Abstract
Natural killer (NK) T lymphocytes are a subpopulation of T lymphocytes regarded as early regulators of immune responses. The majority of NKT cells are restricted by the CD1d molecule. NKT cells have mostly been studied in one single mouse strain, C57BL/6 (B6), because of the absence of NK1.1 in other common mouse strains, and the lack of other reliable surface markers for CD1d-restricted cells. To investigate NKT cell subsets in a mouse strain of a genetic background different from B6, we have back-crossed the NKT cell marker NK1.1 from the B6 mouse to the BALB/c mouse strain. We show that NKT cells in the congenic BALB.B6-NK1.1b mouse share many characteristics with their B6 counterparts, but seem to be deficient in the functional NKT cell subtype characterized by low interleukin-4 and high interferon-γ production, and surface expression of CD49b but not CD69. Moreover, in the thymus but not the spleen of BALB.B6-NK1.1b mice we find a novel Vα14-Jα18 invariant NKT cell subset which is devoid of a set of NK markers, suggesting that these cells represent a less differentiated NKT cell stage, and carries high levels of the T-cell receptor and uses a skewed T-cell receptor Vβ-repertoire.
Keywords: CD1-restricted/natural killer T cells, cell development/differentiation, T-cell receptor, natural killer receptors
Introduction
Natural killer (NK) T cells were first observed as a population of T lymphocytes expressing the pan-NK cell marker NK1.1, and are therefore often referred to as NKT cells. Further studies showed that they belong to a subset of T cells that has unique characteristics.1 Most NKT cells are restricted to antigens in the form of lipids and glycolipids presented in the context of the CD1d molecule.2 NKT cells have a surface phenotype reminiscent of memory cells, and when activated, it has been shown that the NKT-cell population rapidly secretes large amounts of cytokines3, especially interleukin (IL)-4 and interferon-γ (IFN-γ). Because of their rapid response to activation NKT cells can regulate early immunological events in diverse situations, such as the control of autoimmune diseases in humans and mice4, tumour rejection5 and different infections.6
CD1d restricted T cells can be divided into two categories. One group has a restricted T-cell receptor (TCR) repertoire; the cells express a semi-invariant TCR consisting of a conserved TCRα-chain rearrangement (Vα14-Jα18) (iNKT cells), paired with diverse TCRβ-chains having Vβ8.2, 7 or 2 segments.7 This population is activated by the synthetic ligand α-galactosylceramide (αGalCer) presented on CD1d.8 Humans have a corresponding CD1d-reactive T-cell population using the homologous human TCR segments. The second group of CD1d-reactive T cells has diverse TCR9, 10, 11 but otherwise share the main characteristic features of NKT cells, such as rapid cytokine secretion upon activation and a memory surface phenotype.12, 13 Our previous data in B6 and non-obese diabetic mice suggest that these two subsets are also distinct in terms of functional ability and the display of certain surface markers.12, 13, 14 We term the subsets NKT1 (having diverse TCR and making high amounts of IFN-γ and little IL-4) and NKT2 (characterized by usage of the conserved Vα14-Jα18 TCR α-chain, being high producers of both IL-4 and IFN-γ). These different NKT-cell subsets are likely to play distinct roles in immune responses.
The NK1.1 receptor is only expressed in a few mouse strains, such as C57BL/6 (B6).15 There is currently no reagent available that can reliably detect all CD1d-dependent T cells. Through the development of CD1d-tetramers (CD1d-tet), loaded with the ligand α-GalCer, it is possible to identify the subset of CD1d-restricted T cells using the invariant Vα14-Jα18 TCR regardless of NK1.1 expression16, 17, however, this method does not identify the group of CD1d-restricted T cells with diverse TCR.18, 19 Therefore our knowledge of the entire NKT cell population is, to a large extent, based on the NKT cells found in B6 mice. This is further complicated by the fact that not all NK1.1+ T cells are CD1d-restricted, and not all CD1d-restricted T cells are NK1.1+ (see further below). To be able to study NKT cells in another mouse strain, we have created a congenic mouse, which expresses the NK1.1 antigen from the B6 mouse on the BALB/c genetic background (BALB.B6-NK1.1b, referred to below as BALB.NK mice). In the present study, we have analysed and compared the NKT cell and CD1d-tet+ populations in the congenic BALB.NK and the B6 mouse. We investigate the presence of NKT1 and NKT2 subsets in the spleen, and demonstrate a dominance of NKT2 cells in the BALB.NK spleen. Additionally, we detect a novel NKT-cell subset in the BALB.NK thymus, with a high TCRβ expression and lower NK1.1 levels, and a poor display of NK markers suggesting an incomplete differentiation stage.
Materials and methods
Mice
C57BL/6 (B6) and BALB.B6-NK1.1b (BALB.NK) mice used were 10–20 weeks old. The congenic mice used in the experiments were homozygous for NK1.1 expression, and from intercrosses of the seventh and eighth back-cross generation. All mice were maintained in a clean conventional animal facility at Lund University, Sweden.
Genotyping
Nine informative fluorescence labelled microsatellite markers (Interactiva, Ulm, Germany and MWG-Biotech, Germany) were used for genotyping of the congenic fragment on chromosome 6. Polymerase chain reactions (PCRs) were performed with 5 ng of DNA in a reaction volume of 10 µl containing: 10 mm Tris-HCl, pH 9·0, 50 mm KCl, 1·5 mm MgCl2, 1·5 pmol of the respective forward and reversed primer, 0·4 mm dinucleotide triphosphates (Advanced Biotechnologies, Epsom, UK), and 0·25 U Taq DNA polymerase (Amersham Pharmacia Biotech, Uppsala, Sweden). The following conditions were used for amplification of DNA: denaturation at 94° for 3 min, annealing at 56° for 45 s, polymerization at 72° for 1 min, followed by 31 cycles of 94° for 30 s, 56° for 45 s, and 72° for 1 min. The final cycle ended by elongation at 72° for 7 min. The PCR products were analysed on a Megabace 1000 (Amersham Pharmacia Biotech), according to the manufacturer's protocol.
Cell preparation
Where indicated, B cells were depleted from spleen cell suspensions using AutoMACS® separation and anti-B220 microbeads (Miltenyi Biotec, Bergisch, Germany), according to the manufacturer's instructions. Alternatively B cells were depleted by panning on plates coated with rabbit anti-mouse immunoglobulin. After B-cell depletion the cell suspensions had a contamination of 0·5–2·5% B220+ cells. To enrich for DN T cells, B-cell depleted spleen cells were further depleted of CD4+ and CD8+ cells using GK1.5 (anti-CD4) and YTS169.4 (anti-CD8α) antibodies and anti-ratκ-microbeads (Miltenyi Biotech) using a magnetic-activated cell sorting magnet (Miltenyi Biotech). The resulting population contained 6–16% TCRβ+ cells, a total of 1,5–4% contaminating CD4+ or CD8+ cells and 5% B220+ cells.
Flow cytometry
Before staining the cells were incubated with 2.4G2 (anti-CD16/CD32) antibody to block non-specific binding. The following antibodies were obtained from PharMingen (San Diego, CA): TCRβ–allophycocyanin (APC), TCRβ–fluoroscein isothiocyanate (FITC), TCRβ–biotin (H57-597), NK1.1–APC, NK1.1–phycoerythrin (PE), NK1.1–biotin (PK136), CD4–PE, CD4–APC (RM4-5), CD4–PerCP (RM4-3), CD8–PE, CD8–PerCP (53-6.7), CD69–FITC, CD69–PE (H1.2F3), CD5–PE (53-7.3), CD25–FITC (7D4), CD122–FITC, CD122–PE (TM-β1), CD19–FITC (1D3), B220–PE (RA3-6B2), CD49b–FITC, CD49b–PE, CD49b–biotin (DX5), IL-2–PE, IL-4–PE, IL-4–APC, IL-10–PE, IFN-γ–FITC, IFN-γ–PE, IFN-γ–APC, anti-mouse IgG1–PE (A85-1), anti-mouse IgG1–biotin, streptavidin–PE, streptavidin–PerCP and streptavidin–APC. Streptavidin–PE Texas Red (PETR) was obtained from Caltag (San Francisco, CA). CD4-Red613 (H129.19) and streptavidin-Red613 were obtained from Gibco (BRL, Life Technologies, Paisley, UK). NKG2D-biotin (MI-6) was kindly provided by Dr D.H. Raulet.20 CD4–FITC, CD4–Cy5 (GK1.5), CD8–FITC (YTS169.4), CD62L–biotin (Mel-14), heat stable antigen (HSA)–FITC (M1/69), B220–Cy5 (RA3-6B2), Ly49C/I–Cy5 (5E6) and Vβ8.2–biotin (F23.2) have been purified and conjugated according to standard procedures in the laboratory. To detect canonical TCR-carrying T lymphocytes, PE-labelled CD1d-tetramers (kindly provided by Dr M. Kronenberg) loaded with αGalCer (Kirin Brewery Co., Gunma, Japan) were used in most experiments. Due to recent availability of a commercial reagent, we have in the latter experiments used CD1d-IgG1 dimers (PharMingen) loaded with αGalCer (Kirin Brewery Co) with comparable results. Unloaded CD1d-tetramers and dimers were used as background controls. The stained cells were analysed by four colour flow cytometry on a FACSCalibur using the CELLQuest software (Becton Dickinson, San Jose, CA). Fluorescence is displayed on a log scale, showing four decades.
In vitro stimulation of cells for assessment of cytokine production
B-depleted spleen cells or total thymocytes were stimulated for 4 hr with 50 ng/ml phorbol-12 myristate-13 acetate, 500 ng/ml ionomycin in the presence 10 µg/ml brefeldin A (Sigma, St Louis, MO), in RPMI-1640 medium supplemented with 10% fetal calf serum, 20 µm mercaptoethanol, 10 mm Hepes buffer and 1 mm sodium pyruvate. The cells were harvested and stained in different combinations for surface expression of TCRβ, NK1.1, CD69, and CD49b as described above. After surface staining the cells were fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS), and kept over night at 4°. The cell membranes were permeabilized in PBS with 0·5% saponin, 1% bovine serum albumin, 0·1% NaN3 and stained for intracellular IL-4, IL-10, IL-2 and IFN-γ. Cells were incubated for 30 min with the respective antibody diluted in the permeabilization solution. The cells were washed and analysed as described above.
Results
The congenic BALB.NK mouse
The gene encoding the NK1.1 surface marker (Nk1.1 or Nkrp1-c) is located in the NK gene complex on chromosome 6.21 To obtain a BALB/c mouse that expresses the NK1.1 receptor, we have introduced a genetic fragment containing Nkrp1-c from mice onto the BALB/c genetic background. B6 and BALB/c mice were crossed and the progeny further back-crossed to BALB/c mice, selecting mice in each generation for NK1.1 expression. Heterozygous mice at the seventh and eighth back-cross generation were intercrossed to obtain NK1.1b homozygous mice, named BALB.B6-NK1.1b (BALB.NK), used in this investigation. To estimate the size of the B6 derived congenic segment of chromosome 6 present in BALB.NK mice of the eighth generation intercross we used a number of microsatellite markers. In the area of the NK complex, the microsatellite markers D6Mit39, D6Mit54 and D6Mit135 were of B6 origin, while D6Mit230 and D6Mit290 were derived from BALB/c (Fig. 1a). Thus, the congenic fragment extends 22–37 megabase pair (Mbp) and is located between D6Mit230 and D6Mit 290, illustrated by the chromosomal area in white (Fig. 1a). Other studies22, 23 have used a different congenic BALB/c mouse which also expresses the NK1.1 receptor from the B6 mouse. Compared to the BALB.NK mouse used in our studies, the other congenic mouse (originally selected for cytomegalovirus resistance) has a B6 derived fragment that extends further on the telomeric side and a shorter distance on the centromeric side of the NK complex.24
Figure 1.
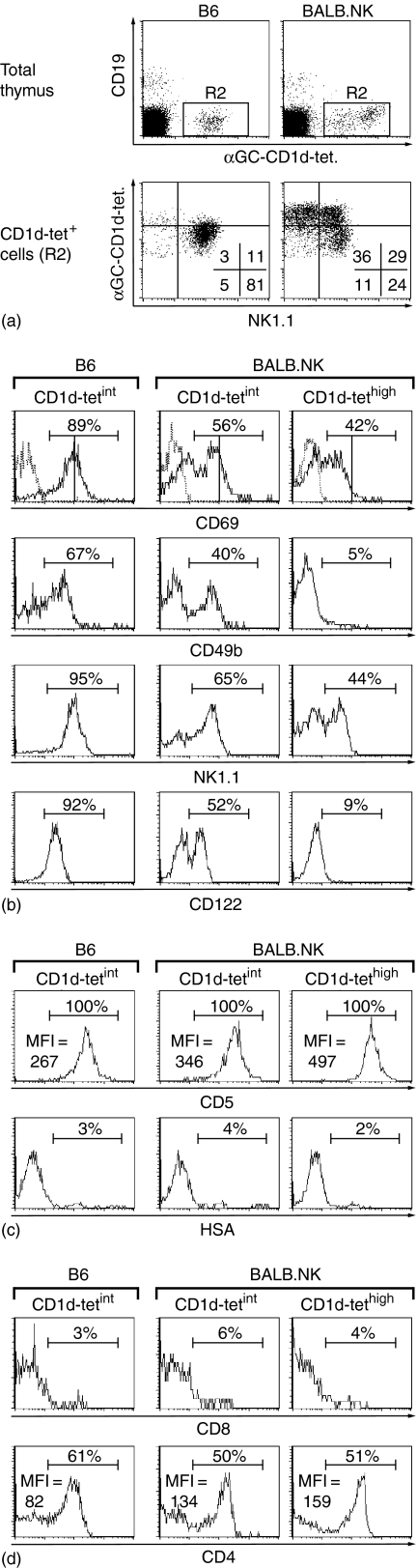
Map of the mouse chromosome 6 genetic fragment containing the NK gene cluster in the BALB.NK congenic strain.(a)BALB.NK mice were typed for the microsatellite markers shown on the map and alleles inherited from C57BL/6 (b) and BALB/c (c) are indicated to the right.(b)NKT cell populations in BALB.NK thymus can be divided into TCRint and TCRhigh cells. Thymocytes were stained with antibodies to TCRβ, NK1.1, CD4 and CD8. NKT cells were gated as shown, and displayed for CD4 and CD8 expression. Frequencies of cells in the respective quadrants are indicated in the upper right corners of the CD4/CD8 plots. The gate used for TCRint NKT cells in BALB.NK thymus is the same as that used for B6 NKT cells. The data is representative of several experiments.
NKT cell populations in the thymus
B6 and BALB.NK had similar frequencies of NK1.1+ TCRβ+ cells in the thymus (0·9 ± 0·2% and 0·9 ± 0·1%, respectively, for total NKT populations, n = 6; Fig. 1b). However, in the BALB.NK thymus NK1.1+ TCRβ+ cells could be divided into two populations with regard to the level of TCRβ and NK1.1 expression (Fig. 1b). One was characterized by intermediate (int) expression of TCRβ and high expression of NK1.1, constituted approximately 60% of the total BALB.NK NKT population and overlapped with the thymic NKT-cell subset in B6 mice, and the second population had high levels of TCRβ and intermediate levels of NK1.1, here referred to as TCRβint and TCRβhigh NKT cells, respectively. The B6 NKT cells had somewhat higher NK1.1 levels than the BALB.NK TCRβint NKT cells (MFI approximately 180 and 100 for B6 and BALB.NK populations, respectively). Both TCRβint populations contained very few CD8+ cells, and the ratio of CD4+ to double negative (DN) cells was slightly lower for BALB.NK cells (Fig. 1b, lower panels).
A unique NKT cell subpopulation in the thymus of BALB.NK mice
We next compared the surface phenotype of the B6 thymic TCRβint NKT cells with the two BALB.NK NKT subsets (Fig. 2). The two TCRβint NKT-cell subsets shared expression pattern of the NK-cell associated markers CD122, CD49b, NKG2D, and Ly49C/I (Fig. 2a). In contrast, the display of these receptors on TCRβhigh BALB.NK NKT cells was strikingly lower in all cases, suggesting that the cells may represent a less differentiated stage. Expression of CD5 but not HSA on TCRint and TCRhigh cells demonstrated, however, that the cells were not immature thymocytes. Instead, the BALB.NK TCRβhigh NKT cells might represent an earlier stage in the differentiation of NKT cells. Ly49C/I is expressed by the majority of thymic NKT cells in adult B6 mice25 and several NK cell markers are up-regulated during NKT cell differentiation.26, 27 Further analysis demonstrated that B6 and BALB.NK TCRβint NKT cells were CD69+ CD62Llow and 40–50% of the cells had CD4 on the surface (Fig. 2c). Similarly, TCRβhigh cells were CD62L– and positive for CD69; however, the level of CD69 was slightly lower, and the frequency of CD4+ cells was reduced (Fig. 2c). Finally we investigated the TCR usage in the thymic NKT cell subsets (Fig. 2d). Binding of αGalCer loaded CD1d-tetramers (CD1d-tet) to cells indicates display of the invariant Vα14-Jα18 TCR, which preferentially pairs with TCR Vβ8.2/7 or 2-chains. Just as among TCRβint cells, the absolute majority (95%) of TCRβhigh NKT cells bound the CD1d-tet. As expected, TCRβhigh cells were also CD1d-tethigh. Surprisingly, around 70% of the TCRβhigh cells expressed the Vβ8·2-chain, while the 45–50% of the cells in the TCRβint subsets used this Vβ-chain, demonstrating a shift in the TCR β-chain repertoire between TCRβint and TCRβhigh NKT cells.
Figure 2.
NKT cell populations in BALB.NK thymus can be divided into TCRint and TCRhigh cells. B6 NKT cells, BALB.NK TCRint and TCRhigh NKT cells, gated as in Figure 1, were displayed for the expression of (a)CD122, CD49b, NKG2D and Ly49C/I (b) CD5 and HSA (C) CD69, CD62L and CD4, and (d)the binding to αGC-loaded CD1d-tetramer and the expression of Vβ8·2. The numbers refer to frequencies of positive cells and mean fluorescence intensity (MFI) as defined by the bars. One representative background control stain is indicated by a dotted line in the upper right histogram plot. The data is representative of at least three experiments, except the NKG2D staining, which has been done in two experiments.
To analyse invariant CD1d-restricted T cells in the thymi regardless of NK1.1 expression, we used the CD1d-tet. BALB.NK thymic cells could be divided into CD1d-tetint and CD1d-tethigh cells, while CD1d-tethigh cells were not observed among B6 thymocytes (Fig. 3). In comparison to B6 CD1d-tetint cells, fewer BALB.NK CD1d-tetint cells expressed CD69, CD49b, NK1.1 and CD122 (Fig. 3b). In contrast, while 40–45% of the BALB.NK CD1d-tethigh population expressed NK1.1 and CD69, none of these cells were positive for CD49b or CD122. All BALB.NK CD1d-tethigh cells were CD5+ HSA– (Fig. 3c), and CD8 expression was very infrequent in all three subsets (Fig. 3d). Moreover, approximately half of the CD1d-tethigh population was CD4+, which was higher than found for the TCRβhigh subset, suggesting that a larger proportion of the NK1.1– CD1d-tet+ cells were CD4+.
Figure 3.
Surface phenotype on αGC-loaded CD1d-tetramer+ (CD1d-tet+) thymic cells. (a) CD1d-tet+ cells (including both tethigh and tetint) in B6 and BALB.NK thymus were gated out (R2, upper plots) and their binding intensity of CD1d-tet is displayed against NK1.1 expression (lower plots). Frequencies of cells in the respective quadrants are indicated in the lower right corner of the plot. B6 CD1d-tetint and BALB.NK CD1d-tetint and CD1d-tethigh thymocytes were displayed in histograms for the expression of (b)CD69, CD49b, NK1.1 and CD122 (c)CD5 and HSA and d)CD8 and CD4. The numbers refer to frequencies of positive cells and mean fluorescence intensity (MFI) as defined by the bars. One representative background control stain is indicated by a dotted line in the upper histogram plot. The data is representative of at least two experiments.
NKT cell populations in the spleen of B6 and BALB.NK mice
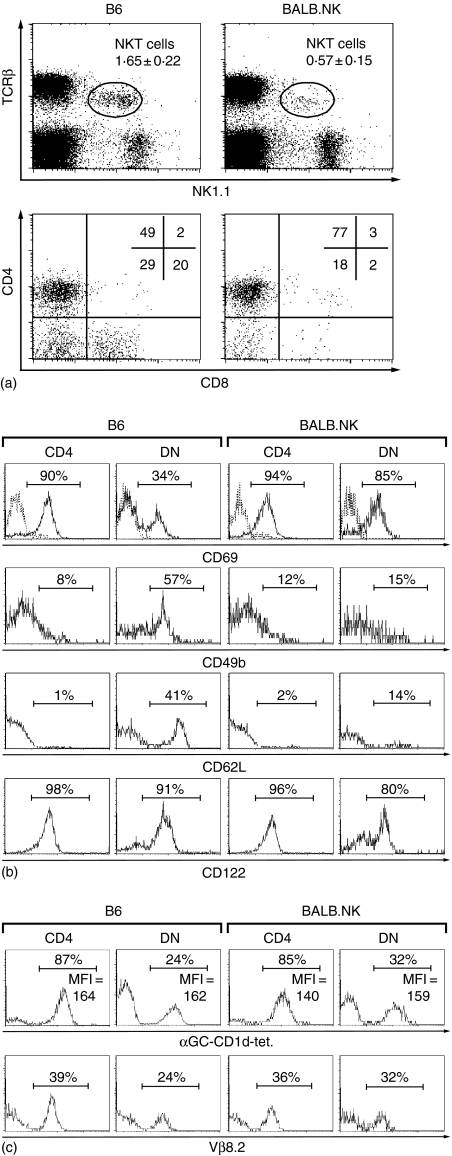
We next compared NKT cells in the spleen of B6 and BALB.NK mice (Fig. 4). Splenic BALB.NK NKT cells had the usual intermediate levels of TCR expression with no signs of a TCRβhigh subset, and the population was about one-third the size of the splenic B6 NKT cell population (1·7 ± 0·2 and 0·6 ± 0·2, respectively, n = 9). The level of NK1.1 expression on the splenic BALB.NK NKT cells was slightly lower than on B6 NKT cells, with a mean fluorescence intensity (MFI) of around 50 and 75, respectively, while no difference was observed for NK1.1 levels on NK cells in the two strains. In both strains, the majority of the NKT cells were CD4+, and the ratios of CD4+ to DN cells were approximately 2 : 1 and 4 : 1 in B6 and BALB.NK, respectively (Fig. 4a, lower dot-plots). Among B6 splenocytes, but not BALB.NK splenocytes, there was a distinct population of CD8+ NKT cells, cells which are thought to represent memory cells restricted by conventional major histocompatibility complex class I molecules.28
Figure 4.
NKT cell populations in BALB.NK spleen is dominated by NKT2 cells. B6 and BALB.NK splenocytes were stained for TCRβ, NK1.1, CD4 and CD8. Cells were gated as shown in (a), and displayed for CD4 and CD8 expression. Frequencies of NKT cells in the upper plot are the mean and SD of nine experiments. Frequencies of cells in the respective quadrants are indicated in the upper right corners of the CD4/CD8 plots. The gate used for NKT cells in BALB.NK spleen is the same as that used for B6 spleen. B6 and BALB.NK splenocytes were first depleted of Ig+ cells. To gate out CD4+ NKT cells (CD4+ TCRβ+ NK1.1+), cells were stained for TCRβ, NK1.1, CD4 and indicated markers. To gate out DN NKT cells (CD4– CD8– TCRβ+ NK1.1+), cells were stained for TCRβ, NK1.1, CD4, CD8 (CD4 and CD8 were detected in the same fluorescence channel) and indicated markers. CD4+ and DN NKT cells were analysed for the expression of (b)CD69, CD49b, CD62L and CD122 (c) the binding to αGC-loaded CD1d-tetramer and the expression of Vβ8·2. The numbers refer to frequencies of positive cells and mean fluorescence intensity (MFI) as defined by the bars. One representative background control stain is indicated by a dotted line in the upper histogram plot. The data is representative of at least three experiments.
Surface phenotype indicates a high proportion of NKT2 cells in the spleen of BALB.NK mice
We compared the subset composition of NKT cells in the two mouse strains in terms of surface phenotype, functional ability and TCR usage. B6 and BALB.NK CD4+ NKT cells had very similar surface phenotypes (Fig. 4b), indicative of a dominance of NKT2 cells, which we have defined as CD69+ CD49b–/low CD62Llow and CD122+.12, 13 Approximately 85% of the CD4+ cells were CD1d-tet+ while 35–40% of the cells expressed the Vβ8·2-chain, similar to what was seen in the thymic TCRβint populations. DN NKT cells in B6 mice have previously been shown to contain a relatively large proportion of NKT1 cells,13 characterized by low CD69 and high CD49b expression and preferential usage of diverse non-Vα14 TCR. In contrast, DN NKT cells in BALB.NK spleen were similar in surface marker expression to the CD4+ NKT cells, suggesting a higher proportion of NKT2-like cells also among non-invariant DN NKT cells in this strain. Thus, there was not only a lower frequency of DN NKT cells in the BALB.NK spleen, but these also appear to contain more cells of the NKT2 subtype.
Cytokine secretion indicates NKT2 dominance among splenic BALB.NK NKT cells
Among total splenic NKT cells in B6 mice, we determined a higher frequency of IFN-γ- than IL-4-producing cells. Activated BALB.NK NKT cells included equal numbers of IFN-γ and IL-4 containing cells (Fig. 5a). When comparing CD4+ and DN NKT cells from B6 mice, the DN cells were weaker IL-4 and stronger IFN-γ producers, a difference that was much less pronounced in the BALB.NK populations (Fig. 5b, c). IL-2 was induced in 40–50% of CD4+ and DN cells from both strains, while IL-10 was not significantly produced by any of the subsets. The low percentage of CD69–CD49b+ (NKT113) cells among splenic BALB.NK NKT cells confirms the dominance of NKT2 cells in this strain (Fig. 6a). A high proportion of CD69+ CD49b–/low NKT cells from both mice were IL-4 producers, while CD49b+ cells were IL-4– (Fig. 6b). IFN-γ was found in CD69+ as well as CD49b+ cells of both strains (Fig. 6c). Thus, the relative lack of CD69– CD49b+ cells in BALB.NK as compared to B6 spleen correlates with an increased IL-4- and decreased frequency of IFN-γ-producing cells.
Figure 5.
Cytokine profile of splenic B6 and BALB.NK NKT cells. Spleen cells were polyclonally stimulated as described in Materials and methods and stained for surface markers and intracellular cytokines. (a)IL-4, IFNγ and IL-10 production among total splenic NKT cells in B6 (white bars) and BALB.NK (black bars). IL-4, IFNγ, IL-10 and IL-2 production among splenic CD4+ (b)and DN (c)NKT cells in B6 (white bars) and BALB.NK (black bars). Frequencies are shown as mean and SD (n = 3).
Figure 6.
Splenic BALB.NK NKT cells show a strong NKT2 surface phenotype and cytokine profile. (a)B6 and BALB.NK splenic NKT cells, gated as in Fig. 4, were displayed for CD49b versus CD69 expression. IL-4 (b)and IFNγ (c)production in B6 (left panel) and BALB.NK (right panel) NKT cells versus CD69 and CD49b expression. Frequencies of cells in the respective quadrants are indicated in the upper right corners of the plots. Data is representative of at least three experiments.
Surface phenotype and cytokine profile of CD4+ and DN CD1d-tet+ splenic NKT cells
The CD4 coreceptor divides human invariant NKT cells into two subsets, which have different surface phenotypes and distinct cytokine secretion profiles upon activation, reminiscent of the characteristics of NKT1 and NKT2 cells, with the exception of preferential TCR usage in the murine subsets. We therefore first compared the surface phenotype of invariant CD1d-restricted NKT cell populations in the two mouse strains (Fig. 6a). As in the thymus, there was a higher frequency of CD1d-tet+ cells that did not express the NK1.1 marker in BALB.NK compared to B6 spleen, confirming previous studies.22 The surface expression of CD49b on splenic CD1d-tet+ cells was similar in both strains with slightly higher levels on DN cells than on CD4+ cells. Among CD1d-tet+ cells in B6 spleen there was a high frequency of CD69+ cells, while somewhat fewer BALB.NK CD1d-tet+ cells displayed this marker. CD4+ and DN CD1d-tet+ cells from both strains were CD122+ and CD25–. We further compared IL-4 and IFN-γ production in B6 and BALB.NK αGalCer loaded CD1d-dimer+ (CD1d-di+) cells. IFN-γ-producing cells were somewhat more frequent than IL-4 among CD1d-di+ cells of both strains (Fig. 7b). Both CD1d-di+ subsets in B6 mice produced IL-4 and IFN-γ (Fig. 7c). The same was true for IFN-γ production by BALB.NK CD1d-di+ cells, while a higher fraction of CD4+ cells than DN cells made IL-4 in these mice. Thus, CD4+ and DN invariant Vα14 CD1d-restricted cells in both mice largely shared the features investigated here.
Figure 7.
Surface phenotype and cytokine profile among αGC loaded CD1d-tetramer+ (CD1d-tet+) spleen cells. B6 and BALB.NK Ig– splenocytes were stained with αGC loaded CD1d-tetramers and the indicated markers. (a) Surface expression of NK1.1, CD49b, CD69, CD122 and CD25 on CD4+ and DN CD1d-tet+ splenic cells. Numbers refer to frequencies of positive cells as defined by the bars. One representative background control stain is indicated by a dotted line in the CD69 histogram plot. Ig– spleen cells were polyclonally activated as described in Materials and Methods and stained with αGC-loaded CD1d-dimers (CD1d-di+) and antibodies against CD4 and intracellular cytokines. (b)The IL-4 and IFN-γ production among total CD1d-di+ cells in B6 (white bars) and BALB.NK (black bars) mice. (c)IL-4 and IFN-γ are plotted against CD4 to compare the profile of CD4+ and DN CD1d-di+ cells in B6 (upper plots) and BALB.NK (lower plots) mice. Data in (b) is the mean of two experiments and data in (a) and (c) is representative of two experiments.
Discussion
To broaden the knowledge on NKT-cell subsets, we have studied a congenic mouse strain that expresses the NK1.1 surface marker from B6 mice on a BALB/c genetic background. Investigation of the thymi of the two strains did not show any difference in the size of the total NKT cell population, in agreement with previous studies of different strain of NK1.1 congenic BALB/c mice.22, 23, 24 However, we could define two distinct NKT subsets in the BALB.NK thymus, with intermediate and high TCRβ expression, respectively, which appeared to be present also in the above investigation.22 The thymic TCRβint NKT cells in the BALB.NK and B6 mice were similar with regards to all the surface markers investigated, while the novel TCRβhigh expressing BALB.NK NKT cells were different from the TCRβint NKT cells in several aspects.
The majority of the BALB.NK thymic TCRβhigh NKT cells had a high level of binding to αGalCer loaded CD1d-tetramer, cells which were not detected in B6 thymus, nor in the spleen of either strain. Within the CD1d-tethigh population around 50% of the cells were CD4+, while the NK1.1+ TCRβhigh population contained only 18% CD4+ cells, indicating the presence of CD4+NK1.1– cells within the CD1d-tethigh population. Our subsequent analysis suggested that the CD1d-tethigh cells might belong to an early stage in the NKT cell development. Despite their mature (HSA–) phenotype, there were striking differences between the TCRβhigh and TCRβint NKT cells, as well as between the CD1d-tetint and CD1d-tethigh subsets. The TCRhigh/CD1d-tethigh cells were essentially negative for NK cell markers such as Ly49C/I, NKG2D and CD49b, although 40% of CD1d-tethigh cells did display NK1.1. NK1.1 is thought to be up-regulated at a relatively late stage in NKT cell differentiation.26, 27 Surprisingly, CD1d-tethigh cells were negative for CD122, a molecule known to be required for efficient NK1.1+ TCRβ+ thymocyte development.29 The CD1d-tethigh cells were also reminiscent of the thymic population found in mice lacking the cytokine receptor common γ-chain (γc° mice).30 γc° mice have a normal proportion of Vβ8+ thymocytes and a normal quantity of thymic Vα14-Jα18 message. However, the cells lack NK1.1 and Ly49 receptors and have reduced levels of CD122, and, further, lack peripheral NKT cells, suggesting a thymic block in NKT differentiation. Thus, the CD1d-tethigh cells in BALB.NK mice might not have received appropriate signals for NKT cell differentiation, including the up-regulation of CD122.
The surface phenotype of CD1d-tethigh cells initially suggested to us that the cells could represent a precursor stage to the TCRβint NKT cells. However, a comparison of the Vβ-chain usage among TCRhigh and TCRint cells showed a bias towards higher Vβ8.2 expression in the TCRhigh and CD1d-tethigh population. The TCR Vβ-segment used by Vα14-Jα18 CD1d-restricted cells influences the avidity for αGalCer-loaded CD1d31 suggesting that there could similarly be an effect on the avidity to CD1d+ endogenous ligand. Therefore, it is possible that CD1d-tethigh and CD1d-tetint cells display TCR repertoires that differ in avidity to the thymic CD1d-complex during selection and differentiation, leading subsequently to the distinct phenotypic profiles of the two subsets. Although CD1d-tethigh cells could not be found in the B6 thymus, the cells may not be unique for BALB.NK mice as a similar population might be present in DBA/2 mice.32 Further analysis of the CD1d-tethigh and CD1d-tetint populations in the BALB.NK thymus could help to elucidate questions regarding thymic selection and final maturation of NKT cells.
When splenic NKT cells were compared we found that the NKT cell population in the BALB.NK mouse was reduced and contained fewer DN cells than B6 splenic NKT cells. The CD1d-tet+ cells in both mice were of the NKT2 type, characterized by CD69 expression and a high IL-4 and IFN-γ production upon activation. In contrast, CD49b+ cells of the NKT1 type, prominent among B6 DN splenic NKT cells, were less frequent in BALB.NK mice. Instead, DN NKT cells in BALB.NK spleen, which just like the B6 counterpart consisted of 70–75% CD1d-tet negative cells, had the characteristics of NKT2 rather than NKT1 cells. Thus, the analysis of BALB.NK mice confirms the association of the CD69+ CD49b– phenotype and IL-4 production, and absence of IL-4 production in CD49b+ NKT cells, demonstrated before in B6 mice.13 However, while in B6 spleen the NKT1 and NKT2 phenotypes were associated with diverse and invariant TCR, respectively, NKT cells with both types of TCR displayed NKT2 features in BALB.NK mice. We have suggested that NKT1 and NKT2 cells serve different functions in the immune system. This might suggest that the NKT population in BALB.NK mice have a more limited functional repertoire, a feature that can be important in the early stages of an immune response. It should be noted, though, that the dominance of NK1.1– CD1d-tet+ cells in BALB.NK spleen indicates that an enlarged NK1.1– NKT1-like subset with diverse TCR could not be excluded in these mice.
Previous reports have shown that the CD4+ and DN subpopulations of human NKT cells, detected with αGalCer loaded CD1d-tetramers, have different surface marker expression and functional ability.33, 34 The expression of NK-cell associated markers was in many cases only observed on DN cells. Both CD4+ and DN cells were capable of T helper 1 (Th1) cytokine production, such as IFN-γ and tumour necrosis factor-α, while Th2 cytokines, such as IL-4 and IL-13, were exclusively secreted by human CD4+ cells. In both B6 and BALB.NK spleen the only difference detected between CD4+ and DN CD1d-di+ cells were slightly higher expression of NK-associated markers on the DN population. Further, in both strains 30% of the DN cells were induced to produce IL-4, although the frequency was higher among CD4+ cells. CD25 expression was exclusively found on CD4+ human CD1d-tet+ cells34 but this marker was not found on any of the CD1d-tet+ populations in B6 and BALB.NK spleen. As suggested before1 this indicates that the functional division of human NKT cells into a CD4+ and DN subpopulation may not be applied to the mouse NKT cells. Taken together, our data demonstrate that there are differences in NKT cell and CD1d-tet+ subsets between distinct mouse strains, and emphasize the importance of the continued analysis of these cells in more than one mouse strain.
Acknowledgments
This work was supported by grants from the Swedish Cancer Foundation, the Swedish Research Council and the following foundations: King Gustaf V 80th Jubilee Fund, Greta och Johan Kock, Alfred Österlund, Crafoord and the Medical Faculty of Lund University to SC. M Stenström was supported by stiftelsen Goljes Minne, the Swedish Foundation for Strategic Research and the Medical Faculty at Lund University, and M Sköld by Stiftelsen Lars Hiertas minne and the Medical Faculty at Lund University.
Abbreviations
- αGalCer
α-galactosylceramide
- BALB.B6-Nk1.1b
BALB.NK
- di
dimer
- DN
double negative
- NKT
natural killer T cell
- tet
tetramer
References
- 1.Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity and function. Annu Rev Immunol. 1997;15:535–62. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 2.Porcelli SA, Modlin RL. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimoto T, Paul WE. CD4pos, NK1.1pos T cells promptly produce interleukin 4 in response to in vivo challenge with anti-CD3. J Exp Med. 1994;179(4):1285–95. doi: 10.1084/jem.179.4.1285. 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond KJ, Kronenberg M. Natural killer T cells: natural or unnatural regulators of autoimmunity? Curr Opin Immunol. 2003;15(6):683–9. doi: 10.1016/j.coi.2003.09.014. 10.1016/j.coi.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Smyth MJ, Crowe NY, Hayakawa Y, Takeda K, Yagita H, Godfrey DI. NKT cells – conductors of tumor immunity? Curr Opin Immunol. 2002;14(2):165–71. doi: 10.1016/s0952-7915(02)00316-3. 10.1016/S0952-7915(02)00316-3. [DOI] [PubMed] [Google Scholar]
- 6.Skold M, Behar SM. Role of CD1d-restricted NKT cells in microbial immunity. Infect Immun. 2003;71(10):5447–55. doi: 10.1128/IAI.71.10.5447-5455.2003. 10.1128/IAI.71.10.5447-5455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268(5212):863–5. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 8.Kawano T, Cui J, Koezuka Y, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–9. doi: 10.1126/science.278.5343.1626. 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 9.Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182(4):993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behar SM, Podrebarac TA, Roy CJ, Wang CR, Brenner MB. Diverse TCRs recognize murine CD1. J Immunol. 1999;162(1):161–7. [PubMed] [Google Scholar]
- 11.Park SH, Roark JH, Bendelac A. Tissue-specific recognition of mouse CD1 molecules. J Immunol. 1998;160(7):3128–34. [PubMed] [Google Scholar]
- 12.Behar SM, Cardell S. Diverse CD1d-restricted T cells: diverse phenotypes, and diverse functions. Semin Immunol. 2000;12(6):551–60. doi: 10.1006/smim.2000.0273. [DOI] [PubMed] [Google Scholar]
- 13.Stenstrom M, Skold M, Ericsson A, Beaudoin L, Sidobre S, Kronenberg M, Lehuen A, Cardell S. Surface receptors identify mouse NK1.1+ T cell subsets distinguished by function and T cell receptor type. Eur J Immunol. 2004;34(1):56–65. doi: 10.1002/eji.200323963. 10.1002/eji.200323963. [DOI] [PubMed] [Google Scholar]
- 14.Duarte N, Stenstrom M, Campino S, Bergman ML, Lundholm M, Holmberg D, Cardell SL. Prevention of diabetes in nonobese diabetic mice mediated by CD1d-restricted nonclassical NKT Cells. J Immunol. 2004;173(5):3112–8. doi: 10.4049/jimmunol.173.5.3112. [DOI] [PubMed] [Google Scholar]
- 15.Giorda R, Weisberg EP, Ip TK, Trucco M. Genomic structure and strain-specific expression of the natural killer cell receptor NKR-P1. J Immunol. 1992;149(6):1957–63. [PubMed] [Google Scholar]
- 16.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192(5):741–54. doi: 10.1084/jem.192.5.741. 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191(11):1895–903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gumperz JE, Roy C, Makowska A, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12(2):211–21. doi: 10.1016/s1074-7613(00)80174-0. 10.1016/S1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 19.Makowska A, Kawano T, Taniguchi M, Cardell S. Differences in the ligand specificity between CD1d-restricted T cells with limited and diverse T-cell receptor repertoire. Scand J Immunol. 2000;52(1):71–9. doi: 10.1046/j.1365-3083.2000.00754.x. 10.1046/j.1365-3083.2000.00754.x. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17(1):19–29. doi: 10.1016/s1074-7613(02)00333-3. 10.1016/S1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama WM, Ryan JC, Hunter JJ, Smith HR, Stark M, Seaman WE. cDNA cloning of mouse NKR-P1 and genetic linkage with LY-49. Identification of a natural killer cell gene complex on mouse chromosome 6. J Immunol. 1991;147(9):3229–36. [PubMed] [Google Scholar]
- 22.Hammond KJ, Pellicci DG, Poulton LD, Naidenko OV, Scalzo AA, Baxter AG. Godfrey DI. CD1d-restricted NKT cells: an interstrain comparison. J Immunol. 2001;167(3):1164–73. doi: 10.4049/jimmunol.167.3.1164. [DOI] [PubMed] [Google Scholar]
- 23.Poulton LD, Smyth MJ, Hawke CG, Silveira P, Shepherd D, Naidenko OV, Godfrey DI, Baxter AG. Cytometric and functional analyses of NK and NKT cell deficiencies in NOD mice. Int Immunol. 2001;13(7):887–96. doi: 10.1093/intimm/13.7.887. 10.1093/intimm/13.7.887. [DOI] [PubMed] [Google Scholar]
- 24.Scalzo AA, Lyons PA, Fitzgerald NA, Forbes CA, Shellam GR. The BALB.B6-Cmv1r mouse: a strain congenic for Cmv1 and the NK gene complex. Immunogenetics. 1995;41(2–3):148–51. doi: 10.1007/BF00182328. [DOI] [PubMed] [Google Scholar]
- 25.Skold M, Stenstrom M, Sidobre S, Hoglund P, Kronenberg M, Cardell S. MHC-dependent and -independent modulation of endogenous Ly49 receptors on NK1.1+ T lymphocytes directed by T-cell receptor type. Immunology. 2003;110(3):313–21. doi: 10.1046/j.1365-2567.2003.01741.x. 10.1046/j.1365-2567.2003.01741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296(5567):553–5. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 27.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ. Godfrey DI. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1 (–) CD4 (+) CD1d-dependent precursor stage. J Exp Med. 2002;195(7):835–44. doi: 10.1084/jem.20011544. 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coles MC, McMahon CW, Takizawa H, Raulet DH. Memory CD8 T lymphocytes express inhibitory MHC-specific Ly49 receptors. Eur J Immunol. 2000;30(1):236–44. doi: 10.1002/1521-4141(200001)30:1<236::AID-IMMU236>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 29.Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15 receptor beta-chain in natural killer 1.1+ T cell receptor-alpha beta+ cell development. J Immunol. 1997;159(12):5931–5. [PubMed] [Google Scholar]
- 30.Lantz O, Sharara LI, Tilloy F, Andersson A, DiSanto JP. Lineage relationships and differentiation of natural killer (NK) T cells: intrathymic selection and interleukin (IL)-4 production in the absence of NKR-P1 and Ly49 molecules. J Exp Med. 1997;185(8):1395–401. doi: 10.1084/jem.185.8.1395. 10.1084/jem.185.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumann J, Voyle RB, Wei BY, MacDonald HR. Cutting edge: influence of the TCR V beta domain on the avidity of CD1d: alpha-galactosylceramide binding by invariant V alpha 14 NKT cells. J Immunol. 2003;170(12):5815–9. doi: 10.4049/jimmunol.170.12.5815. [DOI] [PubMed] [Google Scholar]
- 32.Yagi J, Dianzani U, Kato H, Okamoto T, Katsurada T, Buonfiglio D, Miyoshi-Akiyama T, Uchiyama T. Identification of a new type of invariant V alpha 14+ T cells and responsiveness to a superantigen, Yersinia pseudotuberculosis-derived mitogen. J Immunol. 1999;163(6):3083–91. [PubMed] [Google Scholar]
- 33.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195(5):625–36. doi: 10.1084/jem.20011786. 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V (alpha) 24 natural killer T cells. J Exp Med. 2002;195(5):637–41. doi: 10.1084/jem.20011908. 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]