Abstract
Natural killer (NK) cells in rhesus macaques have been variably defined as CD3− CD16+ or CD3− CD8+, although only limited efforts have been made to validate these definitions rigorously. To better understand the role of NK cells in macaque disease models, we undertook a multiparameter analysis of macaque NK cells employing four-colour flow cytometry and a panel of lineage-specific and non-lineage-specific lymphocyte markers. Using this approach, we identified two distinct populations of candidate NK cells: a major CD8bright CD16+ population and a minor CD8bright CD16− population. Further analysis of the major and minor NK cell populations revealed the expression of multiple markers characteristic of NK cells, including CD2, CD7, CD16, CD161, NKG2A and granzyme B. In addition, a CD56+ subset of cells within the minor rhesus NK population was identified which expressed chemokine and lymph node homing receptors similar to those expressed by the CD56bright NK cell population identified in humans. Cytolytic assays confirmed that the phenotypically defined rhesus NK cells lysed NK-susceptible target cells. Our observations support the existence of several distinct subpopulations of rhesus macaque NK cells, which have significant phenotypic and functional similarities to their human counterparts. These improved immunophenotypic definitions of macaque NK cells should facilitate future analysis of innate immune responses in rhesus macaques and the role of NK cells in AIDS pathogenesis in Simian immunodeficiency virus (SIV)-infected macaques.
Keywords: natural killer cells, non-human primates, AIDS, cytotoxicity
Introduction
Natural killer (NK) cells are a vital component of the innate immune system, which can mediate both effector and regulatory functions. As effector cells, NK cells play key roles in host immune responses against tumours, viruses, intracellular bacteria and parasites.1 NK cells also appear to play an important role in the establishment of maternal-foetal tolerance, a mechanism directed by inhibitory NK receptors such as CD94/NKG2A,2 which recognizes the non-classical major histocompatibility complex (MHC) class I molecule human leucocyte antigen G (HLA-G).3 Through the secretion of cytokines and chemokines, NK cells can also modulate antigen presentation and the subsequent induction of antigen-specific immune responses, and thus serve as an important link between innate and adaptive immune responses.4–6
An increasing body of evidence suggests that NK cells may mediate potent inhibition of viral replication. In mice, depletion of NK cells increases susceptibility to a variety of different viral infections, including influenza and cytomegalovirus (CMV).4,7,8 A major genetic determinant of susceptibility to CMV infection in mice is the Cmv1 gene,9 which encodes the murine NK activation receptor Ly49H which recognizes the CMV m157 protein.10,11 Congenital deficiency of NK cells in humans has been associated with increased susceptibility to herpesvirus infections.12 Further evidence of the importance of NK cells in host defence against viral infections comes from the observation that multiple viruses have evolved a diverse array of mechanisms to avoid recognition of virus-infected cells by NK cells.13
Human NK cells have traditionally been defined based on their expression of CD16 and CD56. Two major populations of human NK cells have been identified: a major population of CD16bright CD56dim cells, which constitute about 90% of the total NK cell population, and a minor population of CD16–/dim CD56bright cells, which comprise about 10% of the total NK cell population.6 The CD16bright CD56dim major NK subset is naturally highly cytotoxic, but possesses little cytokine activity. These cells express high levels of immunoglobulin (Ig)-like NK receptors and contain 10-fold more perforin and granzyme A than the CD16–/dim CD56bright minor NK cell population. In contrast, the CD16–/dim CD56bright minor NK subset produces relatively large amounts of cytokines such as interferon (IFN)-γ and tumour necrosis factor (TNF)-α once activated, but has low natural cytotoxic activity. CD16–/dim CD56bright NK cells also express homing molecules and chemokine receptors such as CD62L and CCR7 which enable them to home to secondary lymphoid organs through high endothelial venules.14,15
In contrast, rhesus NK cells have been less clearly defined. Previous publications using two or three-colour flow cytometric analysis have variably defined rhesus NK cells as being CD3– CD16+16,17 or CD3– CD8+.18 CD56 has been reported not to be expressed on rhesus peripheral blood NK cells by some investigators19 but not others,20,21 and CD56+ NK cells have been identified in rhesus macaque placenta tissue.22 Only limited functional analysis of these different putative NK populations has been reported, and, to date, there has been no comprehensive multicolour phenotypic analysis of rhesus NK cells. To address these limitations, we carried out a comprehensive multiparametric immunophenotypic analysis of rhesus macaque NK cells utilizing a panel of lineage-specific and non-lineage-specific lymphocyte markers. We present evidence that the phenotype of rhesus NK cells is more complex than previously appreciated. Our data suggest that several distinct populations of rhesus NK cells exist: a major NK cell population that is CD3– CD8bright CD16+ CD20–/dim, a minor population that is CD3– CD8bright CD16– CD20–/dim, and a subset of CD16–/dim CD56bright cells within the minor NK population that appears to correspond to the CD56bright human NK cell population. Purified CD3– CD8bright CD20–/dim NK cells exhibited potent cytolytic activity against NK-susceptible target cells. Taken together, our results identify multiple NK cell subsets in rhesus macaques, and they provide the basis for future experiments examining the role of NK cells in the context of Simian immunodeficiency virus (SIV) infection and other macaque models of disease.
Materials and methods
Animals
Rhesus macaques used in the study were housed at the New England Primate Research Center (NEPRC) at Harvard Medical School (Southborough, MA). Animals were maintained in accordance with the guidelines of the local institutional animal use committees, federal and state law, American Association for Accreditation of Laboratory Animal Care (AALAAC) regulations, and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
Human blood donors
Heparinized blood samples were obtained from healthy volunteers under protocols approved by the local human studies institutional review board.
Antibodies and flow cytometric analysis
The following directly conjugated mouse antihuman monoclonal antibodies (mAbs) known to cross-react with rhesus monkey antigens were obtained from BD Biosciences (San Diego, CA): CD2 PE (RPA-2·10); CD3 FITC or PE (SP34); CD7 PE (M-T701); CD8α PerCP or APC (SK1 and SK2); CD14 FITC (M5E2), CD16 FITC or PE (3G8), CD19 PE (HIB19), CD20 PerCP (L27/2H7), CD21 PE (B-ly4), CD40 FITC (5C3), CD56 PE (B159 and MY31), CD62L FITC (SK11), CD66b FITC (G10F5), CD79a PE (HM47), CD161 PE (DX12), IgM FITC (G20-127), CCR7 Pure (2H4), CXCR3 PE (1C6/CXCR3), and CXCR4 PE (12G5). NKG2A PE (Z199), CD56 PE or APC (N901; NKH-1), CD11a FITC (25·3) and CD8β PE (2ST8·5H7) were purchased from Beckman/Coulter (Miami, FL), and granzyme B PE (GB12) was purchased from Caltag (Burlingame, CA). The CD8β mAb 2ST8·5H7 recognizes an epitope that is either specific for CD8β or a conformational determinant of the CD8αβ heterodimer.23,24 As an additional control to evaluate the significance of low-level expression of selected molecules on subpopulations of cells (e.g. CD20 on NK cells and CD8 on B cells) a flow minus one (FMO) control was performed. The FMO control consisted of the omission of the antibody of interest from a typical four-colour tube, thus allowing background fluorescence in the empty channel to be assessed on the subpopulation defined by the three remaining colours.25 In general, at least 50 000 lymphocyte events were acquired. Samples were analysed using a FACSCalibur (BD Biosciences). Files were analysed using either CELLQuest or Paint-A-Gate software (BD Biosciences).
Isolation of peripheral blood mononuclear cells (PBMC)
PBMC were isolated from fresh heparinized blood by centrifugation over a Ficoll-sodium diatrizoate (ICN Biomedicals, Aurora, OH) gradient. Cells were washed twice in 1 × phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS) (Sigma, St. Louis, MO), and resuspended at a concentration of 5 × 107 cells/ml.
Purification of NK cells
NK cells were isolated, according to the manufacturer's instructions, from PBMC by negative selection using a StemSep NK enrichment custom cocktail (Stem Cell Technologies, Vancouver, Canada) containing human-specific anti-CD3, anti-CD14, anti-CD40, and anti-CD66e tetrameric antibody complexes known to cross-react with rhesus macaque antigens. Purified cells were then cultured overnight in a 24-well plate (Costar, Corning, NY) at a concentration of 2 × 106 cells per well in 2 ml of RPMI 1640 medium supplemented with 10% FBS, 10 mm HEPES, 2 mm l-glutamine, 50 IU/ml penicillin and 50 µg/ml streptomycin (R10). The purity of the cell populations was determined by flow cytometric analysis of the cells for their expression of CD3, CD8 and CD20. NK cells (CD3– CD8bright CD20–/dim) isolated in this fashion were consistently >97% pure.
Chromium release assay
Target cells used for chromium release assays consisted of autologous PBMC, K562 cells, and 721·221 cells. K562 cells are a human erythroleukaemia cell line that lacks surface MHC class I molecules, while 721·221 cells are an Epstein–Barr virus (EBV)-transformed human B-cell line that also lacks MHC class I molecules. Briefly, target cells were labelled with 50–100 µCi 51Cr (Perkin-Elmer, Boston, MA) for 1 hr, washed twice with cold RPMI, and then dispensed in triplicate (104 cells/well) for each effector:target (E:T) ratio into 96-well U-bottom plates (Costar). Chromium release was assayed after a 5-hr incubation at 37° in a 5% CO2 incubator. Plates were spun at 200 g for 10 min at 4°, after which 30 µl of supernatant was harvested from each well into individual wells of a LumaPlate-96 (Packard, Meriden, CT), and allowed to dry overnight. Emitted radioactivity was measured in a 1450 MicroBeta Plus Liquid Scintillation Counter (Wallac, Turku, Finland). Spontaneous release was measured from wells containing only target cells and medium. Maximum release was measured from wells containing target cells and 0·1% Triton X-100 (Sigma, St. Louis, MO). The per cent specific cytotoxicity was calculated as follows: [(experimental release – spontaneous release)/(maximum release – spontaneous release)] × 100. Spontaneous release of target cells was <20% in all assays. Effector-to-target ratios for which background lysis of control targets exceeded 20% were excluded from analysis. Based on examination of lysis of autologous targets, NK cell specific lysis of greater than 5% seen at more than one E:T ratio was interpreted as significant. A human NK cell clone (YTS)26 was used as a positive control for analysis of NK cell-mediated lysis.
Results
Identification of candidate rhesus NK cell populations using cluster analysis
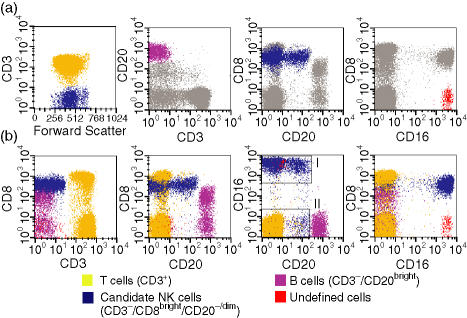
Previous studies have variably defined rhesus macaque NK cells as being CD3– CD16+16,17 or CD3– CD8+.18 However, none of these definitions has been rigorously validated using multiparameter flow cytometric analysis. As an initial step to better characterize NK cells in rhesus macaques, we identified candidate populations of NK cells based on cluster analysis,27 a process that allows identification of distinct populations of cells based on the simultaneous analysis of multiple parameters. Using a four-colour analysis of PBMC stained with antibodies to CD3, CD8α, CD16 and CD20, we first identified populations of T cells and B cells based on their bright expression of CD3 and CD20, respectively (Fig. 1a). Two distinct populations of candidate NK lymphocytes remained: one was CD3– CD8bright CD20–/dim and the other was CD3– CD8– CD16bright (Fig. 1a). Comparison of these different populations of cells with respect to each of these markers produced several notable findings. First, the CD8bright CD20–/dim population (shown in blue in Fig. 1) expressed levels of CD8α that were as bright as those found on CD8+ T cells (Fig. 1b, second panel). These cells also had a bimodal expression of CD16. The majority of CD8bright CD20–/dim cells were CD16bright (subpopulation I), while a smaller population was CD16– (subpopulation II). Subpopulation I (CD8bright CD16+) contained both CD20– and CD20dim populations (Fig. 1b), while CD20 expression on subpopulation II (CD8bright CD16–) varied from CD20– to CD20dim in different animals (Fig. 1b and data not shown). Secondly, a significant proportion of the CD20bright B-cell population expressed low to medium levels of CD8α, which in some cases overlapped significantly with levels found on CD8+ T cells and the CD3– CD8bright CD20–/dim candidate NK population (Fig. 1b, second panel). Finally, as the CD3– CD8– CD16bright population did not express high levels of CD20 and was negative for all markers except CD16, we considered the possibility that these cells (marked in red in Fig. 1) might represent an additional candidate NK cell population.
Figure 1.
Immunophenotypic identification of rhesus macaque natural killer (NK) cells using four-colour flow cytometric analysis.(a) Gating strategy used to identify T cells, B cells and candidate NK cell populations using Paint-A-Gate (BD Biosciences). First, CD3+ cells were displayed in yellow; secondly, CD3– cells were displayed in blue; thirdly, CD20bright cells were displayed in violet, and finally, CD8– CD16+ cells were displayed in red, and by default the CD3– CD8bright CD20–/dim population remained blue. Note that the top panels illustrate the sequential gating process used by Paint-A-Gate; thus, the blue cell population in the first panel on the left represents CD3– cells and not the candidate NK cell population. The grey arrow depicts lymphocyte populations excluded from the indicated gated (coloured) population in each panel.(b) Multicolour phenotypic analysis of T-cell, B-cell and candidate NK cell populations. Note the existence of two CD3– CD8brightCD20–/dim cell populations, one CD16bright (I), and the other CD16–/dim (II), both depicted in blue.
CD3− CD8+ lymphocytes contain both candidate NK cells and CD20bright B cells
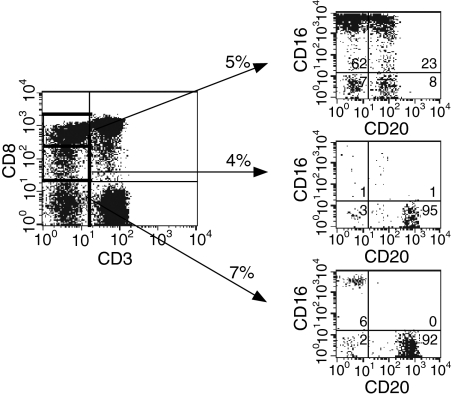
The identification of a CD20bright cell population, presumptively identified as B cells, that expressed low but significant levels of CD8α was somewhat unexpected. This finding suggested that the CD3– CD8+ population, which has been suggested by some investigators to consist predominantly of NK cells,18 contained both NK cells and B cells that varied in their expression of CD8α. We therefore carried out additional studies to subdivide the CD3– CD8+ population. As suggested by our initial analysis in Fig. 1, the population of CD3– CD8+ cells contained several subpopulations of cells defined on the basis of expression of CD16 and CD20. Subdivision of the CD3– population into CD8bright, CD8dim and CD8– subpopulations revealed differential distribution of the candidate NK and B-cell populations in each of these three populations (Fig. 2). CD3– CD8bright cells were enriched for the candidate CD16bright CD20–/dim NK population but also had significant percentages of CD16– CD20– and CD16– CD20dim cells. Analysis of CD8dim or CD8– CD3– PBMC revealed cells that were > 90% CD20bright, and these cells represented largely pure (> 90%) B-cell populations. Conversely, analysis of CD3– CD20bright cells revealed cells that were CD16– and either CD8– or CD8dim, while analysis of CD3– CD20–/dim cells demonstrated the expected candidate NK populations (data not shown).
Figure 2.
Differential distribution of CD8α on candidate rhesus natural killer (NK) and B cells. Whole blood taken from normal rhesus macaques was analysed for surface expression of CD3, CD8, CD16 and CD20. The percentage of cells within a given gate or quadrant is noted. Quadrants were established based on the use of isotype or flow minus one (FMO) controls. Results are representative of 10 macaques.
Validation of CD8bright CD20−/dim cells as NK cells in rhesus macaques
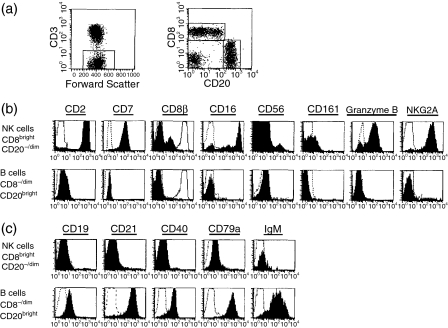
As described above, the use of cluster analysis identified a dominant candidate population of CD8bright CD20–/dim rhesus NK cells (shown in blue in Fig. 1), which could be subdivided into CD16bright and CD16dim subpopulations. We also identified a distinct candidate population of CD8– CD16bright CD20– cells. Of these populations, the CD8bright CD16+ CD20–/dim population represented the dominant population in peripheral blood (mean ± standard deviation = 15·2 ± 9·5%; n = 9), whereas the CD8bright CD16– CD20–/dim and CD8– CD16bright CD20– cells represented relatively minor populations constituting 0·6 ± 0·3% and 0·4 ± 0·3% of normal macaque PBMC, respectively. To evaluate these candidate populations of rhesus NK cells more critically, we examined the expression of a panel of lineage-specific and non-lineage-specific markers characteristic of NK cells on these cells, as well as on the CD8–/dim CD20bright B-cell population. The gating strategy used for analysis is shown in Fig. 3(a). After gating on CD3– cells, CD8bright CD20–/dim and CD8–/dimCD20bright populations were identified. Analysis of the CD8– CD16bright CD20– cells was carried out separately. As shown in Fig. 3(b), the candidate NK population of CD8bright CD20–/dim cells expressed multiple molecules found on human NK cells, including CD2, CD7, CD16, NKG2A and granzyme B.28,29 However, a subset of CD8bright CD20–/dim cells (5–30% of these cells) was identified that did not express CD16 (Fig. 3b). Staining of CD3– CD8bright CD20–/dim cells with a CD8β-specific mAb23,24 revealed dim staining on a variable subpopulation (18·1 ± 7·5%, n = 12) of cells, whereas CD3+ CD8+ T cells were CD8βbright (Fig. 3b); thus, the majority of rhesus NK cells expressed the CD8αα homodimer. Only about 35–65% of CD8bright CD20–/dim cells expressed CD161. This finding may be a consequence of the limited cross-reactivity of the human CD161-specific antibody with its rhesus homologue, as expression of CD161 on this population was monomorphic, and analysis of other populations of lymphocytes did not reveal any CD161bright cells (data not shown). Analysis of CD56 expression revealed bright expression on about 2–7% of the CD8bright CD20–/dim cells. In contrast, no significant expression of any characteristic NK molecules was observed on the CD8–/dim CD20bright B-cell population (Fig. 3b). No significant expression of CD8β was observed on CD20bright B cells (Fig. 3b), confirming that the CD8dim B-cell population expresses the CD8αα homodimer. Analysis of expression of the panel of NK and B-cell markers examined here revealed no significant expression of any of these markers on the CD8α–CD16bright population (data not shown).
Figure 3.
Analysis of CD8bright CD20–/dim and CD8–/dim CD20bright cells using lineage-specific and non-lineage-specific markers. (a) Gating strategy used to define natural killer (NK) cells (CD3–CD8bright CD20–/dim) and B cells (CD3– CD8–/dim CD20bright). Phenotypic markers that characterize NK cells (b) or B cells (c) were used to validate both the CD3– CD8bright CD20–/dim NK cell definition and the CD3– CD8–/dim CD20bright B-cell definition. Histograms for each marker (filled histogram) are shown with the respective isotype-matched control antibody (dotted lines). The CD8β histograms also show CD8β expression on CD3+ CD8α+ T cells (solid line, unfilled histograms). The results shown are representative of 10–12 macaques.
In the light of the unexpected identification of CD8 on the CD20bright population, we also wished to verify phenotypically that this population represented a true B-cell population. Analysis of characteristic B-cell markers such as CD19, CD21, CD40, CD79a and IgM revealed expression of these markers on the candidate CD8–/dim CD20bright B-cell population, but not on the candidate CD8brightCD20–/dim NK population (Fig. 3c). Expression of B-cell markers was similar on the CD8– and CD8dim populations of CD20bright B cells (not shown).
Taken together, these findings clearly validate the working definition of rhesus macaque NK cells as CD3–CD8bright CD20–/dim cells.
Characterization of rhesus macaque NK subpopulations defined by the expression of CD16 and CD56
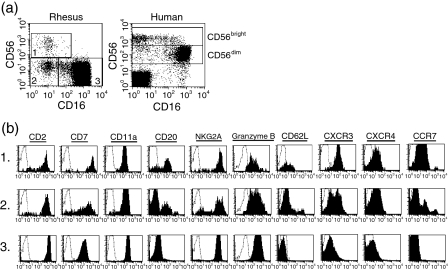
The observation that expression of CD16 and CD56 was not uniform among CD8bright CD20–/dim cells suggested the possibility that subpopulations of rhesus macaque NK cells may exist, similar to those described in humans.6,30 To address this question in more detail, we first obtained a purified population of NK cells by depleting T cells, B cells, monocytes, and granulocytes from PBMC using a cocktail of antibodies (CD3, CD14, CD40, and CD66e), and then analysed expression of a panel of markers on CD8+ cells. Analysis of CD16 and CD56 on this purified population of NK cells revealed three distinct populations of CD8bright CD20–/dim cells: (1) CD16– CD56bright, (2) CD16– CD56–, and (3) CD16+ CD56–(Fig. 4a). In purified NK cells, these populations accounted for 1·8 ± 0·9%, 3·9 ± 2·0% and 92·5 ± 4·8% (mean ± SD, n = 6) of all cells, respectively. Comparison of all three populations of rhesus NK cells with human NK cells with respect to expression of CD16 and CD56 revealed clear similarities. In both species, there was a dominant population of CD16bright cells with low or undetectable CD56 expression and a minor population of CD56bright cells that were CD16–/dim. The predominant difference between the two species was in the decreased expression of CD56 on the major CD16+ NK cell population in macaques.
Figure 4.
Expression of cell surface molecules on subpopulations of rhesus natural killer (NK) cells defined by expression of CD16 and CD56. (a) Analysis of CD16 and CD56 expression on purified NK cells from the peripheral blood of a normal monkey (obtained by immunodepletion of CD3+, CD14+, CD40+ and CD66e+ lymphocytes). Expression of CD16 and CD56 in CD3-depleted lymphocytes from peripheral blood of a normal human donor is shown for comparison. (b) Expression of cell surface markers on subpopulations of rhesus NK cells. Analysis of CD16 and CD56 was initially carried out on CD8α+ NK cells, then subdivided into three populations: (1) CD16– CD56bright, (2) CD16– CD56–, and (3) CD16+ CD56–, as shown in (a), and finally evaluated for the indicated cell surface molecules (b). The number in the left-hand margin indicates the subpopulation analysed. Histograms for each marker (solid lines) are shown with the respective isotype-matched control antibody (dotted lines). These data are representative of four macaques.
Analysis of the expression of homing molecules and chemokine receptors on subpopulations of human NK cells has demonstrated distinct patterns of expression.6,14,30 To better define the subpopulations of rhesus NK cells and to determine whether these subsets paralleled their human counterparts, we examined expression of a panel of markers, including adhesion molecules and chemokine receptors such as CD11a, CD62L, CXCR3, CXCR4 and CCR7, on rhesus NK cell populations defined based on their expression of CD16 and CD56. All three rhesus NK subpopulations expressed similar levels of CD2, CD11a and NKG2A (Fig. 4b). However, differences were observed with respect to expression of the other markers examined in these subpopulations, and in general the two minor CD16– subpopulations (both CD56– and CD56+) appeared similar to one another and distinct from the CD16bright population. Rhesus CD16– NK cells expressed higher levels of CD7, CD20, CD62L, CXCR3, CXCR4 and CCR7 than CD16bright NK cells, and lower levels of granzyme B. This pattern of expression is remarkably similar to that previously reported for the human CD16–/dim CD56bright NK population, which expresses increased levels of CD62L, CXCR3 and CCR7 as compared with CD16bright CD56dim NK cells.14 However, there is a bimodal distribution of CD62L and CXCR3, and even a trimodal distribution of CCR7 within the minor populations, which may indicate that these populations can be subdivided even further. The heterogeneity within these minor populations may reflect differences in the ability of the subsets to home to secondary lymphoid tissue, and/or it may signify different activation states.
Lytic activity of rhesus NK cells
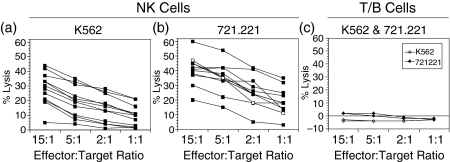
One of the cardinal features of NK cells is their ability to mediate MHC-unrestricted lysis of susceptible target cells, an activity in humans that is mediated predominantly by the dominant CD16bright population.15,31 As another approach to validate our identification of rhesus NK cells as CD8bright CD20–/dim, we examined the ability of rhesus NK cells to lyse classical NK-sensitive target cells such as K562 and 721·221 cells, both of which are MHC class I negative/low. NK cells were purified from PBMC by negative selection with CD3, CD14, CD40, and CD66e-specific antibodies. The resultant NK cell population (which was comprised of > 97% pure CD3– CD8bright CD20–/dim cells) was able to lyse both of the NK-sensitive target cell lines tested, with generally higher levels of lysis observed using 721·221 cells (Figs 5a and b). No significant lysis by NK cells was observed using 51Cr-labelled autologous PBMC as target cells (data not shown). No significant lytic activity against NK-susceptible target cells was observed using PBMC depleted of CD16+ cells and containing > 90% T and B cells (Fig. 5c). Levels of lysis of 721·221 cells at an E:T ratio of 1 : 1 frequently exceeded 20%, and were generally comparable to or higher to that mediated by a human NK cell clone, YTS (Fig. 5b). Finally, depletion of CD16+ cells from PBMC abrogated lysis of NK-sensitive target cells (data not shown).
Figure 5.
Functional assessment of natural killer (NK) cells in normal rhesus macaques using 51Cr release assays. Purified CD3– CD8bright CD20–/dim NK cells (> 97% pure) were used as effector cells in (a) (n = 12) and (b) (n = 10), while CD16-depleted PBMC from two normal macaques were used as effector cells in (c). K562 cells (a and c) and 721·221 cells (b and c) were used as target cells at varying E:T ratios as indicated. Lysis mediated by the human NK cell clone YTS against 721·221 cells is shown for comparison (open squares) in (b).
Discussion
Despite multiple reports on rhesus macaque NK cells, there has been no comprehensive effort to characterize NK cells in this species. We therefore undertook a multiparametric analysis to determine the various subpopulations of rhesus NK cells. Our results demonstrate that NK cells in rhesus macaques are CD3– CD8bright CD20–/dim, and that this population can be further subdivided into major (CD8bright CD16+ CD20–/dim) and minor (CD8bright CD16– CD20–/dim) populations. We have also shown the existence of a subset of macaque CD16– CD56+ NK cells that appears to correspond to an analogous human CD56+ NK subpopulation. Both the major and minor NK populations express lineage-specific and non-lineage-specific markers that have previously been detected on human NK cells such as CD2, CD7, CD16, NKG2A and granzyme B. In addition, as also observed in humans, within the CD16–/dim CD56+ minor NK cell population, a subset of cells expressed homing molecules (CD62L and CCR7) that facilitate homing of lymphocytes to lymph nodes through high endothelial venules. Furthermore, purified CD3– CD8bright CD20–/dim NK cells lysed NK-sensitive targets with efficiencies comparable to that of a human NK cell clone.
Previous studies have proposed a variety of different phenotypic definitions to identify rhesus macaque NK cells. The most commonly used rhesus NK cell definitions have included CD3– CD8+, CD3– CD16+ or CD3– CD8+CD16+,16–19 none of which accurately identifies all of the subpopulations of rhesus NK cells. While the CD3– CD8+ definition identifies all of the subpopulations of NK cells, it also includes contaminating B cells that express low levels of CD8α. Both the CD3– CD16+ and the CD3– CD8+ CD16+ NK cell definitions accurately identify the major macaque NK cell population, but exclude both of the minor NK populations that we have identified (CD16– CD56+ and CD16– CD56–). In addition, the CD3– CD16+ definition also includes an additional CD16+ cell population of unknown origin. Analysis of this CD3– CD8α– CD16+ population revealed no significant expression of any of a panel of 11 different NK or B-cell markers. In contrast to these previously used definitions of rhesus NK cells, the CD3– CD8+ CD20–/dim phenotypic definition that we report here (as gated in Fig. 3a) appears to be the most broad and accurate definition that can be used to identify rhesus NK cells. This definition includes both the major and minor NK cell populations, as well as the previously unrecognized minor subpopulation of CD16– CD56+ cells.
A previous report concluded that CD56 was not expressed on rhesus NK cells and was found predominantly on rhesus monocytes.19 Our data demonstrate that CD56 is indeed expressed on a subpopulation of rhesus NK cells, a finding supported by the fact that rhesus decidual NK cells have also been reported to express CD56 on their surface22 and by other previous reports.20,21 Purified CD56+ NK cells within the lymphocyte scatter gates did not express the monocyte marker CD14, whereas CD14+ monocytes were CD56bright (data not shown). Purified CD56+ NK cells also expressed lineage-specific and non-lineage-specific NK cell markers, homing molecules, and chemokine receptors similar to their human NK cell counterparts. Therefore, multiple lines of evidence suggest that the rhesus CD56+ lymphocytes represent bona fide NK cells.
Our results clearly demonstrate the expression of the CD8α homodimer on a subset of B cells. Expression of low levels of CD8αα on B cells was validated by the use of several different CD8α mAbs, the use of FMO controls, the expression of B-cell markers such as CD79a and CD40 on this population, and the lack of any staining with a CD8β-specific monoclonal antibody. In some rhesus macaques, CD8α B cells constituted up to 79% of the CD3– CD8+ lymphocytes. The expression of CD8α on human B cells has also been reported in pathological states such as B-cell chronic lymphocytic leukaemia (CLL) or HIV-1 infection.32–34 In addition, Schlesinger and colleagues noted that a small subset of B cells from HIV-negative individuals expressed CD8.34 The biological function and significance of these cells are uncertain, but their elevated frequency in disease states such as HIV-1 infection or malignancy suggests that expression is associated with lymphocyte activation.
In humans, two distinct subsets of NK cells have been described. The CD16bright CD56dim NK cells are the major NK population whose major functional activity is cell lysis, with only moderate levels of cytokine production; the minor NK cell population consists of CD16–/dimCD56bright cells, whose primary function is cytokine production, and possesses little lytic activity. In our study, we were able to show that the major rhesus NK cell population (CD16+ cells) was primarily responsible for the potent cytolytic activity against NK-sensitive targets K562 and 721·221 target cells, as revealed by the inability of CD16-depleted PBMC to lyse these target cells efficiently. We were also able to demonstrate that a subset of cells within the minor CD16– CD56–/bright population of macaque NK cells expressed molecules such as CD62L and CCR7 which may facilitate homing to lymph nodes, similar to the human CD16– CD56bright NK population. The identification of a CD56bright subset of NK cells in macaques reinforces the utility of this species as a valuable animal model that can be used for studying innate immune responses in humans, since a murine homologue of the CD56bright NK subset has not yet been identified.6 One important difference to note between the two species is in the utility of CD8 in identifying NK cells. In humans, NK cells express variable levels of CD8αα,35,38 while rhesus macaque NK cells are uniformly CD8ααbright. Interestingly, we observed dim staining with a CD8β-specific monoclonal antibody on about 18% of rhesus NK cells. Similar findings have been reported on CD8α-expressing human NK cells.35 Because CD8αα and CD8αβ differ in regard to function, especially with respect to signalling properties,39 these different subsets of NK cells may have distinct functional properties. One important consequence of the high levels of CD8α expression on rhesus NK cells is that it may be necessary to re-evaluate the specificity of anti-CD8α monoclonal antibodies as a method for depleting cytotoxic T lymphocytes in the rhesus macaque model. Our data suggest that depletion strategies utilizing anti-CD8α antibodies are also likely to result in removal of the major and minor NK cell populations, which may complicate the interpretation of the findings of these studies.40,41
In summary, our data identify several distinct subpopulations of rhesus NK cells and suggest that these cells possess both phenotypic and functional characteristics similar to those of their human counterparts. In the light of the widespread utilization of rhesus macaques as a model for human immune responses, this information should facilitate efforts to modulate the innate immune response and to better understand the role of NK cells in the pathogenesis of viral infections.
Acknowledgments
We would like to give special thanks to Stephen Dewhurst from the University of Rochester for his unconditional support, critical evaluation of the manuscript, encouragement, and guidance. We would also like to thank Amitinder Kaur for helpful discussions, George Cohen from the New England Primate Research Center for providing the YTS clone, and Kristen Toohey for her technical assistance with figure preparation. This work was supported in part by National Institute of Health grants F31 AI010536, P51 RR00168, R01 AI45314 and AI62412. R.L.W is a trainee in the Medical Scientist Training Program funded by NIH grant T32 G07356.
Abbreviations
- AALAAC
American Association for Accreditation of Laboratory Animal Care
- CMV
cytomegalovirus
- EBV
Epstein–Barr virus
- FBS
fetal bovine serum
- NIH
National Institutes of Health
- SIV
Simian immunodeficiency virus
References
- 1.Kiessling R, Klein E, Wigzell H. ‘Natural’ killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5:112–7. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- 2.Ponte M, Cantoni C, Biassoni R, et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci USA. 1999;96:5674–9. doi: 10.1073/pnas.96.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parham P. Immunology: keeping mother at bay. Curr Biol. 1996;6:638–41. doi: 10.1016/s0960-9822(09)00436-9. [DOI] [PubMed] [Google Scholar]
- 4.Biron CA. Initial and innate responses to viral infections – pattern setting in immunity or disease. Curr Opin Microbiol. 1999;2:374–81. doi: 10.1016/s1369-5274(99)80066-6. [DOI] [PubMed] [Google Scholar]
- 5.Moretta A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nature Rev Immunol. 2002;2:957–64. doi: 10.1038/nri956. [DOI] [PubMed] [Google Scholar]
- 6.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–40. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 7.Stein-Streilein J, Guffee J. In vivo treatment of mice and hamsters with antibodies to asialo GM1 increases morbidity and mortality to pulmonary influenza infection. J Immunol. 1986;136:1435–41. [PubMed] [Google Scholar]
- 8.Bukowski JF, Woda BA, Habu S, Okumura K, Welsh RM. Natural killer cell depletion enhances virus synthesis and virus-induced hepatitis in vivo. J Immunol. 1983;131:1531–8. [PubMed] [Google Scholar]
- 9.Scalzo AA, Fitzgerald NA, Wallace CR, Gibbons AE, Smart YC, Burton RC, Shellam GR. The effect of the CMV-1 resistance gene, which is linked to the natural killer cell gene complex, is mediated by natural killer cells. J Immunol. 1992;149:581–9. [PubMed] [Google Scholar]
- 10.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–6. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 11.Daniels KA, Devora G, Lai WC, O'Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Eng J Med. 1989;320:1731–5. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 13.Orange JS, Fassett MS, Koopman LA, Boyson JE, Strominger JL. Viral evasion of natural killer cells. Nat Immunol. 2002;3:1006–12. doi: 10.1038/ni1102-1006. [DOI] [PubMed] [Google Scholar]
- 14.Campbell JJ, Qin S, Unutmaz D, Soler D, Murphy KE, Hodge MR, Wu L, Butcher EC. Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J Immunol. 2001;166:6477–82. doi: 10.4049/jimmunol.166.11.6477. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, Sykora KW, Schmidt RE. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol. 2001;31:3121–6. doi: 10.1002/1521-4141(2001010)31:10<3121::aid-immu3121>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Carver FM, Thomas JM. Natural killer cells in rhesus monkeys: properties of effector cells which lyse Raji targets. Cell Immunol. 1988;117:56–69. doi: 10.1016/0008-8749(88)90076-7. [DOI] [PubMed] [Google Scholar]
- 17.Giavedoni LD, Velasquillo MC, Parodi LM, Hubbard GB, Hodara VL. Cytokine expression, natural killer cell activation, and phenotypic changes in lymphoid cells from rhesus macaques during acute infection with pathogenic simian immunodeficiency virus. J Virol. 2000;74:1648–57. doi: 10.1128/jvi.74.4.1648-1657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibegbu C, Brodie-Hill A, Kourtis AP, Carter A, McClure H, Chen ZW, Nahmias AJ. Use of human CD3 monoclonal antibody for accurate CD4+ and CD8+ lymphocyte determinations in macaques: phenotypic characterization of the CD3– CD8+ cell subset. J Med Primatol. 2001;30:291–8. doi: 10.1034/j.1600-0684.2001.300601.x. [DOI] [PubMed] [Google Scholar]
- 19.Carter DL, Shieh TM, Blosser RL, Chadwick KR, Margolick JB, Hildreth JE, Clements JE, Zink MC. CD56 identifies monocytes and not natural killer cells in rhesus macaques. Cytometry. 1999;37:41–50. [PubMed] [Google Scholar]
- 20.Savary CA, Litzova E, Jackson HJ, Jardine JH, Ang KK. Analysis of interleukin-2 activated killer cells of rhesus monkeys: striking resemblance to the human system. J Leuk Bio. 1993;54:307–13. doi: 10.1002/jlb.54.4.307. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed-Ansari A, Brodie AR, Fultz PN, Anderson DC, Sell KW, McClure HM. Flow microfluorometric analysis of peripheral blood mononuclear cells from nonhuman primates: correlation of phenotype with immune function. Am J Primatol. 1989;17:107–31. doi: 10.1002/ajp.1350170202. [DOI] [PubMed] [Google Scholar]
- 22.Slukvin II, Watkins DI, Golos TG. Phenotypic and functional characterization of rhesus monkey decidual lymphocytes: rhesus decidual large granular lymphocytes express CD56 and have cytolytic activity. J Reprod Immunol. 2001;50:57–79. doi: 10.1016/s0165-0378(00)00090-5. [DOI] [PubMed] [Google Scholar]
- 23.DiSanto JP, Knowles RW, Flomenberg N. The human Lyt-3 molecule requires CD8 for cell surface expression. Embo J. 1988;7:3465–70. doi: 10.1002/j.1460-2075.1988.tb03221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norment AM, Littman DR. A second subunit of CD8 is expressed in human T cells. Embo J. 1988;7:3433–9. doi: 10.1002/j.1460-2075.1988.tb03217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 26.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–71. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 27.Terstappen LWMM, Buescher S, Nguyen M, Reading C. Differentiation and maturation of growth factor expanded human hematopoietic progenitors assessed by multidimensional flow cytometry. Leukemia. 1992;6:1001–10. [PubMed] [Google Scholar]
- 28.Lanier LL. Follow the leader: NK cell receptors for classical and nonclassical MHC class I. Cell. 1998;92:705–7. doi: 10.1016/s0092-8674(00)81398-7. [DOI] [PubMed] [Google Scholar]
- 29.Schanberg LE, Lee DM, Fleenor DE, Ware RE, Patel DD, Haynes BF, Kaufman RE. Characterization of human CD7 transgenic mice. J Immunol. 1995;155:2407–18. [PubMed] [Google Scholar]
- 30.Sedlmayr P, Schallhammer L, Hammer A, Wilders-Truschnig M, Wintersteiger R, Dohr G. Differential phenotypic properties of human peripheral blood CD56dim+ and CD56bright+ natural killer cell subpopulations. Int Arch Allergy Immunol. 1996;110:308–13. doi: 10.1159/000237321. [DOI] [PubMed] [Google Scholar]
- 31.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, Carson WE, Caligiuri MA. Human natural killer cells: a unique innate immunoregulatory role for the CD56 (bright) subset. Blood. 2001;97:3146–51. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 32.Farkas R, Ben-Efraim S, Manor Y, Zan-Bar I, Klajman A. Appearance of the T-cell marker CD8 on B chronic lymphatic leukemia cells in long-term cultures. Cancer Immunol Immunother. 1991;34:181–5. doi: 10.1007/BF01742310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koelliker DD, Steele PE, Hurtubise PE, Flessa HC, Sheng YP, Swerdlow SH. CD8-positive B-cell chronic lymphocytic leukemia. A report of two cases. Am J Clin Pathol. 1994;102:212–6. doi: 10.1093/ajcp/102.2.212. [DOI] [PubMed] [Google Scholar]
- 34.Schlesinger M, Rabinowitz R, Levy P, Maayan S. The expression of CD8 on B lymphocytes in HIV-infected individuals. Immunol Lett. 1996;50:23–7. doi: 10.1016/0165-2478(96)02510-2. [DOI] [PubMed] [Google Scholar]
- 35.Baume DM, Caligiuri MA, Manley TJ, Daley JF, Ritz J. Differential expression of CD8 alpha and CD8 beta associated with MHC-restricted and non-MHC-restricted cytolytic effector cells. Cell Immunol. 1990;131:352–65. doi: 10.1016/0008-8749(90)90260-x. [DOI] [PubMed] [Google Scholar]
- 36.Contini P, Ghio M, Poggi A, Filaci G, Indiveri F, Ferrone S, Puppo F. Soluble HLA-A, -B, -C and -G molecules induce apoptosis in T and NK CD8+ cells and inhibit cytotoxic T cell activity through CD8 ligation. Eur J Immunol. 2003;33:125–34. doi: 10.1002/immu.200390015. [DOI] [PubMed] [Google Scholar]
- 37.Mansour I, Doinel C, Rouger P. CD16+ NK cells decrease in all stages of HIV infection through a selective depletion of the CD16+CD8+CD3– subset. AIDS Res Hum Retroviruses. 1990;6:1451–7. doi: 10.1089/aid.1990.6.1451. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz JE, Forman MA, Lifton MA, et al. Expression of the CD8alpha beta-heterodimer on CD8(+) T lymphocytes in peripheral blood lymphocytes of human immunodeficiency virus– and human immunodeficiency virus+ individuals. Blood. 1998;92:198–206. [PubMed] [Google Scholar]
- 39.Gangadharan D, Cheroutre H. The CD8 isoform CD8αα is not a functional homologue of the TCR co-receptor CD8αβ. Curr Opin Immunol. 2004;16:264–70. doi: 10.1016/j.coi.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Jin X, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–8. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitz JE, Simon MA, Kuroda MJ, et al. A nonhuman primate model for the selective elimination of CD8+ lymphocytes using a mouse-human chimeric monoclonal antibody. Am J Pathol. 1999;154:1923–32. doi: 10.1016/S0002-9440(10)65450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]