels of the eluted samples were imaged by autoradiography and phosphorimager analysis for quantification, as shown in (d). Calreticulin (b) and MHC class I heavy chains (c) were immunoprecipitated from the same lysates to demonstrate equivalent protein synthesis levels and free heavy chain pools. Quantification in (d) shows the reduced MHC class I and β2m association with tapasin in .174 cells, but calreticulin and free heavy chain pools are similar in .174 and 45.1 cells.
This reduced level of MHC class I associated with tapasin in .174 cells is not a result of lower tapasin levels because 45.1 and .174 have similar synthesis rates, degradation rates and steady-state levels (Figs 1a and 2). Nor is this reduced MHC class I interaction the result of a smaller pool of newly synthesized class I heavy chains because these two cell lines have similar levels of newly synthesized free heavy chains (Fig. 3c). The levels of newly synthesized calreticulin were also identical in 45.1 and .174 cells (Fig. 3b). Therefore, in the absence of TAP, MHC class I molecules remain associated with tapasin for extended time-periods and both molecules degrade with similar kinetics. In contrast, the availability of TAP-translocated peptides in 45.1 appears to induce the dissociation of tapasin and MHC class I molecules, making tapasin available for additional interactions. Together, these data indicate that in wild-type cells a single tapasin molecule can be reused, catalytically facilitating the loading of many MHC class I molecules sequentially.
Tapasin and MHC class I molecules interact in a 1: 1 stoichiometry, without evidence of higher-order tapasin multimers
Purification of the TAP complex from wild-type cells has indicated a stoichiometry of four tapasin, MHC class I and calreticulin molecules for every TAP molecule. We utilized chemical cross-linking to verify this stoichiometry and to determine whether tapasin is capable of forming multimers when it is not associated with TAP. In Daudi.β2m cells, where the complete TAP–tapasin–MHC class I–CRT–ERp57 loading complex exists, the detergent Triton-X-100 dissociates tapasin–MHC class I complexes from TAP. We reasoned that by using chemical cross-linkers in Triton-X-100 detergent extracts, the stoichiometry of tapasin to class I can be examined in the absence of the co-ordinating effect of TAP. As class I interaction with tapasin does not occur in Daudi cells, Daudi cells were used as a control to determine which higher order complexes of tapasin were dependent on an interaction with MHC class I.
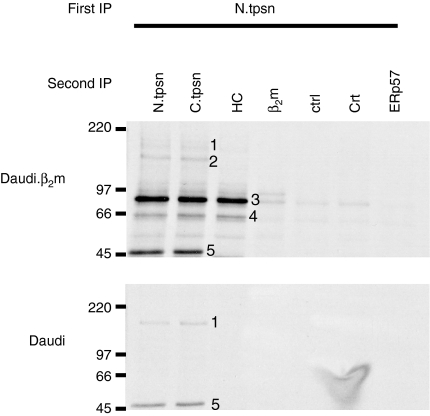
Daudi.β2m cells were radiolabelled, cross-linked with EGS during Triton extraction and then the postnuclear supernatants were subject to immunoprecipitation of tapasin via its N terminus. Bound proteins were eluted, and reimmunoprecipitation by using C-terminal tapasin-specific antisera revealed five different protein species from Daudi.β2m cells (numbered 1–5 in Fig. 4, upper panel). These protein species ranged from 46 000 to 170 000 in apparent MW. The same procedure in Daudi cells revealed only two of these species, at 170 000 and 46 000 MW (species number 1 and species number 5, respectively, Fig. 4, lower panel).
Figure 4.
Cross-linking reveals a 1: 1 tapasin: major histocompatibility complex (MHC) class I stoichiometry. Daudi and Daudi.β2m (β2m-restored) cells were metabolically labelled for 1–2 hr. Proteins were cross-linked by using ethylene glycol bis[succinimidyl succinate] (EGS) during lysis in Triton-X-100-containing buffer. (a) After quenching the remaining cross-linker with excess glycine, precleared lysates were then subjected to tapasin immunoprecipitation (IP) using antisera specific for the N-terminus (N.tpsn). Tapasin and bound proteins were eluted by boiling in the presence of sodium dodecyl sulphate (SDS) and reimmunoprecipitated for using tapasin antisera specific for the N-terminal (N.tpsn) or C-terminal (C.tpsn) peptides, 3B10.7 (HC), β2m-specific antisera (β2m), control (ctrl), calreticulin-specific antisera (Crt), or ERp57-specific antisera (ERp57). Second immunoprecipitates were eluted in reducing Laemmli sample buffer and separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). Five radiolabelled protein species (numbered 1–5) are observed in Daudi.β2m cells, but only two of these species (numbers 1 and 5) are observed in Daudi cells. β2m, β2-microglobulin.
Species number 5, which migrates at 46 000 MW, represents free tapasin that has not been cross-linked to any other protein. Species number 1, at 170 000 MW, is only observed with tapasin-specific antibodies. This species does not include MHC class I heavy chains as it is observed in Daudi cells, which do not have an appreciable tapasin–MHC class I interaction, and it is not immunoprecipitated by the heavy chain-specific antibody, 3B10.7. Although it is possible that the 170 000 -MW species represents tapasin trimers or tetramers, we consider this unlikely because we would expect cross-linking efficiencies to be incomplete, resulting in the detection of each of the intermediate species (i.e. dimers, trimers and tetramers) rather than only the final multimerized product. Therefore, we suggest that species number 1, at 170 000 MW, represents tapasin cross-linked to some currently unidentified protein(s) whose cumulative MW is predicted to be ≈ 120 000.
Given the TAP: tapasin stoichiometry of 1: 4, if the formation of tapasin dimers or tetramers occurred prior to the association with TAP, we might expect to observe these complexes in Triton extracts of Daudi cells as Triton breaks the interaction between tapasin and TAP. However, EGS cross-linking of Daudi cell lysates reveals no higher-order tapasin multimers (Fig. 4, lower panel), indicating that interaction with TAP probably co-ordinates the assembly of four individual tapasin molecules.
Species 2, 3 and 4 are dependent on MHC class I interaction as they appear in Daudi.β2m cells, but not in Daudi cells (Fig. 4: compare the upper panel with the lower panel). Cross-linked species 2 migrates at ≈ 130 000 MW and contains tapasin. However, the identities of the remaining proteins in this complex are currently unclear. Cross-linked species 3 and 4, at 90 000 and 66 000 MW, contain three epitopes: an N-terminal tapasin epitope; a C-terminal tapasin epitope; and an MHC class I heavy chain epitope. Species 3 migrates at the expected MW (≈ 90 000) for a tapasin–MHC class I complex (46 000 + 44 000) in a 1: 1 ratio. Species 4 exhibits a faster mobility (at 66 000 MW), but contains the same tapasin and MHC class I epitopes. Therefore, we suggest that species 4 is also a tapasin–MHC class I complex present in a 1: 1 ratio, but whose conformation is altered such that it migrates faster in an SDS–polyacrylamide gel. Together these data argue that tapasin and MHC class I molecules exist in a 1: 1 stoichiometry and that at least two different conformers of this complex can be detected.
Discussion
The interactions within the MHC class I loading complex are intricate and critical for the efficient assembly of MHC class I molecules with their peptide ligands. Tapasin plays an essential role in the stabilization of this complex. In the current report, we have presented evidence arguing that the stability of tapasin itself is not altered by interaction with MHC class I molecules or TAP. Strikingly, we find that in the absence of peptide, newly synthesized MHC class I molecules and newly synthesized tapasin molecules interact stably with each other for several hours. In wild-type cells, however, MHC class I molecules dissociate from tapasin, and the same tapasin molecule is recycled to assist the folding of another MHC class I molecule. We also utilized chemical cross-linking to demonstrate that each tapasin molecule interacts with a single MHC class I molecule, and this complex can be detected in two different conformations.
Given that tapasin promotes the stability of both TAP and MHC class I molecules, we examined whether these interactions also had a reciprocal effect on tapasin stability. In the absence of either MHC class I interaction or TAP interaction, tapasin levels and/or stability are unaffected. However, in each of these deficient cell types, tapasin still interacts with the other components that may promote its stability. For example, in TAP-deficient .174 cells, tapasin still interacts (quite stably) with MHC class I molecules, and this interaction may promote tapasin stability. In Daudi cells, where the interaction with MHC class I molecules is not appreciable, tapasin interacts with TAP, which may promote tapasin stability. Therefore, tapasin stability may be aided by multiple interactions, any one of which may be sufficient. Further studies of tapasin stability in the absence of all interacting partners are required to address these issues.
We show here that tapasin and MHC class I heavy chains interact in a prolonged and stable manner in TAP-deficient .174 cells. These observations are consistent with reports that tapasin retains MHC class I heavy chains in the ER of invertebrate cells.28 Although the MHC class I molecules in .174 cells do not bind peptide, they are more stable compared to cells lacking tapasin.29 We show that this stability is probably caused by prolonged interaction with tapasin until both molecules are degraded.
By comparing the kinetics of the MHC class I interaction with tapasin, we demonstrate that the stable interaction in TAP-deficient .174 cells sequesters all previously synthesized tapasin molecules. However, in wild-type cells, previously synthesized tapasin molecules that have already assisted in the folding and export of at least one MHC class I molecule, are available for interaction with newly synthesized MHC class I molecules.
Together these data demonstrate that tapasin is limiting for association with MHC class I heavy chains, and that MHC class I heavy chains are synthesized in excess. Therefore, tapasin appears to be the limiting factor controlling surface expression of MHC class I molecules in these B-cell lines. If this were true in other cell types, we would predict that surface MHC class I expression could be directly correlated to tapasin protein levels.
Previous biochemical purification of TAP and its associated molecules has revealed that four MHC class I molecules and four tapasin molecules were present for each TAP molecule. Therefore, it appears that each TAP1/2 heterodimer co-ordinates four tapasin molecules. This stoichiometry may be achieved by the interaction of one tapasin tetramer with one site on TAP, or alternatively, two dimers of tapasin may exist and interact with two sites on TAP (i.e. one site on TAP1, and one site on TAP2). Lastly, it is possible that four independent binding sites exist on TAP (i.e. two sites on TAP1 and two sites on TAP2). Support for the latter two mechanisms of tapasin association with TAP comes from studies demonstrating that tapasin can interact independently with each subunit of TAP.26, 30 These data suggest that there must be at least two binding sites on each TAP molecule. Using the homo-bifunctional chemical cross-linker EGS, we have shown a 1: 1 ratio of tapasin to MHC class I molecules without evidence for higher-order tapasin dimers, trimers or tetramers. We observe two forms of this tapasin–MHC class I complex that migrate differentially by SDS-polyacrylamide gel electrophoresis (SDS–PAGE). These two forms probably represent different conformations of the tapasin–MHC class I complex. Therefore, although the interactions within the peptide-loading complex are numerous and complex, we have detected discrete interactions by using chemical cross-linking. This approach has also revealed two high-molecular-weight cross-linked species (species 1 at ≈ 170 000 MW and species 2 at 130 000 MW) that contain tapasin. Further studies are required to determine whether these species contain molecules known to interact with tapasin or novel proteins that are currently not known to interact with tapasin.
Together, our studies show that tapasin is a relatively stable protein, the half-life of which is not affected by the absence of either MHC class I or TAP interaction. Each tapasin molecule interacts with a single MHC class I molecule to assist folding. Then, tapasin is recycled to assist in the folding of another MHC molecule. Tapasin appears to be the limiting factor in the surface expression of MHC class I molecules which may render the tapasin locus ideal for mutation by tumour cells and the tapasin protein a target for viral proteins to suppress MHC class I expression and the immune response.
Acknowledgments
We thank David Peaper and Dr Pamela Wearsch for helpful discussion and critical reading of the manuscript. This work was supported, in part, by the Howard Hughes Medical Institute.
References
- 1.Ortmann B, Copeman J, Lehner PJ, et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–9. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 2.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–71. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 3.Stoltze L, Schirle M, Schwarz G, et al. Two new proteases in the MHC class I processing pathway. Nat Immunol. 2000;1:413–8. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 4.Spies T, DeMars R. Restored expression of major histocompatibility class I molecules by gene transfer of a putative peptide transporter. Nature. 1991;351:323–4. doi: 10.1038/351323a0. [DOI] [PubMed] [Google Scholar]
- 5.Kelly A, Powis SH, Kerr LA, et al. Assembly and function of the two ABC transporter proteins encoded in the human major histocompatibility complex. Nature. 1992;355:641–4. doi: 10.1038/355641a0. [DOI] [PubMed] [Google Scholar]
- 6.Ahluwalia N, Bergeron JJ, Wada I, Degen E, Williams DB. The P88 molecular chaperone is identical to the endoplasmic reticulum membrane protein, calnexin. J Biol Chem. 1992;267:10914–8. [PubMed] [Google Scholar]
- 7.David V, Hochstenbach F, Rajagopalan S, Brenner MB. Interaction with newly synthesized and retained proteins in the endoplasmic reticulum suggests a chaperone function for human integral membrane protein IP90 (calnexin) J Biol Chem. 1993;268:9585–92. [PubMed] [Google Scholar]
- 8.Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5:103–14. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 9.Diedrich G, Bangia N, Pan M, Cresswell P. A role for calnexin in the assembly of the MHC class I loading complex in the endoplasmic reticulum. J Immunol. 2001;166:1703–9. doi: 10.4049/jimmunol.166.3.1703. [DOI] [PubMed] [Google Scholar]
- 10.Grandea AG, III, Androlewicz MJ, Athwal RS, Geraghty DE, Spies T. Dependence of peptide binding by MHC class I molecules on their interaction with TAP. Science. 1995;270:105–8. doi: 10.1126/science.270.5233.105. [DOI] [PubMed] [Google Scholar]
- 11.Shepherd JC, Schumacher TN, Ashton-Rickardt PG, Imaeda S, Ploegh HL, Janeway CA, Jr, Tonegawa S. TAP1-dependent peptide translocation in vitro is ATP dependent and peptide selective. Cell. 1993;74:577–84. doi: 10.1016/0092-8674(93)80058-m. [DOI] [PubMed] [Google Scholar]
- 12.Androlewicz MJ, Anderson KS, Cresswell P. Evidence that transporters associated with antigen processing translocate a major histocompatibility complex class I-binding peptide into the endoplasmic reticulum in an ATP-dependent manner. Proc Natl Acad Sci USA. 1993;90:9130–4. doi: 10.1073/pnas.90.19.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ortmann B, Androlewicz MJ, Cresswell P. MHC class I/Beta 2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 1994;368:864–7. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 14.Suh WK, Mitchell EK, Yang Y, Peterson PA, Waneck GL, Williams DB. MHC class I molecules form ternary complexes with calnexin and TAP and undergo peptide-regulated interaction with TAP via their extracellular domains. J Exp Med. 1996;184:337–48. doi: 10.1084/jem.184.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson JR, Ora A, Van PN, Helenius A. Transient, lectin–like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol Biol Cell. 1995;6:1173–84. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliver JD, van der Wal FJ, Bulleid NJ, High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–8. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- 17.Gao B, Adhikari R, Howarth M, Nakamura K, Gold MC, Hill AB, Knee R, Michalak M, Elliott T. Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity. 2002;16:99–109. doi: 10.1016/s1074-7613(01)00260-6. [DOI] [PubMed] [Google Scholar]
- 18.Chivers PT, Laboissiere MC, Raines RT. The CXXC motif: imperatives for formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–67. [PMC free article] [PubMed] [Google Scholar]
- 19.Molinari M, Helenius A. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature. 1999;402:90–3. doi: 10.1038/47062. [DOI] [PubMed] [Google Scholar]
- 20.Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I–peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 21.Bangia N, Lehner PJ, Hughes EA, Surman M, Cresswell P. The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. Eur J Immunol. 1999;29:1858–70. doi: 10.1002/(SICI)1521-4141(199906)29:06<1858::AID-IMMU1858>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 22.Lehner PJ, Surman MJ, Cresswell P. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line 220. Immunity. 1998;8:221–31. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- 23.Zarling AL, Luckey CJ, Marto JA, et al. Tapasin is a facilitator, not an editor, of class I MHC peptide binding. JImmunol. 2003;171:5287–95. doi: 10.4049/jimmunol.171.10.5287. [DOI] [PubMed] [Google Scholar]
- 24.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–20. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 25.Garbi N, Tiwari N, Momburg F, Hammerling GJ. A major role for tapasin as a stabilizer of the TAP peptide transporter and consequences for MHC class I expression. Eur J Immunol. 2003;33:264–73. doi: 10.1002/immu.200390029. [DOI] [PubMed] [Google Scholar]
- 26.Raghuraman G, Lapinski PE, Raghavan M. Tapasin interacts with the membrane-spanning domains of both TAP subunits and enhances the structural stability of TAP1 × TAP2 complexes. J Biol Chem. 2002;277:41786–94. doi: 10.1074/jbc.M207128200. [DOI] [PubMed] [Google Scholar]
- 27.DeMars R, Chang CC, Shaw S, Reitnauer PJ, Sondel PM. Homozygous deletions that simultaneously eliminate expressions of Class I and Class II antigens of EBV-transformed B-lymphoblastoid cells. I. Reduced proliferative responses of autologous and allogeneic T cells to mutant cells that have decreased expression of class II antigens. Hum Immunol. 1984;11:77–97. doi: 10.1016/0198-8859(84)90047-8. [DOI] [PubMed] [Google Scholar]
- 28.Schoenhals GJ, Krishna RM, Grandea AG, III, Spies T, Peterson PA, Yang Y, Fruh K. Retention of empty MHC class I molecules by tapasin is essential to reconstitute antigen presentation in invertebrate cells. EMBO J. 1999;18:743–53. doi: 10.1093/emboj/18.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandea AG, III, Lehner PJ, Cresswell P, Spies T. Regulation of MHC class I heterodimer stability and interaction with TAP by tapasin. Immunogenetics. 1997;46:477–83. doi: 10.1007/s002510050308. [DOI] [PubMed] [Google Scholar]
- 30.Koch J, Guntrum R, Heintke S, Kyritsis C, Tampe R. Functional dissection of the transmembrane domains of the transporter associated with antigen processing (TAP) J Biol Chem. 2004;279:10142–7. doi: 10.1074/jbc.M312816200. [DOI] [PubMed] [Google Scholar]