Abstract
Despite the current availability of Salmonella vaccines, typhoid fever remains a significant public health problem in developing countries. A greater understanding of T-cell activation and the development of immunological memory during Salmonella infection should lead to the development of more effective prophylactic intervention. Here, we review recent literature on the initiation, expansion and memory development of T-cell responses using the mouse model of typhoid. We pay particular attention to strategies for tracking T-cell responses in vivo and ex vivo, and suggest models to integrate some these studies.
Keywords: T cell, Salmonella, bacteria, memory, vaccination
Introduction
Salmonella enterica are Gram-negative bacteria that infect humans and animals, causing a spectrum of disease ranging from systemic infection to gastroenteritis, depending on the particular bacterial serovar and the host species infected.1 Typhoid fever is a systemic disease caused by Salmonella enterica, serovar typhi, a highly invasive enteric pathogen found almost exclusively in developing counties. According to World Health Organization estimates, the annual global incidence of typhoid fever is around 16 million cases per year, and accounts for 600 000 deaths.
Salmonella enterica serovar typhimurium (hereafter referred to as S. typhimurium) infection of susceptible mouse strains causes an invasive systemic disease that is similar in many respects to typhoid fever.2 This model is widely accepted as the best experimental system for studying human typhoid fever and has proved extremely valuable in uncovering mechanisms of innate and acquired immune resistance to intracellular pathogens.
Immune responses to Salmonella in mice
Host defence against S. typhimurium infection requires significant contributions from both the innate and acquired arms of the immune system.1,3 The initial stages of infection are characterized by an innate immune response triggered by host recognition of several microbial structures4 including pathogen-associated molecular patterns such as flagellin, lipopolysaccaride (LPS), and lipoproteins. Each of these bacterial products can induce the production of inflammatory cytokines that are likely to contribute to the initial control of Salmonella infection.5–7
Salmonella infection also induces antigen-specific CD4 T-cell, CD8 T-cell, and B-cell responses, all of which can contribute to protective immunity. Many immunodeficient mouse strains are unable to control the in vivo replication of attenuated Salmonella strains, providing a reasonable model for determining the contribution of different cell types to the primary immune response. For example, nude, T-cell receptor αβ-deficient, and major histocompatibility complex (MHC) class-II deficient, mice all succumb to infection with strains of attenuated Salmonella that are normally eradicated in wild-type mice.8–10 In marked contrast, mice lacking MHC class-I restricted T cells, display no difference, or only a mild defect in the resolution of primary infection with attenuated Salmonella.9,11 Similarly, several groups have reported that mice lacking B cells are able to control primary infection with attenuated Salmonella in a manner similar to wild-type controls.12–14 At face value, these studies would suggest that CD4 T cells are critical for resistance to Salmonella, and that other lymphocyte populations are relatively unimportant. However, these immunodeficient mouse models do not paint a true picture of the complexity of the host immune response to Salmonella. In fact, the true requirements for immunity to murine typhoid are apparent when re-infecting vaccinated mice with virulent strains of Salmonella.
It has been known for some time that vaccination of susceptible mice with attenuated strains of Salmonella provides robust immunity to re-challenge with virulent Salmonella.15 However, it has proved difficult to transfer this protective immunity to naïve recipients unless T cells and serum antibodies are both transferred.16 Similarly, MHC class-I-deficient and B-cell deficient mice resolve initial infection with vaccine strains of Salmonella but, unlike wild-type mice, this provides no protection against re-challenge with virulent Salmonella.11–14 Thus, a greater role for CD8 T cells and B cells is uncovered using a model of re-challenge with virulent Salmonella than is observed when infecting immunodeficient mice with attenuated bacteria. Therefore, despite the simplicity of the model system, it seems unlikely that infection of immunodeficient mice with slow-growing Salmonella provides an accurate model of the immune response to typhoid fever, and results should be interpreted with some caution. Together, the available data using the mouse model point to a central role for CD4 T cells in the development of protective immunity to Salmonella, and an extremely important contributory role for both CD8 T cells and B cells. Indeed, these data nicely parallel work in human typhoid infection, where Salmonella-specific CD8 T-cell and B-cell responses can be detected following exposure to attenuated Salmonella, and are likely to play an important role in mediating protective immunity.17–21
The involvement of T-cell responses in mediating protective immunity to Salmonella infection has been apparent for decades. However, a detailed examination of Salmonella-specific T-cell activation has been lacking, because in large part of restrictions in the tools that are available in this model. The limited number of defined class-I and class-II Salmonella epitopes makes any attempt to examine Salmonella-specific T cells particularly challenging.22 However, a number of recent studies have succeeded in examining the in vivo activation and expansion of CD4 T during Salmonella infection cells by using T-cell receptor (TCR) transgenic adoptive transfer systems.23 Of particular relevance is that some of these studies have examined Salmonella-specific T-cell activation in the intestine, the most likely physiological site of initial activation.
Initial activation of naïve Salmonella-specific T cells
TCR transgenic adoptive transfer systems are an experimental methodology that simply raises the frequency of circulating antigen-specific T cells above the level of detection for flow cytometric and immunohistological analysis. Such systems are arguably the best immunological tools for detailed analysis of naïve T-cell activation in vivo.24 The first study using this approach examined the activation of ovalbumin (OVA)-specific, DO11.10 transgenic CD4 T cells after subcutaneous injection of a Salmonella strain that was engineered to express OVA.25 OVA-specific T cells were found to have proliferated in the local draining lymph node and acquired the ability to secrete interferon-γ (IFN-γ) by 5 days after Salmonella-OVA infection. This study therefore validated the usefulness of this methodology for examining the activation of Salmonella-specific CD4 T cells in vivo.
A similar approach, but using a more physiological route of infection, examined the activation of DO11.10 T cells in the Peyer's patch in response to oral Salmonella–OVA infection.26 In this case, Peyer's patch OVA-specific T cells were found to expand and contract with slightly delayed kinetics compared to previous studies, perhaps reflecting a lower initial antigen load in lymphoid tissue when using the oral route. Immunohistology was used to physically locate OVA-expressing bacteria and activated OVA-specific CD4 T cells in the Peyer's patch. Interestingly, Salmonella remained in the subepithelial dome (SED) region of the tissue, while OVA-specific T cells were located exclusively in the interfollicular (IFR) T-cell area. These data imply that some form of antigen transport, from the SED to the IFR, is required to initiate T-cell activation in the Peyer's patch, as has been suggested in other model systems.27
Despite the utility and adaptability of using heterologous expression systems like Salmonella-OVA to track T-cell activation to microbial infection, there are also some limitations to this approach. The most obvious of these being that OVA is clearly not a natural microbial antigen. Thus, the data may not accurately reflect T-cell activation to a natural bacterial antigen. In order to characterize the response to a natural Salmonella epitope, a TCR transgenic adoptive transfer system for tracking CD4 T-cell responses to Salmonella flagellin was developed.28 In agreement with earlier experiments using Salmonella–OVA,26 Salmonella flagellin-specific T-cell activation was observed in the Peyer's patch after oral infection. Surprisingly, Salmonella-specific CD4 T cells were activated to express surface CD69 within 3 hr of oral infection and produced maximal levels of interleukin-2 (IL-2), 9–12 hr later.28 The rapidity of this T-cell response suggests that the natural process of Salmonella antigen acquisition, processing and presentation can be accomplished within a couple of hrs after oral infection and has implications for the mechanism of antigen presentation to Salmonella-specific T cells in the Peyer's patch.
Previous work suggested that dendritic cells in the Salmonella-infected Peyer's patch may acquire bacterial antigens from infected macrophages that had been induced to undergo apoptosis (Fig. 1a).29 However, it seems unlikely that the process of macrophage infection, apoptosis, engulfment by a myeloid dendritic cell in the SED, and subsequent T-cell activation could be accomplished within the short time frame noted in the flagellin–CD4 system. Therefore, it would be beneficial to consider some alternative models of Salmonella antigen acquisition and presentation in the Peyer's patch.
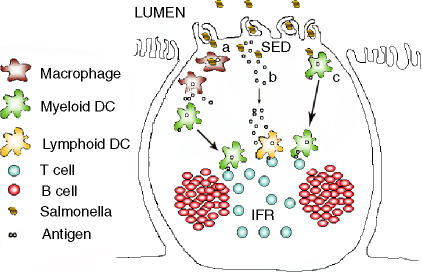
Figure 1.
Possible pathways of antigen presentation in the Peyer's patch following Salmonella infection.(a)Myeloid dendritic cells (DC) lining the subepithelial dome (SED) acquire the Salmonella antigens released by apoptotic macrophages loaded with bacteria and migrate to the interfollicular region (IFR) to effectively present the antigen to T cells.(b)Salmonella entry into the Peyer's patch leads to the release of soluble bacterial antigens into the SED. These proteins quickly disseminate to the IFR in lymph fluid, are acquired by the resident lymphoid DC population, and subsequently presented to the T cells.(c)Myeloid DCs in the SED acquire Salmonella antigens directly following bacterial entry and migrate to the IFR for efficient antigen presentation.
Perhaps the simplest model would be that soluble bacterial antigens are excreted into the SED lymph fluid, and that these flow rapidly to the IFR T-cell area without need for any cellular transport (Fig. 1b). These bacterial antigens could then be processed and presented by the resident lymphoid dendritic cell population in the IFR, already situated in close proximity to naïve T cells.30 This model has the distinct advantage in that it proposes no anatomical scampering around the Peyer's patch by either dendritic or T-cell populations, and therefore has the potential to be more rapid than models involving any cellular chemotaxis. Furthermore, movement of lymph fluid from the SED to IFR would be in general agreement with currently understood models of the anatomy and lymph circulation within the Peyer's patch.31 Adding a slightly unusual possibility to this simple model is the finding that protease digestion of Salmonella flagellin coincidentally gave rise to the exact minimal I-Ab binding epitope recognized by Salmonella flagellin-specific T cells5,32 implying that this particular T-cell epitope is relatively protease resistant. Therefore, although processing by IFR resident dendritic cells seems more likely, is remains possible that Salmonella flagellin is transported to the IFR by lymph, processed by undefined extracellular proteases, to rapidly give rise to the required I-Ab binding epitope recognized by flagellin-specific T cells. Whatever the exact mechanism of peptide generation, the rapid movement of bacterial antigens to the IFR in lymph fluid seems a reasonable model that would explain the kinetics of Salmonella-specific CD4 T-cell activation noted in vivo.28
An alternative possibility is that myeloid dendritic cells within the SED engulf and process Salmonella antigens and subsequently carry them to T cells within the IFR (Fig. 1c). This is similar to model-A except that it requires no intermediary macrophage infection and apoptosis. Indeed we favour this particular model of antigen presentation, as CCR6-deficient mice that lack a myeloid dendritic population underlying the SED also display deficiencies in the activation of Salmonella flagellin-specific T cells in the Peyer's patch (Salazar-Gonzalez and McSorley, unpublished observation). Another study has already reported the relocation of SED dendritic cells to the IFR in response to oral Salmonella infection, although the rapidity of this process was not examined in any detail.33 As flagellin itself has intrinsic proinflammatory properties, it may actually provide the trigger that initiates the exodus of dendritic cells from the SED.5,34–37 In agreement with this hypothesis, flagellin stimulation of intestinal epithelial cells in vitro was found to influence the migration of dendritic cells through production of the chemokine CCL20.38 To follow this idea further, it is worth considering the possibility that flagellin epitopes are preferentially processed and presented compared to other Salmonella proteins which lack proinflammatory properties.
It should be emphasized that each of these models of Salmonella antigen presentation in the Peyer's patch are not mutually exclusive, and the only natural Salmonella epitope that has been examined to date is found within a highly expressed, secreted antigen with unusual pro-inflammatory properties.37,39 Future experiments will determine to what extent some combination of each of these models accounts for Salmonella-specific T-cell activation in the Peyer's Patch.
Expansion of Salmonella-specific effector T cells
Although Salmonella-specific CD4 T-cell activation is quickly initiated in mucosal lymphoid tissues, bacteria are able to escape this lymphoid tissue and penetrate to systemic sites, most notably the spleen, liver, and bone marrow.40 In the face of intracellular microbial replication and dissemination, it is vitally important for the host that Salmonella-specific T cells are promptly expanded and acquire effector functions. A number of recent studies have examined the expansion of polyclonal T-cell responses following Salmonella infection. Experiments by Mittrucker et al. noted the surprising finding that the majority of splenic CD4 and CD8 T cells in Salmonella-infected resistant mice display an activated phenotype.41 Furthermore, many of these cells had gained the capacity to secrete IFN-γ in response to re-stimulation,41 a known property of effector T cells.42 In agreement with these data, other reports have described large numbers of Salmonella-specific CD4 and CD8 T cells following infection of susceptible mice with attenuated Salmonella.43–45 Interestingly, these studies suggest that the peak frequency of Salmonella-specific CD4 T cells may be greater than 50% of all CD4 T cells.44 Although such numbers are not unusual when assessing the CD8 response to viral infection46,47 previous estimates of the clonal burst size of CD4 T cells have been much lower in other models.48–50 Furthermore, one study has reported that CD4 T cells and CD8 T cells have intrinsic differences in their proliferative capacity.51 However, the available data from Salmonella infection demonstrate that massive peak CD4 responses can be detected in vivo. Whether this large response actually represents extensive proliferation of Salmonella-specific naïve T cells, or the recruitment of an unusually large number of pre-existing pathogen-specific T cells, remains to be determined. It is possible that an elevated frequency of Salmonella-specific T cells will be found in the T-cell repertoire of uninfected mice due to cross-reactivity between Salmonella and endogenous gut flora antigens, although this has yet to be demonstrated experimentally.
Numerous studies have described the development of CD4 and CD8 T cells that secrete IFN-γ during Salmonella infection.52,53 Indeed, one characteristic of the large frequency of Salmonella-specific CD4 T cells noted in recent studies is that many of these cells are able to produce IFN-γex vivo.41,43–45 These data underline the importance of IFN-γ production to the generation of protective immunity in this model.9 In addition to the development of effector cytokine production, many of these activated Salmonella-specific T cells acquire the capacity to migrate to non-lymphoid tissues such as the liver,45 a major site of bacterial replication. Thus, during infection, a Salmonella-specific effector population expands rapidly in lymphoid tissues and redistributes effectively to the major non-lymphoid sites of bacterial infection. However, it should be noted that most of these studies have examined the effector function and migration of Salmonella-specific T cells in response to attenuated bacteria, and the T-cell response to virulent bacteria may differ. Indeed, a deficiency in the migration of Salmonella-specific T cells to non-lymphoid tissues was previously noted in mice infected with virulent Salmonella.28 However, it is extremely difficult to examine T-cell effector function and migration when using a mouse model of acute infection that is rapidly fatal. Perhaps future studies using virulent Salmonella and antibiotic treatment will clarify whether T-cell effector function and migration is compromised following infection with virulent bacteria.
Given the reported magnitude of the polyclonal Salmonella-specific T-cell response41,44,45 one might imagine that it would be easy to generate class-I and class-II tetramer reagents to track Salmonella-specific responses in vivo. However, as already noted above, the number of naturally defined class-I and class-II epitopes in the mouse model of Salmonella infection is somewhat limited. For CD4 T cells there are only three defined Salmonella epitopes, I-Ak/FliC 339-50,54 I-Ab/FliC 427-41,32 and I-Ad/SipC 381-94.55 For CD8 T cells, two Kb peptide epitopes from outer membrane protein C56 and an epitope presented by the class-Ib molecule Qa-157 have been described. Aside from the limited number of epitopes to choose from, the frequency of endogenous T-cell responses to each of these defined epitopes is either untested54–57 or found to be surprisingly low.32 Therefore, identification of new class-I and class-II Salmonella epitopes, or better characterization of existing epitopes, is required for the generation of tetramer reagents and a more complete analysis of the endogenous T-cell response to Salmonella infection.
It is not yet clear whether the large polyclonal population of Salmonella-specific T cells consist of a small number of highly expanded clones responding to a few major antigen specificities, or many small pools representing numerous different clonotypes. The antibody response in Salmonella-infected mice is clearly directed against many different target antigens, including flagellin, lipoproteins, and LPS.58 It seems likely that the T-cell response will be similarly diverse. Flagellin remains the most consistently identified target antigen of Salmonella-specific CD4 T cells in vivo, and can confer limited protective immunity when used in a subunit vaccine formulation.32,54,59,60 The most thoroughly studied CD8 T-cell response in the mouse model is directed against a GroEL peptide presented by the MHC class Ib molecule Qa-1.11,57 CD8 T cells responding to GroEL display an interesting cross-reactivity to a peptide derived from mouse heat-shock protein-60, although the functional significance of this cross-reactivity for Salmonella infection is unclear. Recent innovative work has managed to identify five new Salmonella proteins that are controlled by highly expressed, in vivo inducible promoters.61 The rationale for this approach was that by identifying Salmonella proteins with these attributes, the most likely targets for recognition by the adaptive immune response would also be discovered. Indeed, this study demonstrated that two of these proteins, Mig-14 and SseB can mediate protective immunity when used as a subunit vaccine. It therefore seems likely that T cells responding to these antigens will comprise a fraction of the large Salmonella-specific CD4 cell response described above.41,44,45 However, at present the Salmonella target antigens recognized by T cells remain incompletely defined.
The role of B cells in relation to the expansion of Salmonella-specific T cells remains unclear. Aside from the requirement for B cells in mediating protective immunity, it has also been reported that B-cell antigen presentation to T cells can augment the Salmonella-specific CD4 response.12,62 During primary Salmonella infection, Salmonella-specific T cells initially expand within the T-cell area of lymphoid tissues and are only observed to penetrate B-cell follicles at a later stage.23,28 Therefore, anatomically, B-cell antigen presentation to T cells is likely be a secondary event to the initial activation mediated by dendritic cells in the T-cell area of lymphoid tissues. Furthermore, although vaccinated B-cell deficient mice succumb to infection with virulent Salmonella, they also display a reasonable degree of immunity when re-exposed to attenuated Salmonella.13 The most likely explanation for this protective immunity is that a Salmonella-specific effector T-cell response can be generated in the absence of B cells, although it is possible that it may be somewhat compromised when compared to wild-type mice. It would also seem likely that B-cell presentation to T cells would be most clearly observed during a secondary infection, where Salmonella-specific B cells are at an elevated frequency, or following high-dose Salmonella challenge, when bacteria have been found to associate more readily with B cells.43 However, at present these issues remain incompletely resolved experimentally and it is possible that B cells contribute to antigen presentation during Salmonella infection.
Salmonella-specific memory T cells
Long-term immunity to pathogens is believed to reside within the memory pool of T and B lymphocytes that can persist for the lifetime of an individual.63 Despite the current availability of live attenuated, and polysaccaride vaccines, typhoid fever remains a significant health care problem in developing nations. Therefore, a greater understanding of the generation of immunological memory during Salmonella infection is critical to improving current vaccine strategies.
Immunization of both humans and mice with live attenuated Salmonella vaccine strains, leads to the generation of long lasting immunity and resistance to re-challenge.15,64 The ability of expanded polyclonal CD4 and CD8 T cells to secrete IFN-γ in response to Salmonella antigens persists in the mouse model for at least 6 months after vaccination (Srinivasan and McSorley, unpublished observation). Therefore, the generation of immunological memory and protective immunity can be a robust phenomenon after exposure to attenuated Salmonella.
The Salmonella flagellin-specific TCR transgenic adoptive transfer system has been used to examine the development of memory CD4 T cells in Salmonella-infected mice. In other model systems the adoptive transfer of TCR transgenic T cells inhibits the expansion of endogenous T cells with identical peptide/MHC specificity.65 However, the expansion of a large endogenous CD4 response to Salmonella actually inhibited the persistence of the SM1 T cells in vivo.44 The basis of this competition is not yet clear, and may involve competition for T-cell growth or survival factors such as IL-7.66,67 However, we favour a modification of the model of Rollenhagen et al.61 where persistent antigen presentation of Salmonella epitopes in vivo may actually select clonotypes for survival to the memory pool (Fig. 2). It has been reported that flagellin expression is rapidly down-regulated by Salmonella growing in macrophages.68 Therefore, the presentation of this particular Salmonella epitope may be limiting as the infection progresses in vivo. In the face of intense competition with a massive endogenous Salmonella-specific pool directed against other epitopes that are highly expressed in vivo, SM1 T cells may fail to be selected for survival to the memory pool. Experiments are currently underway in our laboratory to test this hypothesis. It should be noted, however, that such a model contrasts with previous work with pathogen-specific CD8 T cells, where the peak expansion frequency of a given T-cell clonotype correlates well with the memory frequency.46
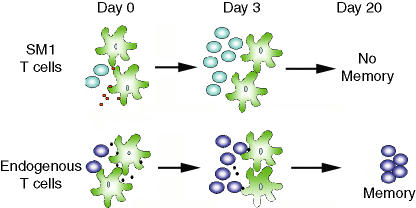
Figure 2.
A model to explain the survival of certain antigen-specific Salmonella-specific memory cells. SM1 cells are initially activated (Day 0), expand (Day 3), but fail to get selected into the memory pool because of an inadequate supply of flagellin peptide as the infection progresses in vivo. Endogenous Salmonella specific T cells directed against other antigenic epitopes expand and are successfully selected to persist because of maintained antigen presentation.
These limited observations on memory T-cell development have some implications for vaccination against typhoid fever. For example, the current dogma that live attenuated Salmonella vaccines will always provide better protective immunity than a subunit vaccine may be incorrect. The success of live attenuated vaccines may have more to do with the selective pressure placed upon the expanded Salmonella-specific T-cell pool, than with the nature of the vaccine itself. In other words, if the antigenic targets of the Salmonella-specific memory T-cell response could be elucidated, immunization with these individual proteins may have the potential to be as protective as a live attenuated strain of Salmonella. Indeed, the recent studies by Rollenhagen et al. appear to validate this hypothesis.61 This point is not trivial from a vaccine standpoint, since the live attenuated typhoid vaccine is not currently licensed in the US for children under the age of 6 years old, primarily because of safety concerns associated with a live vaccine.69,70 Therefore, the generation of a subunit vaccine that could reproduce the efficacy and long-term protection of a live Salmonella vaccine would represent a considerable advance in vaccine development, as it would be possible to target the most vulnerable demographic with prophylactic intervention.
Evasion of Salmonella-specific T cell responses
Intracellular pathogens have developed numerous strategies to gain access to the host and to survive under the constant glare of a hostile immune system. Salmonella have been variously described to actively interfere with antigen processing and presentation71–74 induce apoptosis of antigen-presenting cells29,75–79 and to generate an immunosuppressive environment in vivo.80–82
The fact that Salmonella can infect dendritic cells of diverse origin43,79,83–86 may indicate that the bacteria have the capacity to directly inhibit the priming of naïve T cells. Indeed, one elegant study recently identified the Salmonella yej operon as encoding a bacterial transporter system that interferes with MHC class-I antigen presentation in macrophages.73 Although the exact mechanism of this interference is not understood, it was proposed that this transporter system might prevent peptide loading of phagosomal MHC class I molecules by flooding the vacuole with competing short peptides. Another recent report demonstrated that Salmonella can avoid lysosomal degradation and impair the antigen presentation properties of dendritic cells.74 Interestingly, this process was blocked in the presence of Salmonella-specific immunoglobulin G, which binds FcgR on dendritic cells, and effectively targets bacteria to lysosomes. These data may shed light on the requirement for serum antibody in protective immunity to Salmonella infection. The role of antibody may actually have less to do with the opsonization and clearance of extracellular bacteria87 and more to do with the inhibition of a specific bacterial process that can interfere with naïve T-cell activation.
The notion that Salmonella may interfere with the activation of naïve T cells fits nicely with data using the Salmonella flagellin-specific adoptive transfer system. Although Salmonella-specific T cells are efficiently activated following infection, this activation is highly dose dependent.88 Indeed, flagellin specific SM1 cells were found to be totally unresponsive after low dose infection with virulent Salmonella, despite extensive bacterial replication in vivo.88 Infection with a live vaccine strain of Salmonella also demonstrates the same sensitivity to challenge dose (Srinivasan and McSorley, unpublished data). These data are reminiscent of human vaccine trials with attenuated Salmonella that are usually poorly immunogenic unless administered in multiple doses with large numbers of bacteria.64,89 It is possible that natural exposure to low numbers of bacteria avoid detection by the adaptive immune system, and that this can be attributed to some of the evasion mechanisms discussed above. If this turns out to be the case, the evasion properties of Salmonella vaccine strains clearly need to be elucidated and inhibited, as has already been described in the case of the yej operon.73
Conclusion
Salmonella induce rapid and robust T-cell activation following infection of the mammalian host. The exact processes involved in the initial activation, expansion, and memory development of Salmonella-specific T cells are beginning to be unravelled. It seems likely that the speed and magnitude of the Salmonella-specific T cell effector response has been underestimated and that this particular model may be particularly suited to understanding T-cell activation in response to infection. However, clearer identification of target antigens and an understanding of bacterial processes to inhibit T-cell activation are required in the future. Addressing these issues is likely to lead to significant improvements in current typhoid vaccine formulations, or the generation of novel typhoid vaccines that will be safer and more immunogenic than those currently available.
References
- 1.Jones BD, Falkow S. Salmonellosis: host immune responses and bacterial virulence determinants. Annu Rev Immunol. 1996;14:533–61. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 2.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 2001;3:1335–44. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 3.Mittrucker HW, Kaufmann SH. Immune response to infection with Salmonella typhimurium in mice. J Leukoc Biol. 2000;67:457–63. doi: 10.1002/jlb.67.4.457. [DOI] [PubMed] [Google Scholar]
- 4.Wick MJ. Living in the danger zone: innate immunity to Salmonella. Curr Opin Microbiol. 2004;7:51–7. doi: 10.1016/j.mib.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Gewirtz AT, Simon PO, Schmitt CK, Taylor LJ, Hagedorn CH, O'Brien AD, Neish AS, Madara JL. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J Clin Invest. 2001;107:99–109. doi: 10.1172/JCI10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tapping RI, Akashi S, Miyake K, Godowski PJ, Tobias PS. Toll-like receptor 4, but not toll-like receptor 2, is a signaling receptor for Escherichia and Salmonella lipopolysaccharides. J Immunol. 2000;165:5780–7. doi: 10.4049/jimmunol.165.10.5780. [DOI] [PubMed] [Google Scholar]
- 7.Sha J, Fadl AA, Klimpel GR, Niesel DW, Popov VL, Chopra AK. The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect Immun. 2004;72:3987–4003. doi: 10.1128/IAI.72.7.3987-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha K, Mastroeni P, Harrison J, de Hormaeche RD, Hormaeche CE. Salmonella typhimurium aroA, htrA, and AroD htrA mutants cause progressive infections in athymic (nu/nu) BALB/c mice. Infect Immun. 1997;65:1566–9. doi: 10.1128/iai.65.4.1566-1569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA-infection in gene-targeted immunodeficient mice. major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol. 1996;156:3321–6. [PubMed] [Google Scholar]
- 10.Weintraub BC, Eckmann L, Okamoto S, Hense M, Hedrick SM, Fierer J. Role of alphabeta and gammadelta T cells in the host response to Salmonella infection as demonstrated in T-cell-receptor-deficient mice of defined Ity genotypes. Infect Immun. 1997;65:2306–12. doi: 10.1128/iai.65.6.2306-2312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo WF, Ong H, Metcalf ES, Soloski MJ. T cell responses to Gram-negative intracellular bacterial pathogens. a role for CD8+ T cells in immunity to Salmonella infection and the involvement of MHC class Ib molecules. J Immunol. 1999;162:5398–406. [PubMed] [Google Scholar]
- 12.Mastroeni P, Simmons C, Fowler R, Hormaeche CE, Dougan G. Igh-6 (–/–) (B-cell-deficient) mice fail to mount solid aquired resistance to oral challenge with virulent Salmonella enterica serovar typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect Immun. 2000;68:46–53. doi: 10.1128/iai.68.1.46-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McSorley SJ, Jenkins MK. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:3344–8. doi: 10.1128/iai.68.6.3344-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittrucker HW, Raupach B, Kohler A, Kaufmann SH. Cutting edge. role of B lymphocytes in protective immunity against Salmonella typhimurium infection. J Immunol. 2000;164:1648–52. doi: 10.4049/jimmunol.164.4.1648. [DOI] [PubMed] [Google Scholar]
- 15.Hoiseth SK, Stocker BAD. Aromatic-dependent Salmonellatyphimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–9. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 16.Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Adoptive transfer of immunity to oral challenge with virulent Salmonella in innately susceptible BALB/c mice requires both immune serum and T cells. Infect Immun. 1993;61:3981–4. doi: 10.1128/iai.61.9.3981-3984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sztein MB, Tanner MK, Polotsky Y, Orenstein JM, Levine MM. Cytotoxic T lymphocytes after oral immunization with attenuated vaccine strains of Salmonella typhi in humans. J Immunol. 1995;155:3987–93. [PubMed] [Google Scholar]
- 18.Salerno-Goncalves R, Pasetti MF, Sztein MB. Characterization of CD8 (+) effector T cell responses in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol. 2002;169:2196–203. doi: 10.4049/jimmunol.169.4.2196. [DOI] [PubMed] [Google Scholar]
- 19.Lundin BS, Johansson C, Svennerholm AM. Oral immunization with a Salmonella enterica serovar typhi vaccine induces specific circulating mucosa-homing CD4 + and CD8 + T cells in humans. Infect Immun. 2002;70:5622–7. doi: 10.1128/IAI.70.10.5622-5627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salerno-Goncalves R, Wyant TL, Pasetti MF, Fernandez-Vina M, Tacket CO, Levine MM, Sztein MB. Concomitant induction of CD4+ and CD8+ T cell responses in volunteers immunized with Salmonella enterica serovar typhi strain CVD 908-htrA. J Immunol. 2003;170:2734–41. doi: 10.4049/jimmunol.170.5.2734. [DOI] [PubMed] [Google Scholar]
- 21.Salerno-Goncalves R, Fernandez-Vina M, Lewinsohn DM, Sztein MB. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol. 2004;173:5852–62. doi: 10.4049/jimmunol.173.9.5852. [DOI] [PubMed] [Google Scholar]
- 22.Srinivasan A, McSorley SJ. Visualizing the immune response to pathogens. Curr Opin Immunol. 2004;16:494–8. doi: 10.1016/j.coi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Bumann D. T cell receptor-transgenic mouse models for studying cellular immune responses to Salmonella in vivo. FEMS Immunol Med Microbiol. 2003;37:105–9. doi: 10.1016/S0928-8244(03)00064-6. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, Itano A, Pape KA. In vivo activation of antigen-specific CD4 T cells. Annu Rev Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- 25.Chen ZM, Jenkins MK. Clonal expansion of antigen-specific CD4 T cells following infection with Salmonella typhimurium is similar in susceptible (Itys) and resistant (Ityr) BALB/c mice. Infect Immun. 1999;67:2025–9. doi: 10.1128/iai.67.4.2025-2029.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bumann D. In vivo visualization of bacterial colonization, antigen expression and specific T-cell induction following oral administration of live recombinant Salmonella enterica serovar typhimurium. Infect Immun. 2001;69:4618–26. doi: 10.1128/IAI.69.7.4618-4626.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwasaki A, Kelsall BL. Mucosal immunity and inflammation. I. Mucosal dendritic cells: their specialized role in initiating T cell responses. Am J Physiol. 1999;276:G1074–8. doi: 10.1152/ajpgi.1999.276.5.G1074. [DOI] [PubMed] [Google Scholar]
- 28.McSorley SJ, Asch S, Costalonga M, Rieinhardt RL, Jenkins MK. Tracking Salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–77. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 29.Yrlid U, Wick MJ. Salmonella-induced apoptosis of infected macrophages results in presentation of a bacteria-encoded antigen after uptake by bystander dendritic cells. J Exp Med. 2000;191:613–23. doi: 10.1084/jem.191.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelsall BL, Strober W. Distinct populations of dendritic cells are present in the subepithelial dome and T cell regions of the murine Peyer's patch. J Exp Med. 1996;183:237–47. doi: 10.1084/jem.183.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelsall BL, Strober W. Gut-associated lymphoid tissue. In: Ogra PL, Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, editors. Mucosal Immunology. London: Academic Press; 1999. pp. 293–318. [Google Scholar]
- 32.McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J Immunol. 2000;164:986–93. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- 33.Shreedhar VK, Kelsall BL, Neutra MR. Cholera toxin induces migration of dendritic cells from the subepithelial dome region to T- and B-cell areas of Peyer's patches. Infect Immun. 2003;71:504–9. doi: 10.1128/IAI.71.1.504-509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyant TL, Tanner MK, Sztein MB. Salmonella typhi flagella are potent inducers of proinflammatory cytokine secretion by human monocytes. Infect Immun. 1999;67:3619–24. doi: 10.1128/iai.67.7.3619-3624.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gewirtz AT, Nava TA, Lyons S, Godowski PJ, Madera JL. Cutting Edge: Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–5. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 37.McSorley SJ, Ehst BD, Yu Y, Gewirtz AT. Bacterial flagellin is an effective adjuvant for CD4 T cells in vivo. J Immunol. 2002;169:3914. doi: 10.4049/jimmunol.169.7.3914. [DOI] [PubMed] [Google Scholar]
- 38.Sierro F, Dubois B, Coste A, Kaiserlian D, Kraehenbuhl JP, Sirard JC. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc Natl Acad Sci USA. 2001;98:13722–7. doi: 10.1073/pnas.241308598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macnab RM. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–58. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 40.Carter PB, Collins FM. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mittrucker H, Kohler A, Kaufmann SH. Characterization of the murine T-lymphocyte response to Salmonella enterica serovar typhimurium infection. Infect Immun. 2002;70:199–203. doi: 10.1128/IAI.70.1.199-203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–23. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 43.Yrlid U, Svensson M, Hakansson A, Chambers BJ, Ljunggren HG, Wick MJ. In vivo activation of dendritic cells and T cells during Salmonella enterica serovar Typhimurium infection. Infect Immun. 2001;69:5726–35. doi: 10.1128/IAI.69.9.5726-5735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasan A, Foley J, McSorley SJ. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated salmonella causes interclonal competition. J Immunol. 2004;172:6884–93. doi: 10.4049/jimmunol.172.11.6884. [DOI] [PubMed] [Google Scholar]
- 45.Kirby AC, Sundquist M, Wick MJ. In vivo compartmentalization of functionally distinct, rapidly responsive antigen-specific T-cell populations in DNA-immunized or Salmonella enterica serovar Typhimurium-infected mice. Infect Immun. 2004;72:6390–400. doi: 10.1128/IAI.72.11.6390-6400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJD, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–87. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 47.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–75. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varga SM, Welsh RM. Stability of virus-specific CD4+ T cell frequencies from acute infection into long term memory. J Immunol. 1998;161:367–74. [PubMed] [Google Scholar]
- 49.Whitmire JK, Murali-Krishna K, Altman J, Ahmed R. Antiviral CD4 and CD8 T-cell memory: differences in the size of the response and activation requirements. Philos Trans R Soc Lond B Biol Sci. 2000;355:373–9. doi: 10.1098/rstb.2000.0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med. 2001;7:913–9. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 51.Foulds KE, Zenewicz LA, Shedlock DJ, Jiang J, Troy AE, Shen H. Cutting edge. CD4 and CD8 T cells are intrinsically different in their proliferative responses. J Immunol. 2002;168:1528–32. doi: 10.4049/jimmunol.168.4.1528. [DOI] [PubMed] [Google Scholar]
- 52.Kaufmann SH. Immunity to intracellular bacteria. Annu Rev Immunol. 1993;11:129–63. doi: 10.1146/annurev.iy.11.040193.001021. [DOI] [PubMed] [Google Scholar]
- 53.Mastroeni P. Immunity to systemic Salmonella infections. Curr Mol Med. 2002;2:393–406. doi: 10.2174/1566524023362492. [DOI] [PubMed] [Google Scholar]
- 54.Cookson BT, Bevan MJ. Identification of a natural T cell epitope presented by Salmonella-infected macrophages and recognised by T cells from orally immunised mice. J Immunol. 1997;158:4310–9. [PubMed] [Google Scholar]
- 55.Musson JA, Hayward RD, Delvig AA, Hormaeche CE, Koronakis V, Robinson JH. Processing of viable Salmonella typhimurium for presentation of a CD4 T cell epitope from the Salmonella invasion protein C (SipC) Eur J Immunol. 2002;32:2664–71. doi: 10.1002/1521-4141(200209)32:9<2664::AID-IMMU2664>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 56.Diaz-Quinonez A, Martin-Orozco N, Isibasi A, Ortiz-Navarrete V. Two Salmonella OmpC K (b) -restricted epitopes for CD8+-T-cell recognition. Infect Immun. 2004;72:3059–62. doi: 10.1128/IAI.72.5.3059-3062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo WF, Woods AS, DeCloux A, Cotter RJ, Metcalf ES, Soloski MJ. Molecular mimicry mediated by MHC class Ib molecules after infection with gram-negative pathogens. Nat Med. 2000;6:215–8. doi: 10.1038/72329. [DOI] [PubMed] [Google Scholar]
- 58.Brown A, Hormaeche CE. The antibody response to salmonellae in mice and humans studied by immunoblots and ELISA. Microb Pathog. 1989;6:445–54. doi: 10.1016/0882-4010(89)90086-7. [DOI] [PubMed] [Google Scholar]
- 59.Sztein MB, Wasserman SS, Tacket CO, Edelman R, Hone D, Lindberg AA, Levine MM. Cytokine production patterns and lymphoproliferative responses in volunteers orally immunised with attenuated vaccine strains of Salmonella typhi. J Infect Dis. 1994;170:1508–17. doi: 10.1093/infdis/170.6.1508. [DOI] [PubMed] [Google Scholar]
- 60.Strindelius L, Degling Wikingsson L, Sjoholm I. Extracellular antigens from Salmonella enteritidis induce effective immune response in mice after oral vaccination. Infect Immun. 2002;70:1434–42. doi: 10.1128/IAI.70.3.1434-1442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rollenhagen C, Sorensen M, Rizos K, Hurvitz R, Bumann D. Antigen selection based on expression levels during infection facilitates vaccine development for an intracellular pathogen. Proc Natl Acad Sci USA. 2004;101:8739–44. doi: 10.1073/pnas.0401283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ugrinovic S, Menager N, Goh N, Mastroeni P. Characterization and development of T-Cell immune responses in B-cell-deficient (Igh-6 (–/–) mice with Salmonella enterica serovar Typhimurium infection. Infect Immun. 2003;71:6808–19. doi: 10.1128/IAI.71.12.6808-6819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 64.Levine MM, Ferreccio C, Black RE, Germanier R. Large-scale field trial of Ty21a live oral typhoid vaccine in enteric-coated capsule formulation. Lancet. 1987;1:1049–52. doi: 10.1016/s0140-6736(87)90480-6. [DOI] [PubMed] [Google Scholar]
- 65.Blattman JN, Anita R, Sourdive DJD, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med. 2002;195:657–64. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–8. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 67.Huster KM, Busch V, Schiemann M, Linkemann K, Kerksiek KM, Wagner H, Busch DH. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc Natl Acad Sci USA. 2004;101:5610–5. doi: 10.1073/pnas.0308054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–18. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- 69.Typhoid immunization. Recommendations of the Immunization Practices Advisory Committee (ACIP) MMWR Recomm Rep. 1990;39:1–5. [PubMed] [Google Scholar]
- 70.ACIP Updates. Recommendations for the use of antiviral agents in influenza. Advisory Committee on Immunization Practices. Am Fam Physician. 1995;52(659–660):663. [PubMed] [Google Scholar]
- 71.Pryjma J, Baran J, Ernst M, Woloszyn M, Flad HD. Altered antigen-presenting capacity of human monocytes after phagocytosis of bacteria. Infect Immun. 1994;62:1961–7. doi: 10.1128/iai.62.5.1961-1967.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitchell EK, Mastroeni P, Kelly AP, Trowsdale J. Inhibition of cell surface MHC class II expression by Salmonella. Eur J Immunol. 2004;34:2559–67. doi: 10.1002/eji.200425314. [DOI] [PubMed] [Google Scholar]
- 73.Qimron U, Madar N, Mittrucker HW, et al. Identification of Salmonella typhimurium genes responsible for interference with peptide presentation on MHC class I molecules: Deltayej Salmonella mutants induce superior CD8 T-cell responses. Cell Microbiol. 2004;6:1057–70. doi: 10.1111/j.1462-5822.2004.00418.x. [DOI] [PubMed] [Google Scholar]
- 74.Tobar JA, Gonzalez PA, Kalergis AM. Salmonella escape from antigen presentation can be overcome by targeting bacteria to Fcgamma receptors on dendritic cells. J Immunol. 2004;173:4058–65. doi: 10.4049/jimmunol.173.6.4058. [DOI] [PubMed] [Google Scholar]
- 75.Chen LM, Kaniga K, Galan JE. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–15. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 76.Monack DM, Raupach B, Hromockyj AE, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–8. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol. 2000;38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- 79.van der Velden AW, Velasquez M, Starnbach MN. Salmonella rapidly kill dendritic cells via a caspase-1-dependent mechanism. J Immunol. 2003;171:6742–9. doi: 10.4049/jimmunol.171.12.6742. [DOI] [PubMed] [Google Scholar]
- 80.al-Ramadi BK, Chen YW, Meissler JJ, Jr, Eisenstein TK. Immunosuppression induced by attenuated Salmonella. Reversal by IL-4. J Immunol. 1991;147:1954–61. [PubMed] [Google Scholar]
- 81.al-Ramadi BK, Meissler JJ, Jr, Huang D, Eisenstein TK. Immunosuppression induced by nitric oxide and its inhibition by interleukin-4. Eur J Immunol. 1992;22:2249–54. doi: 10.1002/eji.1830220911. [DOI] [PubMed] [Google Scholar]
- 82.Schwacha MG, Meissler JJ, Jr, Eisenstein TK. Salmonella typhimurium infection in mice induces nitric oxide-mediated immunosuppression through a natural killer cell-dependent pathway. Infect Immun. 1998;66:5862–6. doi: 10.1128/iai.66.12.5862-5866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hopkins SA, Niedergang F, Corthesy-Theulaz IE, Kraehenbuhl JP. A recombinant Salmonella typhimurium vaccine strain is taken up and survives within murine Peyer's patch dendritic cells. Cell Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-5822.2000.00035.x. [DOI] [PubMed] [Google Scholar]
- 84.Yrlid U, Wick MJ. Antigen presentation capacity and cytokine production by murine splenic dendritic cell subsets upon Salmonella encounter. J Immunol. 2002;169:108–16. doi: 10.4049/jimmunol.169.1.108. [DOI] [PubMed] [Google Scholar]
- 85.Jantsch J, Cheminay C, Chakravortty D, Lindig T, Hein J, Hensel M. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell Microbiol. 2003;5:933–45. doi: 10.1046/j.1462-5822.2003.00334.x. [DOI] [PubMed] [Google Scholar]
- 86.Johansson C, Wick MJ. Liver dendritic cells present bacterial antigens and produce cytokines upon Salmonella encounter. J Immunol. 2004;172:2496–503. doi: 10.4049/jimmunol.172.4.2496. [DOI] [PubMed] [Google Scholar]
- 87.Hsu HS. Pathogenesis and immunity in murine salmonellosis. Microbiol Rev. 1989;53:390–409. doi: 10.1128/mr.53.4.390-409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Srinivasan A, Foley J, Ravindran R, McSorley SJ. Low-dose salmonella infection evades activation of flagellin-specific CD4 T cells. J Immunol. 2004;173:4091–9. doi: 10.4049/jimmunol.173.6.4091. [DOI] [PubMed] [Google Scholar]
- 89.Ferreccio C, Levine MM, Rodriguez H, Contreras R. Comparative efficacy of two, three, or four doses of TY21a live oral typhoid vaccine in enteric-coated capsules: a field trial in an endemic area. J Infect Dis. 1989;159:766–9. doi: 10.1093/infdis/159.4.766. [DOI] [PubMed] [Google Scholar]