Abstract
The small GTPase accelerators regulator of G protein signalling (RGS) proteins are important regulators of proximal signalling from G protein coupled receptors. Although natural killer (NK) cells express a number of G-protein coupled receptors, expression of RGS proteins has not been investigated. We analysed the expression of RGS proteins in rat NK cells, and detected mRNA for RGS1, RGS2, RGS5, RGS8, RGS16, and RGS18. Interestingly, when we included a panel of different leucocyte subsets, we found that RGS8 was selectively expressed by NK cells. NK cells are under control of both activating and inhibitory receptors and, utilizing a xenogeneic system where the mouse activating Ly49D or inhibitory Ly49A receptors were transfected into the rat RNK-16 cell line, the potential regulation of RGS proteins by single NK cell receptors was studied. We found that ligation of Ly49D led to a rapid and transient increase in message for RGS2, while Ly49A ligation up-regulated RGS2, RGS16, and RGS18 mRNA. Both receptors also induced a prolonged increase in RGS2 endogenous protein levels. These findings suggest that RGS proteins may be influenced by or involved in NK cell receptor events, suggesting a crosstalk between G-protein coupled receptors and NK cell receptors.
Keywords: NK cells, RGS, chemokine, Ly49
Introduction
The natural killer (NK) cell is a critical component of the immune response against infected or neoplastic cells.1–3 In rodents, NK cell activity is restricted by opposing signals from activating and inhibitory receptors of the C-type lectin like receptor families, such as the Ly49, NKR-P1, and CD94/NKG2-families.4 The mouse inhibitory Ly49A and activating Ly49D receptors specifically recognize the MHC class I molecule H2-Dd.5,6 We recently demonstrated that ligation through Ly49D or Ly49A can modulate chemotaxis of NK cells in response to the chemokines CXCL10 or CXCL12, indicating that Ly49 receptors can crosstalk with chemokine receptors.7 Chemokine receptors belong to the G-protein coupled receptor family, and signal through G proteins upon binding of chemokines. In this process guanosine diphosphate (GDP) is exchanged for guanosine triphosphate (GTP) in the Gα subunit, allowing activation of the G protein, and dissociation of the signalling effectors Gα and Gβγ subunits. However, the Gα subunit possesses an intrinsic GTPase activity, which brings it back to the GDP bound state, allowing reassociation of the Gβγ subunit and thereby limiting its own activity. This process can be tightly regulated by a family of proteins called regulators of G protein signalling (RGS).
RGS proteins bind to GTP-bound forms of Gα subunits, of either the Gαi or Gαq subfamilies, and can accelerate their return to a GDP-bound state by a 100-fold, facilitating an inhibition of the signal. This interaction occurs through the RGS domain, a highly conserved region of about 120 amino acids. Most RGS proteins are relatively small, consisting mainly of the RGS domain. However, their short N- and C-terminal ends provide a level of signalling specificity.8 Most RGS proteins are predicted to be either cytosolic or nuclear, although their sites of action are presumed to be adjacent to G proteins at the plasma membrane (reviewed in 9 and 10). Despite considerable knowledge about the in vitro activities of RGS proteins, much is still unknown about their function.
RGS proteins are expressed by most cells and tissues. They are particularly abundant in the brain, but are also found in the heart, in the liver, and in lymphoid tissue.11 RGS proteins have been classified into six subfamilies. Members of the B/R4 subfamily show some preference for peripheral tissues, such as leucocytes. B cells are particularly well studied, and express a number of B/R4 subfamily RGS proteins, in particular RGS1 and RGS13.12–15 Expression of RGS proteins in monocytes and dendritic cells was also recently demonstrated.16 RGS proteins have not previously been characterized in NK cells, and we chose to study the expression of seven RGS proteins of the B/R4 subfamily in NK cells. Previous studies have shown that the B-cell receptor or Toll-like receptors (TLR) can modulate the expression and function of RGS proteins.12,16 Similarly, we assessed in this study whether stimulation through Ly49D or Ly49A affected RGS expression.
Materials and methods
Animals
Eight to 12-week-old female rats of the PVG.7B strain (which possesses a ‘non-immunogenic’ CD45 allotype, RT7b, but is otherwise interchangeable with the standard PVG strain RT7a) have been maintained at the Institute of Basic Medical Sciences for more than 20 generations. Rats were maintained under conventional conditions and regularly screened for common pathogens. The animals were housed in compliance with guidelines set by the Experimental Animal Board under the Ministry of Agriculture of Norway.
Reagents
RGS2 antibody (Ab9963) was purchased from AbCam (Cambridge, UK) and mouse monoclonal HA.11 antibody was from Nordic Biosite (Taby, Sweden). Monoclonal rat antibody towards Ly49D and Ly49A (12A8, rat immunoglobulin G (IgG)2a), monoclonal mouse 3.2.3 (anti-NKRP1A/B), and monoclonal mouse G4.18 (anti-CD3) were produced from their respective hybridoma cell lines. Secondary F(ab′)2 anti-rat F(ab′)2 was from Jackson Immunolabs (West Grove, PA).
Cells and cell lines
Rat mononuclear cells were generated by fractionating spleen cells over Lymphoprep (Nycomed Pharma, Oslo, Norway). NK cells were positively selected from the mononuclear cells using 3.2.3-coated M280 magnetic Dynabeads (Dynal Biotech, Oslo, Norway). These freshly isolated NK cells were either used directly, or cultured for 7 days in complete RPMI (cRPMI; 10% fetal calf serum, 2 mm l-glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 1 mm Na pyruvate, and 25 µm 2-mercaptoethanol, all from Invitrogen, Breda, Netherlands) supplemented with rat recombinant interleukin-2 (rIL-2, obtained from dialysed cell culture supernatant of an IL-2 gene-transfected Chinese hamster ovary cell line). T cells or B cells were positively selected from the rat mononuclear cells with either G4.18-coated or IgG-coated magnetic Dynabeads, respectively. T cells were used immediately or cultured for 3 days in cRPMI containing concanavalin A (ConA) or IL-2. RNK-16, a rat leukaemic NK cell line, 293T cells, and the rat macrophage cell line R2-MΦ were cultured in cRPMI. RNK-16.Ly49A or RNK-16.Ly49D stable transfectants were maintained in cRPMI supplemented with 1 mg/ml active G418.
Semiquantitative reverse transcription–polymerase chain reaction (RT–PCR) analysis
Total RNA was extracted using Trizol reagent (Invitrogen), and 1 µg total RNA was subsequently transcribed into cDNA using oligodT primers and the MMLV reverse transcriptase (Promega, Madison, WI). PCR was performed on 0·5 µl cDNA with specific primers for RGS proteins or CD45 as control for RNA quality. In order to semiquantify the expression levels of RGS mRNA, cycles were titrated for each RGS species to avoid saturation. The cDNA panel of rat leucocyte subsets (from PVG rats) used in Fig. 1(b) was a generous gift from Dr E. Dissen, University of Oslo17 and equal amounts of cDNA were subjected to PCR. Primers used were: RGS1: 5′-cgt-cga-caa-atg-cca-gga-atg-ttc-ttt-tc-3′, 5′-ggt-cga-cct-tta-aag-tat-ttg-cct-gaa-gg-3′, Tm 55°, 29 cycles. RGS2: 5′-cgt-cga-cga-gaa-tgc-aaa-gtg-cc-3′, 5′-ggt-cga-ctg-tag-cat-ggg-gct-ccg-3′, Tm 55°, 29 cycles. RGS5: 5′-ggggaattcaaatgtgtaagggactgg-3′, 5′-cccctctagagttgattaactccttataaaactc-3′, Tm 57°, 35 cycles. RGS8: 5′-ctc-gag-atg-tgg-aac-acc-tta-cc-3′, 5′-ctc-gag-act-gag-cct-cct-ctg-gct-ttg-3′, Tm 58°, 30 cycles. RGS13: 5′-ggg-aat-tca-aat-gag-cag-gca-tat-ctg-3′, 5′-ggg-tct-aga-aga-gct-ttg-gga-ttg-aac-3′, Tm 57°, 35 cycles. RGS16: 5′-ggt-cga-cac-cat-gtg-ccg-cac-cct-ag-3′, 5′-gtc-gac-agt-gtg-tga-agg-ctc-agc-3′, Tm 57°, 30 cycles. RGS18: 5′-ggt-cga-caa-tat-gga-tat-gtc-act-g-3′, 5′-cgt-cga-cta-acc-aaa-tgg-caa-c-3′, Tm 55°, 30 cycles. CD45: 5′-cgg-ggt-tgt-tct-gtg-ctc-tgt-tc-3′, 5′-ctt-tgc-tgt-ctt-cct-ggg-ctt-tgt-3′, Tm 67°, 25 cycles. The identity of all RGS species was verified by sequencing (GATC, Konstanz, Germany).
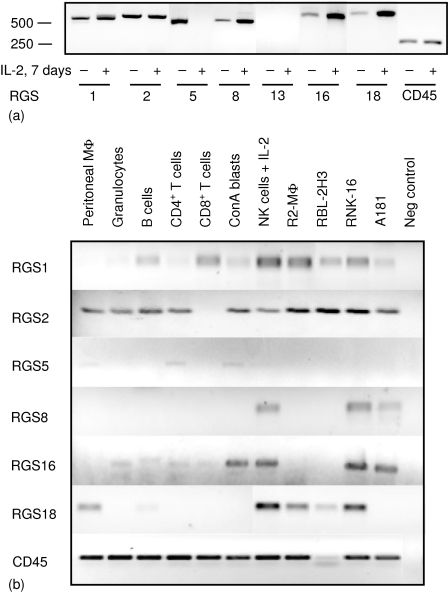
Figure 1.
Expression of RGS mRNA in rat leucocytes. (a) RT—PCR analysis of RGS transcripts performed on total RNA isolated from freshly isolated rat NK cells or rat NK cells cultured for 7 days in the presence of IL-2. Products were visualized by agarose gel electrophoresis, and length of products determined with a 100 bp ladder (Roche, Indianapolis, IN). CD45 was included as a control for equal amounts of cDNA. (b) RT—PCR analysis of RGS in freshly isolated rat haematopoietic cells from PVG rats and cell lines as indicated. R2 is a macrophage cell line, RBL-2H3 is a basophilic leukaemia, RNK-16 and A181 are NK cell lines derived from F344 rats. Negative control represents water subjected to first-strand cDNA synthesis plus PCR. The experiments were repeated twice to ensure reproducibility.
Generation of hemagglutinin-tagged RGS and transfections
Full-length cDNA encoding RGS8 was amplified by PCR from total RNA extracted from RNK-16 cells. Amplified RGS PCR products were cloned into the TA cloning vector pCR2.1 (Invitrogen). After restriction enzyme digestion and agarose gel purification, the RGS insert was further subcloned into the expression vector pECHA containing a C-terminal HA-tag (provided by Dr E. Dissen, University of Oslo18). The RGS8-HA construct in combination with an Ly49D construct was stably transfected into RNK-16 cells as previously described.19 Briefly, RNK-16 cells were electroporated with 10 µg each of ScaI-linearized RGS8-HA and Ly49D plasmids using a Bio-Rad Electroporator (Bio-Rad, Uppsala, Sweden). After electroporation, cells were cultured overnight, and then plated into 96-well plates at 104 cells/well in medium containing 1 mg/ml active G418 (Boehringer Mannheim, Mannheim, Germany). Transfectants were characterized by flow cytometry and Western blotting, and assessed for lytic activity. Cells were grown for at least 2 days without G418 prior to functional studies. 293T cells were transiently transfected with an RGS2-HA construct (generated as described above) using Lipofectamine/Opti-MEM (Invitrogen, San Diego, CA).
Stimulation of cells and lysis
For studying changes in RGS mRNA or protein mediated by Ly49 receptors, RNK-16.Ly49D or RNK-16.Ly49A cells were incubated with 1 µg 12A8 F(ab′)2 antibody per 106 cells for 30 min at 4°, and then added to 24-well plates precoated with secondary anti-rat F(ab′)2 fragments. 1 × 106 cells/sample were stimulated at 37° for 1–18 hr, then directly lysed in Trizol (Invitrogen) for RNA preparation. Alternatively cells were lysed in 30 µl NP-40 lysis buffer (1% NP-40 (Igepal-30, Sigma, St Louis, MO), 25 mm Tris-HCl, 150 mm NaCl, 10 mm NaF, 1 mm phenylmethylsulphonyl fluoride, 1 mm NaVO4) for 30 min on ice, and then centrifuged at 9500 g for 10 min at 4° to remove cell debris prior to electrophoresis.
Western blotting
Whole cell lysates, prepared as above, were separated on 12·5% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gels and electrophoretically transferred to polyvinylidene fluoride membranes. The membranes were blocked in TTBS (10 mm Tris, 154 mm NaCl, 0·05% Tween-20, pH 7·4) containing 5% skimmed milk. Incubation with antibodies for RGS2 (1 : 10 000), or HA (1 : 3000) was performed in blocking solution for 3–18 hr. Membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies (Jackson Immunolabs, 1 : 5000 in TTBS) for 1 hr. Signals were detected with the ECL system (Pierce, Rockford, IL).
Confocal imaging
RNK-16 cells or rat IL-2-activated NK cells were fixed in 4% formaldehyde (in phosphate-buffered saline (PBS)) for 10 min. 5 × 104 cells were cytospun onto microslides coated with poly l-lysine. Cells were permeabilized with 0·2% Triton-X-100 in PBS for 10 min, then incubated with rabbit anti-RGS2 (1 : 1000) or anti-HA (1 : 1000) overnight at room temperature, followed by 1 hr incubation at room temperature with specific Cy3-conjugated secondary antibodies (1 : 100). All antibody solutions were diluted with PBS supplemented with 1% bovine serum albumin. Cover slips were mounted with DakoCytomation Fluorescent Mounting Medium (DakoCytomation, Carpinteria, CA). The cells were examined using a Leica TCS confocal microscope (Leica Microsystems, Heidelberg, Germany). A 100× oil objective was used.
Results
Expression of RGS mRNA in NK cells
The expression of seven RGS proteins of the B/R4 subfamily was studied in NK cells of PVG rats. Total RNA was extracted from both freshly isolated NK cells and NK cells cultured in IL-2 for 7 days, and equal amounts of cDNA were subjected to RT–PCR analysis. Primers for RGS1, RGS2, RGS5, RGS8, and RGS13 were designed from previously published rat sequences, while primers for RGS16 and RGS18, not previously described in rat, were based on mouse sequences assuming that there is high homology between mouse and rat RGS. As members of the RGS protein family are closely related, PCR reactions were run under conditions that yielded only one band. This band was then sequenced to verify the identity of the RGS protein. We detected expression of six of the seven RGS proteins under study in NK cells; RGS1, RGS2, RGS5, RGS8, RGS16, and RGS18 (Fig. 1a). While RGS1 and RGS2 appeared to be similarly expressed in both freshly isolated and IL-2-activated NK cells, RGS5 was only found in freshly isolated cells. We have repeatedly failed to detect RGS5 in IL-2-activated NK cells, using both several different batches of cDNA and increasing the number of PCR cycles. In contrast, RGS8, RGS16, and RGS18 were all expressed at higher amounts in IL-2-activated NK cells, as compared with freshly isolated cells. RGS16 and RGS18 have not previously been described in rat, and we found that their sequences were highly homologous to the published mouse sequences. Rat RGS16 has 91% nucleotide identity with mouse RGS16, while rat RGS18 shares 93% nucleotide identity with mouse RGS18 (GenBank accession numbers AY651775 and AY651776, respectively).
We next compared the expression of these RGS proteins with other rat leucocyte subsets. To our knowledge, no studies have previously addressed RGS expression by rat leucocytes. RT—PCR analysis was performed on a cDNA panel derived from myeloid and lymphoid cells originating from the PVG rat strain, as well as NK cells from the F344 rat strain (RNK-16 and A181 cell lines; Fig. 1b). RGS1 and RGS2 were both expressed in almost all leucocytes tested, confirming previous studies in mouse and human. Curiously, RGS2 appears to be absent in CD8+ T cells. RGS5 was only weakly expressed by CD4+ T cells, ConA blasts and peritoneal macrophages, while no message was found in IL-2-activated NK cells or NK cell lines. RGS16 was detected in all lymphocyte subsets and in granulocytes, but not in macrophages or the basophilic leukaemia RBL-2H3. We detected RGS18 in macrophages, mast cells, B cells, and NK cells but not in T cells or granulocytes. The most surprising finding was the discovery that RGS8, recognized as a brain specific RGS in rat and not previously detected in immune cells, was selectively expressed only in NK cells and not in any other leucocyte subset studied.
Protein expression of RGS2
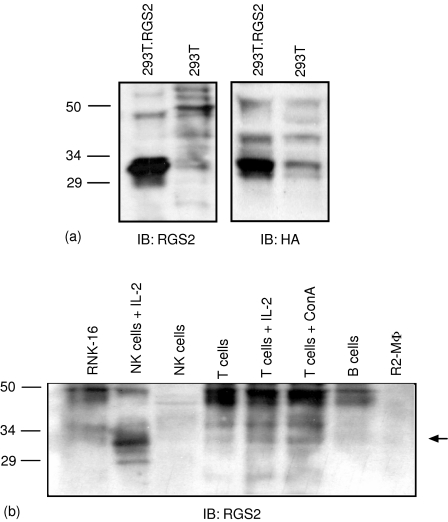
RGS proteins are expressed at low endogenous levels in resting cells, and this fact, combined with the lack of good quality antibodies, makes studies of endogenous RGS proteins difficult. We have had some success using antibodies towards RGS2, and therefore sought to study the protein expression of RGS2 in NK cells. We first determined the approximate size of rat RGS2 by transiently transfecting 293T cells with an RGS2 construct tagged to HA. We detected a band around 33 000 MW, using antibodies towards the HA-tag or RGS2 itself (Fig. 2a). This size corresponds with results from other groups, reporting a single or double band around 30 000MW20,21 although the predicted size is 24 000 MW. We next determined the expression of endogenous RGS2 protein in NK cells, and compared it with other rat leucocyte subsets (Fig. 2b). Although both freshly isolated and IL-2-cultured NK cells, as well as the RNK-16 cell line, expressed RGS2 mRNA, we failed to detect significant levels of RGS2 protein in both freshly isolated NK cells and RNK-16 cells. This also applied for the other leucocyte subsets tested. However, RGS proteins are known to be expressed at almost undetectable levels in resting cells. In contrast, NK cells cultured in the presence of IL-2 expressed high levels of RGS2, indicating that long-term activation of NK cells with IL-2 may up-regulate or stabilize RGS2 protein expression. An increase in RGS2 protein was also observed after activating T cells with either IL-2 or ConA. We consistently detected a band around 45 000 MW with the RGS2 antibody in RNK-16 cells, as well as in other cells tested, which is not present in the IL-2-activated NK cells. This pattern was also observed using another RGS2 antibody obtained from a different supplier (data not shown). We do not know the nature of this band, and it is present both in reduced and non-reduced samples (data not shown). It could either represent a modification of the protein, or a nonspecific band. Further protein analysis of other RGS proteins awaits the availability of RGS antibodies.
Figure 2.
Expression of endogenous RGS2 protein in rat leucocytes. (a) Immunoblotting of transiently transfected RGS2-HA in 293T cells, with HA antibody (right panel) and RGS2 antibody (left panel). Lysates were separated on a 12·5% SDS—PAGE gel. (b) Subsets of rat leucocytes were lysed and separated on a 12·5% SDS—PAGE gel (1·5 × 106 cells/well). Immunoblot analysis of RGS2 was performed with anti-RGS2. The approximate molecular weight of the bands is indicated. The blot is representative of four independent experiments.
Localization of RGS in NK cells
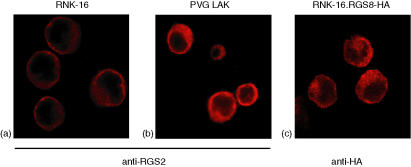
In order to further study RGS proteins in NK cells, we studied their intracellular localization by confocal microscopy. Consistent with the low protein expression of RGS2 detected by Western blotting, RNK-16 cells stained only weakly for RGS2. However, we determined its expression to be predominantly in the cytoplasm (Fig. 3a). The same localization was observed in IL-2-activated NK cells, with a clear localization to the cytoplasm, but not to the nucleus (Fig. 3b). The staining of RGS2 was much brighter in these cells, correlating with the higher amount of RGS2 protein detected by immunoblotting (Fig. 2b). We further investigated the localization of the NK-cell specific RGS8 protein. RGS8 was tagged to HA and stably transfected into RNK-16 cells. RNK-16.RGS8-HA cells were stained with an antibody towards the HA-tag, and we thus determined its localization to be predominantly in the cytoplasm (Fig. 3c), as observed for RGS2.
Figure 3.
Intracellular localization of RGS2 and RGS8. Confocal fluorescence images of (a) RNK-16 cells, and (b) PVG LAK cells stained with anti-RGS2 (1 : 1000), and (c) RNK-16.RGS8-HA cells stained with anti-HA (1 : 1000). Images were collected on a Leica TCS confocal microscope using a 100× oil immersion lens.
Ly49 receptors modulate RGS expression
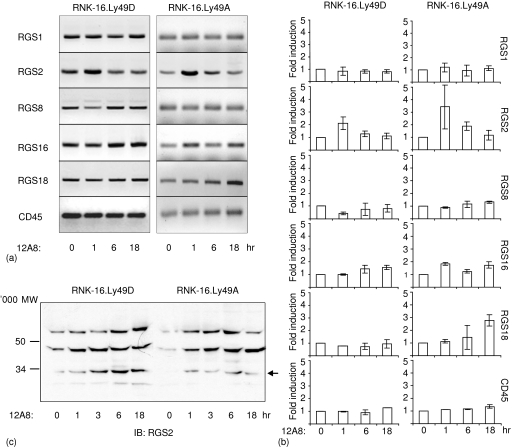
Previous studies have shown that the B-cell receptor or TLR can modulate the expression and function of RGS proteins.12,16 The activating Ly49D and inhibitory Ly49A receptors on NK cells both recognize the major histocompatibility complex class I molecule H2-Dd. However, they elicit different responses upon ligation, either activating (Ly49D) or inhibiting (Ly49A) target cell lysis.5,6,22 We investigated whether the Ly49D or Ly49A receptors could affect RGS mRNA expression in NK cells, using a xenogeneic system where either the mouse receptor Ly49D or Ly49A was stably transfected into the rat NK cell line RNK-16. In this way, we were able to ensure that only the Ly49D or the Ly49A receptor would be triggered. RNK16.Ly49A or RNK16.Ly49D cells were stimulated with crosslinking antibodies for 1–18 hr, after which RNA was extracted. RT—PCR was performed using specific primers for RGS1, RGS2, RGS8, RGS16, and RGS18. We detected a rapid twofold increase of RGS2 message within hour of crosslinking Ly49D (Fig. 4a, b). Apart from RGS2, we detected a reduction of RGS8 message to about half basal levels after 1 hr, which was restored after 6 hr (Fig. 4b). No other significant changes were observed after Ly49D stimulation. As observed for Ly49D, crosslinking the inhibitory Ly49A receptor led to a rapid increase in RGS2 message, which was even higher than after Ly49D crosslinking (Fig. 4a, b). Also, we found a threefold induction of RGS18 mRNA after 18 hr of activation, and a 1·8-fold induction of RGS16 mRNA after 1 and 18 hr (Fig. 4b). No significant changes in mRNA levels were found for RGS1 or the control CD45 after crosslinking either receptor. We also tested whether RGS5, found in freshly isolated NK cells but not in RNK-16 cells, could be up-regulated by triggering the Ly49 receptors. No mRNA for RGS5 was detected (data not shown). The effect of Ly49 activation on RGS2 protein levels was also tested. We detected an increase in the appropriately sized band (∼33 000 MW) beginning 1 hr after crosslinking Ly49D or Ly49A, which was sustained for the duration of activation, up to 18 hr of receptor cross-linking (Fig. 4c). We further tested by confocal microscopy whether stimulation of Ly49D would lead to relocalization of RGS2 from the cytoplasm, but we could detect no such changes (data not shown). Differences in RGS2 protein and mRNA baseline expression were found in RNK-16.Ly49D and RNK-16.Ly49A cells, which we believe could be a result from clonal variations of RNK-16 cells.
Figure 4.
Regulation of RGS proteins by Ly49 receptors. (a) RNK-16.Ly49D or RNK-16.Ly49A cells were stimulated with crosslinking F(ab)2 12A8 antibodies for 0–18 hr. RNA was extracted and subjected to semiquantitative RT—PCR with primers specific for various RGS proteins or CD45. PCR was performed with suboptimal cycles (described in Materials and Methods). PCR products were fractionated by agarose gels followed by ethidium bromide staining. Data shown are representative of five independent experiments. (b) Quantification of relative mRNA levels at the time points shown in (a) and two additional experiments. Relative RGS mRNA levels were determined by Kodak 1D Image Analysis software, and band intensities measured at the 0 hr time point was set to 1. Data show the fold change in RGS mRNA levels relative to the unstimulated cells. Statistical analysis by Student's t-test: RGS2 1 hr Ly49D P ≤ 0·02, RGS2 1 hr Ly49A P ≤ 0·04, RGS8 1 hr Ly49D P ≤ 0·006, RGS16 1 hr Ly49A P ≤ 0·03, RGS18 18 hr P ≤ 0·009. (c) RNK-16.Ly49D or RNK-16.Ly49A cells were stimulated for 0–18 hr with crosslinking of 12A8 F(ab)2 antibodies (12A8), and directly lysed in 2× lysis buffer. RGS2 protein expression was determined by separation on 12·5% SDS—PAGE gel and blotting with anti-RGS2 1 : 1000. Data are representative of three independent experiments.
Discussion
RGS proteins have been identified as proximal regulators of the G-protein coupled receptor signalling pathways. Although NK cells express a number of G-protein coupled receptors, such as receptors for chemokines and lysophospholipids23,24 the expression of RGS proteins in NK cells has not yet been addressed. In this paper, we demonstrate that NK cells express six members of the B/R4 subfamily of RGS proteins; RGS1, RGS2, RGS5, RGS8, RGS16, and RGS18.
We compared the expression of RGS proteins in NK cells with other leucocyte subsets from rat, as RGS proteins have not previously been studied in rat leucocytes. RGS18 was originally reported as a myeloerythroid lineage specific RGS with high expression in megakaryocytes25–27 and no expression in mature lymphoid lineages.26 In addition to macrophages and RBL-2H3, we were also able to detect expression of RGS18 in NK cells and in B cells. Others have also recently detected RGS18 in lymphocyte populations,16,28 indicating that it has a broader expression pattern than first appreciated. The most interesting finding was the selective expression of RGS8 by NK cells but not by any other leucocyte subset examined. RGS8 has previously been identified as a neuronal tissue specific protein, where it controls K+ and Ca2+ currents.28,29 NK cells account for only a small percentage of the total lymphocyte population in blood or spleen, and this may explain the previous failure to detect RGS8 in pooled lymphocytes. The significance of an NK cell specific RGS molecule is not clear at the moment, but studies are underway to further investigate the role of RGS8 in NK cells.
The intracellular localization of RGS proteins varies considerably among family members, and also between different cell types. Localization of individual RGS proteins can not be easily predicted in different cell types, and expression has been demonstrated in the nucleus, in the cytoplasm, in intracellular vesicles and at the plasma membrane.15 Ours is one of few studies that address the localization of endogenous RGS2, and we find a clear cytoplasmic localization of endogenous RGS2 in NK cells. Previous studies of RGS2 localization have indicated a nuclear localization. However, these studies were performed using RGS2-GFP transfectants20 and fusing proteins with GFP may influence subcellular localization.
RGS2 has been described as an early activation gene30 implicated in controlling the levels of the second messenger cAMP.31 In NK cells, the level of cAMP is important for proper cytotoxic responses, with high concentrations of cAMP inhibiting cytotoxicity.32 The rapid up-regulation of RGS2 message and protein by both the inhibitory Ly49A and the activating Ly49D receptor could be an important mechanism to fine tune the cAMP concentration. We have previously shown that Ly49D or Ly49A stimulation induces a decrease in chemotaxis towards the chemokine CXCL12.7 Ligation of the inhibitory Ly49A had a broad influence on RGS mRNA expression, up-regulating RGS2, RGS16 and RGS18 mRNA. It is tempting to speculate that Ly49A can indirectly inhibit GPCR events by modulating the levels of certain RGS proteins. Similar work has shown that activation through the B-cell receptor, T-cell receptor, TLR or the tumour necrosis factor-α receptor, leads to either up-regulation or down-regulation of certain RGS molecules at both the mRNA and protein levels, and may be correlated with chemotactic responses.12,15,16,33 These results, together with our own, suggest that receptors other than G-protein-coupled receptors are able to regulate the expression, and possibly function, of RGS proteins.
In conclusion, we report here the first identification of RGS proteins in NK cells. We demonstrate that both the Ly49D and Ly49A receptors regulate RGS expression. Ly49 receptor-induced changes in RGS molecule expression may function to regulate NK cell responsiveness to lymphoid chemokines and thereby help direct cell-positioning events. In the future, it will be interesting to study whether Ly49 receptors, through their actions on RGS proteins, can modulate not only chemotaxis but also other GPCR mediated events such as Ca2+ flux and cAMP generation, which are important for cytolytic activity.
Acknowledgments
The authors wish to thank Dr Henrik Huitfeldt and Jannicke Widerberg at Laboratory of Toxicopathology, Rikshospitalet University Hospital, University of Oslo for generous help with confocal imaging, and Dr Michael Daws for critically reading through the manuscript.
This work was supported by the Norwegian Cancer Society (B.R., M.I), the Norwegian Research Council (B.R., L.K), Oddrun Mjåland's fund for Cancer Research, National Institutes of Health grant RO1 AI 44126, and by the US Veterans Administration (J.C.R). M.I. is a research fellow of the Norwegian Cancer Society, and L.K. is a PhD student with the Norwegian Research Council.
References
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biron CA. Activation and function of natural killer cell responses during viral infections. Curr Opin Immunol. 1997;9:24–34. doi: 10.1016/s0952-7915(97)80155-0. [DOI] [PubMed] [Google Scholar]
- 3.Bancroft GJ. The role of natural killer cells in innate resistance to infection. Curr Opin Immunol. 1993;5:503–10. doi: 10.1016/0952-7915(93)90030-v. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–16. doi: 10.1038/nri1055. 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura MC, Linnemeyer PA, Niemi EC, Mason LH, Ortaldo JR, Ryan JC, Seaman WE. Mouse Ly-49D recognizes H-2Dd and activates natural killer cell cytotoxicity. J Exp Med. 1999;189:493–500. doi: 10.1084/jem.189.3.493. 10.1084/jem.189.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George TC, Mason LH, Ortaldo JR, Kumar V, Bennett M. Positive recognition of MHC class I molecules by the Ly49D receptor of murine NK cells. J Immunol. 1999;162:2035–43. [PubMed] [Google Scholar]
- 7.Inngjerdingen M, Rolstad B, Ryan JC. Activating and inhibitory Ly49 receptors modulate NK cell chemotaxis to CXC chemokine ligand (CXCL) 10 and CXCL12. J Immunol. 2003;171:2889–95. doi: 10.4049/jimmunol.171.6.2889. [DOI] [PubMed] [Google Scholar]
- 8.Hiol A, Davey PC, Osterhout JL, et al. Palmitoylation regulates regulators of G-protein signaling (RGS) 16 function. J Biol Chem. 2003;278:19301–8. doi: 10.1074/jbc.M210123200. 10.1074/jbc.M210123200. [DOI] [PubMed] [Google Scholar]
- 9.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. The regulator of G protein signaling family. Annu Rev Pharmacol Toxicol. 2000;40:235–71. doi: 10.1146/annurev.pharmtox.40.1.235. 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 10.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins. Regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 11.Kehrl JH. Heterotrimeric G protein signaling. roles in immune function and fine-tuning by RGS proteins. Immunity. 1998;8:1–10. doi: 10.1016/s1074-7613(00)80453-7. 10.1016/S1074-7613(00)80453-7. [DOI] [PubMed] [Google Scholar]
- 12.Reif K, Cyster JG. RGS molecule expression in murine B lymphocytes and ability to down-regulate chemotaxis to lymphoid chemokines. J Immunol. 2000;164:4720–9. doi: 10.4049/jimmunol.164.9.4720. [DOI] [PubMed] [Google Scholar]
- 13.Hong JX, Wilson GL, Fox CH, Kehrl JH. Isolation and characterization of a novel B cell activation gene. J Immunol. 1993;150:3895–904. [PubMed] [Google Scholar]
- 14.Murphy JJ, Norton JD. Multiple signaling pathways mediate anti-Ig and IL-4-induced early response gene expression in human tonsillar B cells. Eur J Immunol. 1993;23:2876–81. doi: 10.1002/eji.1830231122. [DOI] [PubMed] [Google Scholar]
- 15.Shi GX, Harrison K, Wilson GL, Moratz C, Kehrl JH. RGS13 regulates germinal center B lymphocytes responsiveness to CXC chemokine ligand (CXCL) 12 and CXCL13. J Immunol. 2002;169:2507–15. doi: 10.4049/jimmunol.169.5.2507. [DOI] [PubMed] [Google Scholar]
- 16.Shi GX, Harrison K, Han SB, Moratz C, Kehrl JH. Toll-like receptor signaling alters the expression of regulator of G protein signaling proteins in dendritic cells: Implications for G protein-coupled receptor signaling. J Immunol. 2004;172:5175–84. doi: 10.4049/jimmunol.172.9.5175. [DOI] [PubMed] [Google Scholar]
- 17.Berg SF, Fossum S, Dissen E. NILR-1, a novel immunoglobulin-like receptor expressed by neutrophilic granulocytes, is encoded by a leukocyte receptor gene complex on rat chromosome 1. Eur J Immunol. 1999;29:2000–6. doi: 10.1002/(SICI)1521-4141(199906)29:06<2000::AID-IMMU2000>3.0.CO;2-5. 10.1002/(SICI)1521-4141(199906)29:06<2000::AID-IMMU2000>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Westgaard IH, Dissen E, Torgersen KM, Lazetic S, Lanier LL, Phillips JH, Fossum S. The lectin-like receptor KLRE1 inhibits natural killer cell cytotoxicity. J Exp Med. 2003;197:1551–61. doi: 10.1084/jem.20021253. 10.1084/jem.20021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura MC, Niemi EC, Fisher M, Schultz LD, Seaman WE, Ryan JC. Mouse Ly-49A interrupts early signaling events in natural killer cell cytotoxicity and functionally associates with the SHP-1 tyrosine phosphatase. J Exp Med. 1997;185:673–84. doi: 10.1084/jem.185.4.673. 10.1084/jem.185.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowman EP, Campbell JJ, Druey KM, Scheschonka A, Kehrl JH, Butcher EC. Regulation of chemotactic and proadhesive responses to chemoattractant receptors by RGS (regulator of G-protein signaling) family members. J Biol Chem. 1998;273:28040–8. doi: 10.1074/jbc.273.43.28040. [DOI] [PubMed] [Google Scholar]
- 21.Tang M, Wang G, Lu P, et al. Regulator of G-protein signaling-2 mediates vascular smooth muscle relaxation and blood pressure. Nat Med. 2003;9:1506–12. doi: 10.1038/nm958. 10.1038/nm958. [DOI] [PubMed] [Google Scholar]
- 22.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly49+ IL-2-activated natural killer cells. Nature. 1992;66:358–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 23.Inngjerdingen M, Damaj B, Maghazachi AA. Expression and regulation of chemokine receptors in human natural killer cells. Blood. 2004;97:367–75. doi: 10.1182/blood.v97.2.367. [DOI] [PubMed] [Google Scholar]
- 24.Kveberg L, Bryceson Y, Inngjerdingen M, Rolstad B, Maghazachi AA. Sphingosine 1 phosphate induces the chemotaxis of human natural killer cells. Role for heterotrimeric G proteins and phosphoinositide 3 kinases. Eur J Immunol. 2004;32:1856–64. doi: 10.1002/1521-4141(200207)32:7<1856::AID-IMMU1856>3.0.CO;2-B. 10.1002/1521-4141(200207)32:7<1856::AID-IMMU1856>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Nagata Y, Oda M, Nakata H, Shozaki Y, Kozasa T, Todokoro K. A novel regulator of G-protein signaling bearing GAP activity for Galphai and Galphaq in megakaryocytes. Blood. 2001;97:3051–60. doi: 10.1182/blood.v97.10.3051. [DOI] [PubMed] [Google Scholar]
- 26.Yowe D, Weich N, Prabhudas M, et al. RGS18 is a myeloerythroid lineage-specific regulator of G-protein-signalling molecule highly expressed in megakaryocytes. Biochem J. 2001;359:109–18. doi: 10.1042/0264-6021:3590109. 10.1042/0264-6021:3590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagnon AW, Murray DL, Leadley RJ. Cloning and characterization of a novel regulator of G protein signalling in human platelets. Cell Signal. 2002;14:595–606. doi: 10.1016/s0898-6568(02)00012-8. 10.1016/S0898-6568(02)00012-8. [DOI] [PubMed] [Google Scholar]
- 28.Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Mol Brain Res. 2004;122:24–34. doi: 10.1016/j.molbrainres.2003.11.014. 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Saitoh O, Kubo Y, Miyatani Y, Asano T, Nakata H. RGS8 accelerates G-protein-mediated modulation of K+ currents. Nature. 1997;390:525–9. doi: 10.1038/37385. 10.1038/37385. [DOI] [PubMed] [Google Scholar]
- 30.Siderovski DP, Heximer SP, Forsdyke DR. A human gene encoding a putative basic helix-loop-helix phosphoprotein whose mRNA increases rapidly in cycloheximide-treated blood mononuclear cells. DNA Cell Biol. 1994;13:125–47. doi: 10.1089/dna.1994.13.125. [DOI] [PubMed] [Google Scholar]
- 31.Sinnarajah S, Dessauer CW, Srikumar D, et al. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–5. doi: 10.1038/35059104. 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- 32.Torgersen KM, Vaage JT, Levy FO, Hansson V, Rolstad B, Tasken K. Selective activation of cAMP-dependent protein kinase type I inhibits rat natural killer cell cytotoxicity. J Biol Chem. 1997;272:5495–500. doi: 10.1074/jbc.272.9.5495. 10.1074/jbc.272.9.5495. [DOI] [PubMed] [Google Scholar]
- 33.Fong CW, Zhang Y, Neo SY, Lin SC. Specific induction of RGS16 (regulator of G-protein signaling 16) mRNA by protein kinase C in CEM leukaemia cells is mediated via tumour necrosis factor alpha in a calcium-sensitive manner. Biochem J. 2000;352:747–53. 10.1042/0264-6021:3520747. [PMC free article] [PubMed] [Google Scholar]