Abstract
The colonic epithelium provides an interface between the host and micro-organisms colonising the gastrointestinal tract. Molecular recognition of bacteria is facilitated through Toll-like receptors (TLR). The colonic epithelium expresses relatively high levels of mRNA for TLR3 and less for TLR2 and -4. Little is known of the expression patterns and mode of induction of expression for these pattern recognition receptors in human colon. The aim of this study was to investigate their localization in the gut and induction of expression in epithelial cell lines by mucosal bacteria. TLR2 and -4 were expressed only in crypt epithelial cells, expression was lost as the cells matured and moved towards the gut lumen. In contrast, TLR3 was only produced in mature epithelial cells. HT29 and CACO-2 had different levels of expression for TLR1–4. Co-culture of HT29 cells with different mucosal isolates showed that they were highly responsive to bacterial challenge, with up-regulation of mRNA for TLR1–4. In contrast, CACO-2 cells were refractive to bacterial challenge, showing little difference in mRNA levels. TLR3 was induced in HT29 only by Gram-positive commensals with up-regulation of both mRNA and protein and an enhancement of the antiviral immune response. This pattern of expression allows induction of responsiveness to bacteria only by the crypt epithelium so that tolerance to commensal organisms can be maintained. In contrast, mature columnar epithelium is able to respond to viral pathogens, which are not part of the normal gut commensal microbiota.
Keywords: Toll-like receptors, colonic epithelium, mucosal bacteria, tolerance
Introduction
The colonic epithelial surface is constantly exposed to antigens from the normal microbiota, food antigens and pathogenic micro-organisms. Resident bacteria that colonize colonic and rectal mucosae, often occur in microcolonies on the epithelial surface.1,2 The mechanisms for molecular recognition and distinction of these commensal organisms from similar pathogenic species are unclear. Commensal species are known to display many of the same surface components as pathogenic organisms, without initiating an aggressive inflammatory immune response. Only mature epithelial cells that exit the crypts, and form the luminal surface of the mucosa, are in direct contact with the live microbiota, the crypts themselves are not colonized by the commensal flora.2
The abilities of epithelial cells to detect bacterial cellular components requires the expression of pattern recognition receptors (PRR) that recognize repetitive patterns present on Gram-positive and Gram-negative bacteria, fungi, viruses and parasites.3 Toll-like receptors (TLR) are a group of 10 human PRR that are homologous to Drosophila Toll protein.4 Specific ligands or pathogen-associated molecular patterns (PAMPs) have been attributed to a number of human TLR on different cell types.3,4 TLR2 is required for recognition of Gram-positive and mycobacterial PAMPs,5,6 including bacterial lipopeptide,7 lipoteichoic acid (LTA) and peptidoglycan (PGN).8 TLR3 recognizes double stranded viral RNA,9 TLR4 recognizes LPS,10–12 TLR5 is the PPR for flagella13 and TLR9 recognizes bacterial DNA.14 The recognition system is more complicated, however, because certain combinations of TLR are required for the detection of some PAMPs,15 TLR2 in combination with TLR6 or TLR1 is required for recognition of PGN, yeast cell wall zymosan,16 and phenol soluble modulin,17 whereas, TLR2 in association with TLR1 allows recognition of triacylated bacterial lipopeptides.18 This suggests that different TLR expressed on the surface of a cell allows it to respond to any PAMPs using varying PPR combinations. However, not all TLRs are expressed by all cell types, rather they are widely distributed in cells throughout the body, in both immune and non-immune tissues, with varying levels of expression.19 It has been previously reported that the healthy colon has relatively high levels of mRNA for TLR3, -4, -5, and -7 when compared to that of spleen, with TLR3 being the most abundant.19 These levels of expression may change with disease within the colonic mucosa, as there are reports of increased TLR2 and TLR4 during intestinal inflammation.20,21 However, this increase is due expression by the inflammatory infiltrate rather than expression by epithelial cells.21 The exact localization of TLR in human colon is unknown, but investigation of the expression patterns may give an indication of the areas of bacterial recognition in the epithelium, and yield information on how recognition of commensal and pathogen is regulated.
The aim of this investigation was to determine the expression patterns of TLR2, -3, and -4 in healthy colonic epithelia by immunohistochemistry. Then devise a model system to investigate the ability of different types of mucosal bacteria to regulate expression of these pattern recognition molecules in colonic epithelial cell lines.
Materials and methods
Human tissue
Normal human colon and rectal tissue for immunohistochemistry was obtained with ethical approval from the Ninewells Hospital Tissue Bank. The patients gave informed consent and the study was approved by the Tayside Committee on Medical Research Ethics, Dundee.
Immunohistochemistry
Tissue specimens were fixed in formalin and embedded in paraffin. Four-micron thick sections were cut using a microtome (Leica RM 2135) and mounted onto clean glass slides, which had been coated with poly L-lysine to improve tissue adherence. The slides were then dried for 1 hr at 60°, de-paraffined in Histoclear (National Diagnostics, Atlanta, GA) and rehydrated using a graded alcohol series. Endogenous peroxidase activity was blocked by treatment with 1·5% hydrogen peroxide blocking solution. Antigen retrieval was done using 1·5 m citric acid buffer, pH 6·0. Slides were boiled for 15 min, and left to cool in the antigen retrieval buffer, before being transferred to phosphate-buffered saline (PBS), and immunostained. Tissue sections were incubated in normal horse serum 25% avidin blocking agent (Vector Laboratories, Burlingame, CA) to reduce non-specific background staining, and then with 10 µg/ml mouse anti-human TLR3 (Imgenex, San Diego, CA), TLR2 or TLR4 (eBioscence, San Diego, CA) overnight at 4° in a humidified chamber. Controls were incubated with an isotype control immunoglobulin G1 (IgG1) and IgG2a antibody (eBioscience). A 25% (w/v) biotin solution (Vector Laboratories) was included in the primary antibody solution, to reduce non-specific background staining. The slides were washed in PBS and immunohistochemical (IHC) analysis was conducted using a biotin–streptavin immunoperoxidase method (ABC Elite, Vector Laboratories), and diaminobenzidine (DAB), in 0·03% hydrogen peroxide, as chromogenic agent (Dako, Ely, UK), as per the manufacturer's instructions. The slides were counterstained with Mayer's haematoxylin, washed briefly in water, dehydrated through several graded alcohol steps, before being cleared in Histoclear, and finally mounted using DPX mountant (Diachem International Ltd, Skelmersdale, UK). Sections known to stain positively were included in each batch, and negative controls were also prepared by replacing the primary antibody with PBS. The slides were viewed on a Nikon Eclipse E400 binocular microscope.
Bacterial strains and culture conditions
Six different bacterial species (Enterococcus faecalis, Escherichia coli, Peptostreptococcus anaerobius, Bifidobacterium longum, Bifidobacterium bifidum, Bacteroides fragilis) were selected from a pool of isolates obtained from rectal mucosal biopsies for initial studies. A further two E. coli, one Gemminger spp., two P. anaerobius, one Peptostreptococcus prevotii and three E. faecalis were tested to confirm the TLR3 mRNA up-regulation. The organisms were identified by chemotaxonomic methods2,22 and identity was verified by 16S rDNA gene sequence analyses, using pan bacterial specific 16S rDNA primers,23 and were maintained on Wilkins–Chalgren agar plates, in an anaerobic chamber (atmosphere 10% H2, 10% CO2, 80% N2), at 37°. For use in the present study, the bacteria were inoculated into sealed Universal bottles containing prereduced anaerobic cooked meat broth supplemented with 2% fetal calf serum and incubated overnight at 37°.
Tissue culture
The human colon adenocarcinoma cell line HT-29 (ATCC HTB 38) and CACO-2 were cultured in Dulbecco's modified Eagle's medium, supplemented with 10% heat-inactivated fetal calf serum and 2 mm l-glutamine (Invitrogen, Paisley, UK). The cells were grown in 5% CO2 at 37° to confluent monolayers in 24 well plates (Corning Inc., New York, NY). For immuno-fluorescence studies, the cells were grown on sterile microscope slides until they formed confluent monolayers.
Infection of cell monolayers
HT29 and CACO-2 monolayers were incubated with 5 × 106 bacterial cells ml−1 in the same culture medium as described above without fetal calf serum, for three hrs, in a 5% CO2 atmosphere. The bacteria were then aspirated off, and the monolayer collected. After one wash with sterile culture medium, the pellet was snap frozen in liquid nitrogen and stored at −80° for subsequent purification of RNA. Slides for immunohistochemistry were incubated for 18 hr with 5 × 106 bacteria ml−1, washed three times in PBS, air dried, and then stored frozen at −20°.
Co-culture with purified Toll-like receptor ligands
HT29 monolayers were incubated with 10 µg/ml of lipoteichoic acid (LTA) (from Streptococcus faecalis, Sigma, Poole, UK), peptidoglycan (PGN; from Staphylococcus aureus, Fluka, Deisenhofen, Germany) and poly I:C (Sigma) or 50 ng/ml of lipid A (from E. coli, Sigma) for 3 hr as previously described. The cells were harvested as before and snap-frozen for mRNA purification.
RNA and cDNA preparation
RNA was purified using the RNA easy kit (Qiagen, Hilden, Germany) with an initial clean-up step using a Qiashredder column (Qiagen), and an additional step of DNA digestion to ensure no genomic DNA contamination. The samples were reverse transcribed using the AMV reverse transcription kit (Promega, Madison, WI) as per the manufacturer's instructions, and were aliquoted before storage at −80°.
Preparation of DNA standards for quantitation of DNA
Standard amounts of DNA corresponding to the target sequences are needed to do real-time polymerase chain reaction (PCR). This was done by purifying plasmid DNA containing the target sequences. Briefly, cDNA from normal healthy colon, or mononuclear blood cells, was amplified using each PCR primer pair (5′–3′, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward GGAAGGTGAAGGTCGGAGTC, GAPDH reverse TCAGCCTTGACGGTGCCATG, TLR1 forward GAAGATTTCTTGCCACCCTAC, TLR1 reverse GAACACAATGTGCAGACTCTC, TLR2 forward CTGGACAATGCCACATAC, TLR2 reverse CTAATGTAGGTGATCCTG, TLR3 forward CACTATGCTCGATCTTTCCTAC, TLR3 reverse CAATTCAGGTACCTCACATTG, TLR4 forward CAGAACTGCAGGTGCTGG, TLR4 reverse GTTCTCTAGAGATGCTAG, interferon-β forward CGCCGCATTGACCATCTA, interferon-β reverse GACATTAGCCAGGAGGTTCTCA). The product of correct size and sequence was purified using the Qiaquick PCR purification kit (Qiagen) and ligated into a vector using the pGEM-T easy vector system I (Promega). JM109 competent E. coli (Promega) were transformed with each ligated vector, and after overnight incubation, positive colonies were chosen. From each selected colony, the plasmid was purified using the Wizard plus SV miniprep system (Promega). Concentration of the plasmid preparation was determined by agarose gel electrophoresis, with known standards (New England Biolabs, Beverly, MA). The samples were diluted to 1010 molecules µl−1 aliquoted and stored (−80°).
Quantitative real-time PCR
The appropriate plasmid preparation was diluted to give a standard curve of 106−101 molecules µl−1 for all assays except GAPDH, which had a standard curve of 108−101 molecules µl−1. Real-time PCR was done using an iCycler and iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). The annealing temperature was 56° for each reaction. Test samples were added in triplicate at 2 µl per well in a 20 µl total reaction volume. The results are expressed as the average for three separate experiments.
Immunofluorescence
Slides were fixed using ice-cold 50% (v/v) acetone/methanol (VWR International Ltd, Poole, UK) fixative at room temperature for 15 min. The slides were then blocked for 20 min at room temperature in 5% (v/v) normal rabbit serum in PBS. Ten µg ml−1 of the affinity purified goat polyclonal anti-human TLR3 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA) was added in the block buffer, and incubated overnight at 4°. The slides were washed three times in PBS, and fluoroscein isothiocyanate (FITC)-conjugated rabbit anti-goat immunoglobulins (Dako) were added (1/50 dilution in block buffer) for 1 hr at room temperature. The slides were then washed as before, and visualized using a Nikon eclipse E800 upright microscope, attached to a Nikon PCM 2000 confocal system. A 60× Plan Apo immersion lens, with a numerical aperture of 1·4 was used for all of the images. Images were captured and overlaid using C-imaging software (Compix Inc, Cranberry Township, PA). Three-dimensional images were created using ImarisSurpass 3D imaging software (Bitplane AG, Zurich, Switzerland).
Induction of antiviral response
HT29 monolayers were incubated with or without 5 × 106B. bifidum, as above, for 18 hr to induce TLR3 protein up-regulation. The cells were washed and 10 µg/ml of poly I:C was added and incubated for 3 hr. The cells were then harvested as before and levels of mRNA for interferon-β were assessed by quantitative real-time PCR. The results were expressed per 106 molecules of GAPDH as before.
Chemicals
Unless stated otherwise, all chemicals and serological reagents were purchased from Sigma. Bacteriological culture media were obtained from Oxoid (Basingstoke, UK).
Statistics
All results are expressed as a mean plus standard deviation. The statistical analyses are a standard unpaired Student's t-test with a 95% confidence interval.
Results
Expression and localization of TLR2, -3 and -4 in the normal human colon
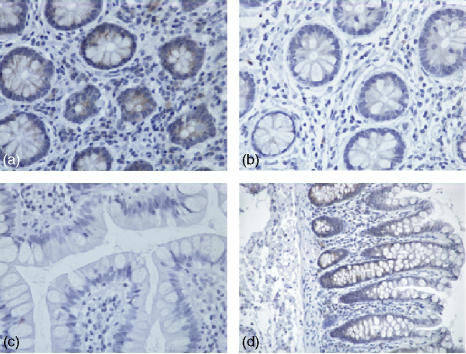
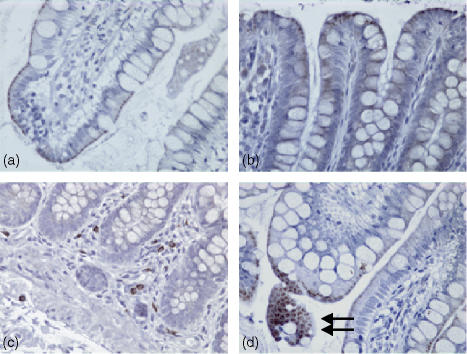
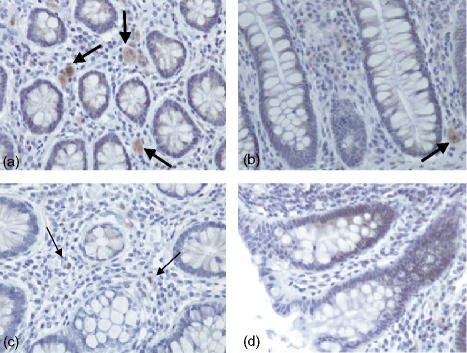
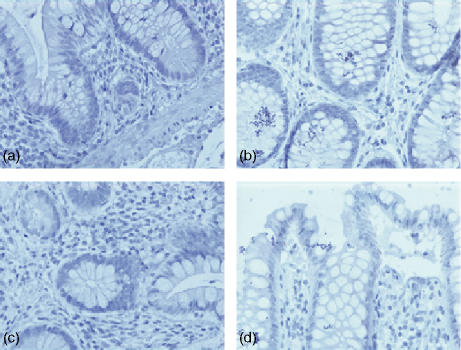
Expression of TLR2 in human colonic tissue was observed in both the epithelium and in the lamina propria. TLR2 expression was confined to crypt cells as shown in (Fig. 1a, d). However, this expression decreased as these cells matured and moved up towards the gut lumen (Fig. 1b, d), so that there was no TLR2 expression at the luminal surface (Fig. 1c). Expression of TLR2 in the lamina propria was sporadic with more positive lymphoid cells close to the crypt rather than the luminal surface of the epithelium. As shown in Fig. 2, expression of TLR3 in normal healthy colonic tissue was localized in the most mature columnar epithelial cells exiting the crypts, and forming the luminal surface of the epithelium. TLR3 was discretely localized just under the luminal surface of these cells, but not on the cell membrane. When the luminal surface was stained (Fig. 2d) and observed from above, TLR3 expression had a ‘crazy paving’ like appearance with the junctions between the cells being negative for TLR3. Expression of TLR3 in the lamina propria was confined to large mononuclear cells (Fig. 2c). The expression pattern for rectal mucosa was very similar, with only the most mature epithelial cells expressing TLR3 protein. TLR4 expression (Fig. 3) was very similar to that of TLR2, being confined to the crypt epithelial cells (Fig. 3a, d) with very strong expression occurring in large mononuclear cells of the lamina propria, being particularly localized near the crypts (Fig. 3a, b). Smaller lymphoid cells also expressed TLR4 throughout the lamina propria (Fig. 3c, d). Figure 4 shows four negative controls demonstrating no positive staining when non-specific isotype control antibodies were used on the tissue sections.
Figure 1.
Immunohistochemical analysis of normal healthy human colon stained with a monoclonal mouse anti-human TLR2 IgG2a antibody, visualized using immunoperoxidase. (a) Transverse section (×40) across the crypt region with staining in the epithelium and lamina propria. (b and c) Respectively, a transverse and longitudinal section of more mature epithelial cells that interface with the gut lumen (×40). (d) A longitudinal section (×20) from crypt to lumen.
Figure 2.
Immunohistochemical analysis of normal healthy human colon stained with a monoclonal mouse antihuman TLR3 IgG1 antibody, visualized using immunoperoxidase. (a and b) Longitudinal sections (×40) through the epithelium at the luminal surface. (c) A longitudinal section (×20) from crypt to lumen, and (d) a longitudinal section with a transverse region (×40) showing staining seen from the gut lumen.
Figure 3.
Immunohistochemical analysis of normal healthy human colon stained with a monoclonal mouse anti-human TLR4 IgG2a antibody, visualized using immunoperoxidase. Expression patterns of human TLR4. (a)A transverse section (×40) across the crypt region with staining in the epithelium and lamina propria. (b) A longitudinal section through the crypt region (×40). (c) (×40) A longitudinal section of more mature epithelial cells. (d) A longitudinal section (×20) from crypt to lumen.
Figure 4.
(a–d) Negative controls, both transverse and longitudinal, of healthy human colon stained with IgG1 and IgG2a isotype controls followed by appropriate secondary antibody plus immunoperoxidase visualization.
mRNA expression in HT29 and CACO-2 cells after coculture with mucosal bacteria
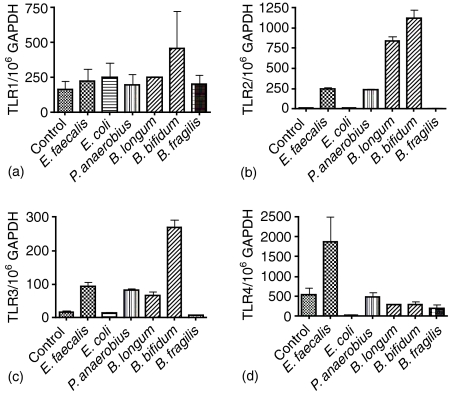
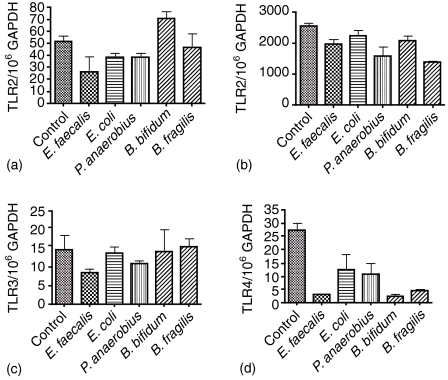
No significant differences in mRNA expression for TLR1 were seen on coculture with any of the test bacteria (Fig. 5a). TLR2 levels were significantly increased on coculture of HT29 with all of the Gram positive species (P <0·0001 for all Gram positive bacteria tested), but not the Gram negative bacteria (Fig. 5b). This pattern was repeated for mRNA expression of TLR3 with the B. bifidum (P = 0·0004) consistently showing the greatest increase in expression over control (Fig. 5c). The increase in TLR3 by Gram-positive organisms was confirmed using a further three peptostreptococci (P. anaerobius A, 652/106 GAPDH, P. anaerobius B 845/106 GAPDH and P. prevotii 206/106 GAPDH), three enterococci (E. faecalis A; 222/106 GAPDH, E. faecalis B; 284/106 GAPDH and E. faecalis C; 309/106 GAPDH) compared to Gram-negative E. coli (A; 68/106 GAPDH, B; 42/106 GAPDH) and one Gemminger sp. (18/106 GAPDH). The control (HT29 cells without bacteria) had 62 molecules of mRNA for TLR3/106 GAPDH. An increase in TLR4 mRNA was only found with the E. faecalis strains (P = 0·1083, not significant) with the E. coli coculture showing a significant reduction in TLR4 mRNA levels (P = 0·0319; Fig. 5d). CACO-2 cells showed no significant increase in mRNA for TLR1 (Fig. 6a), -2 (Fig. 6b), -3 (Fig. 6c) or -4 (Fig. 6d) in any of the coculture experiments. Moreover, there was a general trend towards a reduction in mRNA expression that was particularly evident for TLR2 and -4 (Fig. 6b, d, respectively).
Figure 5.
Levels of gene expression for TLR1 (a), TLR2 (b), TLR3 (c) and TLR4 (d) by HT29 cells in coculture with six different bacterial isolates from the rectal mucosa for three hrs. Results are expressed as a mean plus standard deviation of three different experiments.
Figure 6.
Levels of gene expression for TLR1 (a), TLR2 (b), TLR3 (c) and TLR4 (d) by CACO-2 cells in coculture. See legend to Fig. 5.
mRNA expression levels in response to TLR ligands
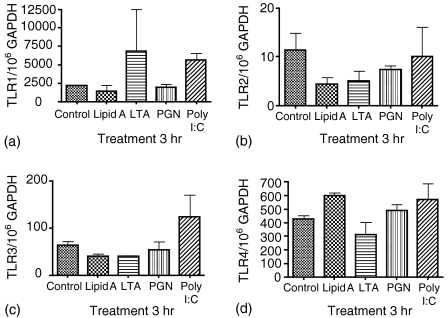
The HT29 cell monolayers showed both positive and negative TLR expression patterns in response to live bacteria. In order to determine if particular determinants were responsible for this effect, purified ligands (lipid A, LTA, PGN and poly I:C) of TLR receptors were cocultured with the HT29 cells (Fig. 7a–d). TLR1 was significantly up-regulated (P = 0·0134) over control by poly I:C but not by the three bacterial components LTA, PGN and lipid A (Fig. 7a). There was no significant difference seen in the expression of TLR2 mRNA in response to the four pathogen associated molecular patterns tested (Fig. 7b). TLR 3 mRNA was not significantly increased by any of the components tested although poly I:C did demonstrate a non-significant increase in TLR3 mRNA (Fig. 7c). Lipid A induced a significant increase in TLR4 mRNA (P = 0·005) with the other three components showing no significant difference over control (Fig. 7d).
Figure 7.
Levels of gene expression for TLR1 (a), TLR2 (b), TLR3 (c) and TLR4 (d) by HT29 cells in coculture with different TLR ligands for 3 hr. Results are expressed as a mean plus standard deviation of three different experiments.
In order to check the activity of these ligands to induce the expected response, peripheral blood mononuclear cells (PBM) were used. A simple coculture experiment was carried out. 5 × 106 PBM were cultured for 3 hr in RPMI 1640 (Invitrogen) with the same concentration of ligand as above. TLR1 mRNA levels were significantly decreased in the PGN treated cells (P = 0·0318) there was no significant difference between the other three ligands and the control cells. TLR2 mRNA was decreased in the PGN treated sample (P = 0·0641). Only poly I:C showed a significant increase in TLR3 mRNA levels (P = 0·0366). Both lipid A and poly I:C demonstrated a significant increase in TLR4 mRNA levels (P = 0·0008 and 0·0084, respectively).
Up-regulation and localization of TLR3 expression in HT29 cells
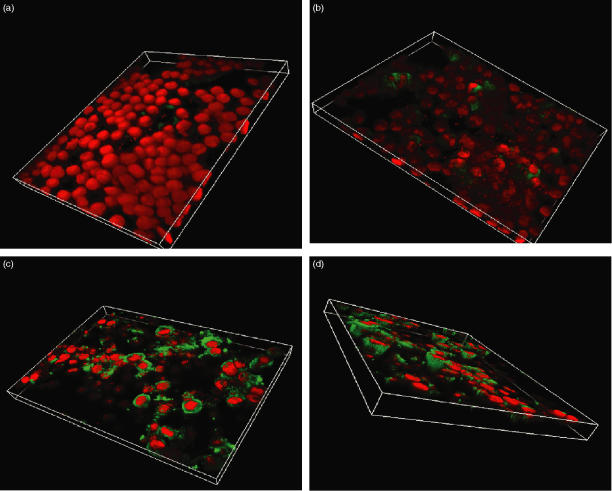
Immunofluorescence of HT29 monolayers cocultured with B. fragilis showed little TLR3 expression (Fig. 8a). When the orientation of the image was reversed, and the monolayer was viewed from below, the bacteria could be clearly identified, as small red bodies (Fig. 8b). There were, however, very low levels of fluorescence, which did not appear to correspond to localization of the bacteria and which was indistinguishable from the isotype control staining (not shown). HT29 cells cocultured with B. bifidum are shown in Fig. 8(c), there was a marked increase in TLR3 protein expression after 18 hr incubation with B. bifidum, with strong staining in the cell cytoplasm. From an oblique view, it is evident that this increased TLR3 expression followed the morphology of the cell (Fig. 8d).
Figure 8.
Confocal microscopy of an 18-hr coculture of HT29 cells with either Bacteroides fragilis (a, b) or Bifidobacterium bifidum (c, d). The cells were stained with goat antihuman TLR3 antibody, followed by a secondary FITC-conjugated rabbit anti-goat immunoglobulin antibody giving green staining where TLR3 is expressed. Cell nuclei have been counterstained in red with propidium iodide.
Enhancement of antiviral response by B. bifidum
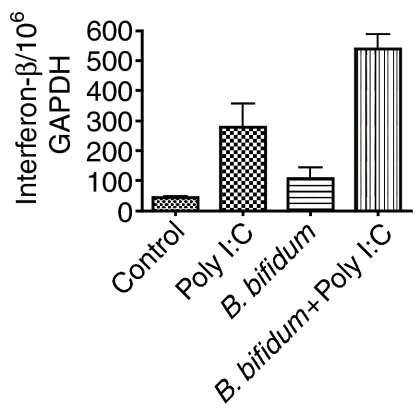
In order to determine if this observed up-regulation of TLR3 by Gram-positive bacteria plays a role in the ability of these cells to respond to viral challenge, a TLR3 stimuli (poly I:C) was used to mimic viral RNA and the level of interferon-β mRNA was measured (Fig. 9). Incubation with B. bifidum alone for 18 hr did not significantly up regulate mRNA for interferon-β. Poly I:C added for 3 hr to control cells alone demonstrated a significant up-regulation of mRNA for interferon-β (P = 0·0393). Poly I:C incubated for 3 hr with HT29 cells previously incubated for 18 hr with B. bifidum showed a significant increase in interferon-β mRNA over the control cells (P = 0·0004) and over the cells incubated with poly I:C alone (P = 0·0438). This demonstrates an enhanced antiviral response in epithelial cells previously exposed to Gram-positive bacteria.
Figure 9.
Levels of mRNA for interferon beta in HT29 cells in coculture with B. bifidum for 18 hr followed by a 3-hr stimulation with poly I:C. The results are expressed as a mean plus standard deviation of three different experiments.
Discussion
The colon and rectum play host to large numbers of different micro-organisms, both resident commensals and transient pathogens, each with their own unique molecular footprint. The first line of contact between the host and the normal microbiota are epithelial cells lining the mucosal surface, which provide a connection with the immune system, via cells in the lamina propria. Correct recognition and appropriate responses towards these normal inhabitants of the gut is essential for its healthy functioning, and maintenance of the mucosal barrier.
Analysis of expression levels for different TLR by colonic intestinal epithelial cells has been controversial. mRNA expression levels (relative to levels in the spleen) has been shown to be highest for TLR3, -4, -5, and -7.19 Expression of TLR2, which recognizes molecular patterns in Gram-positive organisms, has been reported at the level of mRNA, however, protein expression has not been confirmed in colonic epithelial cells.20,24,25 One previous study has shown TLR4 protein only in the crypts of mouse colon with expression levels highest in the more distal part of the colon.25 Both TLR2 and -4 are the main recognition receptors of bacterial surface structures in both Gram-negative and Gram-positive organisms.5–8,10–12 One study in mice with induced dextran sodium sulphate (DSS) colitis demonstrated an increase in mRNA for both TLR2 and -4 and a corresponding increase in TLR4 protein in epithelial cells at the epithelial surface; furthermore this expression would appear to move proximally up the colon with inflammation and was correlated with interleukin-1β expression.25 This change in expression of TLR4 in DSS colitis would allow recognition of Gram-negative bacteria at the luminal surface of the epithelium and not just in the crypts providing a mechanism for bacterial driven intestinal inflammation in this colitis model.25 In our model system we find that a Gram-positive bacteria, E. faecalis, induces up-regulation of TLR4 in HT29 cells. This organism has been implicated in ulcerative colitis2 suggesting a possible mechanism whereby E. faecalis colonization of colitic mucosa could up-regulate TLR4 in the disrupted architecture of the colon and allow recognition of Gram-negative bacteria at a site which would normally be refractive to these organisms, i.e. the luminal surface. Both TLR2 and -4 are only up-regulated by bacteria in HT29 cells and not CACO-2; this has previously been reported by other groups.24–27 Furthermore when we use the bacterial components lipid A, LTA and PGN to induce TLR expression in HT29 cells we see no change in TLR2 expression levels and only lipid A enhancing TLR4 expression levels. This would suggest that single purified components are not sufficient to induce up-regulation of these molecules, either because of their physical characteristics, which will be very different from their characteristics when they are part of a live bacterial surface, or that the observed up-regulation is caused by bacteria interacting with different surface receptors, which then either directly up regulate TLR or do so through cytokine-mediated receptor up-regulation. Furthermore, we need to consider that these cells are not primary cell isolates but are cell lines which are transformed and may have characteristics that are not identical to primary cells.28–30 These cell lines do, however, confer one large advantage over primary isolates as they have never encountered bacterial antigens or live bacteria; therefore, the responses we are measuring can be attributed to a primary response to bacteria. The only human situation where the colon is truly sterile is before birth. Therefore, we can use these cells to determine the effect of initial exposure of colonic epithelial cells of different maturation states to mucosal bacterial challenge. The HT29 cell line can be used as a model for newly formed epithelial crypt cells, as they can be maintained with the morphology and physiology of nascent crypt cells under certain growth conditions.28,31–33 In contrast, CACO-2 epithelial cells are much more mature, like those that are moving towards the luminal surface.33
TLR3 is the most abundant TLR mRNA expressed in the colon,19 and is the PRR for double-stranded viral RNA.9 This high expression in the colon is postulated to provide a first line of defence against virus infection, because all virus infections induce dsRNA at some point in their replication cycle.34 Furthermore, common intestinal viruses such as rotaviruses are themselves dsRNA viruses, providing immediate ligands for TLR3 when they come in contact with epithelial cells. The downstream consequences of ligation of TLR3 are induction of a typical reaction of viral infection, with the expression of type 1 interferons.35–37 It has also been demonstrated that some viruses target the TLR3 signalling cascade to dampen the immune response.38 TLR3 expression is also up-regulated by interferon-β alone.39 Intuitively, the cellular expression pattern of TLR3 demonstrated in this study would suggest that it is expressed to deal with an intracellular pathogen that enters the mucosa from the gut lumen, as its location was primarily in the cell cytoplasm, directly below the brush border. Expression of this protein was restricted to the most mature epithelial cells that had fully achieved their characteristic columnar morphology, which appears as the cells arrive to form the luminal barrier. Nascent cells in the crypt do not express any protein for TLR3. This is in direct contrast to the pattern seen with TLR2 and -4 where protein expression in the epithelium is confined to the crypts, a region normally devoid of commensal microorganisms.2 In the HT29 model, the cells require stimuli to up-regulate TLR2 and TLR3 mRNA from very low levels of expression. After introduction of Gram-positive mucosal isolates levels of mRNA expression significantly increase for both these recognition molecules. This was not surprising for TLR2 because components of Gram-positive bacteria are the reported ligand for this receptor5–8 and ligation of existing TLR2 would induce the observed up-regulation of expression.24 However, in this model the purified TLR2 ligands did not induce TLR2 up-regulation as previously discussed. This up-regulation was not expected for TLR3. Expression of this molecule is essential for maintenance of a functioning mucosa which is able to respond to viral infection and minimize mucosal damage. The normal mucosal microbiota comprises large numbers of Gram-positive and Gram-negative organisms, that exist in close contact with epithelial cells, at the luminal surface. Among these species are a number of organisms with probiotic properties particularly Gram-positive bifidobacteria and lactobacilli; these organisms have varying abilities to induce probiotic effects and it has been shown for lactobacilli that the magnitude of these effects are greatly dependent on the isolate and species chosen.40 These are amongst the first organisms to colonize the colon after birth41 that have been reported to aid the development of a fully mature and correctly functioning epithelial barrier.42–44 We would postulate that on initial exposure of the newborn colon to bacterial colonization by Gram-positive species up-regulation of TLR3 by the nascent epithelium would allow the newborn colon to be immediately able to respond to viral infections. We have demonstrated in our HT29 cells that they can respond to viral challenge as shown by their interferon-β induction in response to poly I:C exposure; however, with an initial exposure to a Gram-positive commensal organism we can demonstrate a significantly enhanced interferon-β response.
The reported unresponsiveness of the intestinal epithelium contrasts with other barrier epithelia, that demonstrate potent responsiveness to bacterial challenge directly through TLR recognition.45–47 It has recently been proposed that TLR may have a dual function in the intestinal epithelium both in recognition of pathogenic organisms and initiation of down stream antimicrobial responses but also as a controller of intestinal homeostatsis.48 Signalling through TLR proceeds through MyD88 and finally results in nuclear factor (NF)-κB activation.3 In MyD88 knockout mice increased proliferation of colonic epithelial cells is observed.48 This is also repeated in a knockout of p50, component of NF-κB, where the colon has extensive proliferative zones and elongated crypts similar to that of MyD88 knockout mice.49 This dysregulation of colonic cell turnover must be due in part to the loss of signalling through TLR via MyD88 and NF-κB in the crypt epithelium. Physiologically these elongated crypts confer a higher susceptibility to damage of barrier function.50,51 This susceptibility to damage could also be induced by expression of TLR2 and -4 at an inappropriate site in the mucosa at the luminal surface where they could interact with the commensal flora. This could arise when perturbation of the mucosal architecture occurs because of chronic inflammatory bowel disease.
In conclusion, we have demonstrated that the maturation stage of epithelial cells greatly affects the way they respond to bacterial stimuli. TLR2 and -4 are expressed only in the crypts of colonic epithelium. These are sterile environments where protection of the epithelial stem cells from bacterial pathogens is required to maintain gut homeostasis. Crypt cells, as modelled by undifferentiated HT29 cells, are alone in their ability to respond to live bacterial stimuli and up-regulate TLR2 and -4. In contrast TLR3 is expressed at the luminal surface conferring protection of the epithelium from viruses which are not members of the normal mucosal microbiota. The expression of TLR3 may be enhanced by interaction of new epithelial cells with Gram-positive commensal organisms, which in turn enhances the antiviral response in the mature epithelium.
Acknowledgments
This work was funded by the Medical Research Council UK.
Abbreviations
- DAB
diaminobenzene
- DC
dendritic cells
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- TLR
toll-like receptor
- PAMPS
pathogen-specific molecular patterns
- PGN
peptidoglycan
- PRR
pattern recognition receptor
References
- 1.Macfarlane S, Hopkins MJ, Macfarlane GT. Bacterial growth and metabolism on surfaces in the large intestine. Microb Ecol Hlth Dis. 2000;2:64–72. [Google Scholar]
- 2.Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin Infect Dis. 2004;38:1690–9. doi: 10.1086/420823. 10.1086/420823. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Kaisho T, Akira S. Toll-like receptors. Ann Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 4.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci U S A. 1998;95:588–93. doi: 10.1073/pnas.95.2.588. 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram negative and gram positive bacterial cell wall components. Immunity. 1999;11:443–51. doi: 10.1016/s1074-7613(00)80119-3. 10.1016/S1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 6.Underhill DM, Ozinsky A, Smith KD, Aderem A. Toll-like receptor 2 mediates mycobacteria-induced proinflammatory signalling in macrophages. Proc Natl Acad Sci USA. 1999;96:14459–63. doi: 10.1073/pnas.96.25.14459. 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aliprantis AO, Yang RB, Mark MR, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–9. doi: 10.1126/science.285.5428.736. 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 8.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J Biol Chem. 1999;274:17406–9. doi: 10.1074/jbc.274.25.17406. 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 9.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kB by Toll-like receptor 3. Nature. 2001;413:732–8. doi: 10.1038/35099560. 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 10.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–82. doi: 10.1084/jem.189.11.1777. 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arbour NC, Lorenz E, Schutte BC, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–91. doi: 10.1038/76048. 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 12.Rhee SH, Hwang D. Murine TOLL-like receptor 4 confers lipopolysaccharide responsiveness as determined by activation of NF kappa B and expression of the inducible cyclooxygenase. J Biol Chem. 2000;275:34035–40. doi: 10.1074/jbc.M007386200. 10.1074/jbc.M007386200. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 14.Hemmi H, Takeuchi O, Kawai T, et al. A toll-like receptor recognises bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 15.Ozinsky A, Smith KD, Hume D, Underhill DM. Co-operative induction of pro-inflammatory signaling by Toll-like receptors. J Endotox Res. 2000;6:393–6. 10.1179/096805100101532333. [PubMed] [Google Scholar]
- 16.Ozinsky A, Underhill DM, Fontenot J, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci USA. 2000;97:13766–71. doi: 10.1073/pnas.250476497. 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajjar AM, O'Mahony DS, Ozinsky A, Underhill DM, Aderem A, Klebanoff SJ, Wilson CB. Cutting edge. functional interactions between toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J Immunol. 2001;166:15–9. doi: 10.4049/jimmunol.166.1.15. [DOI] [PubMed] [Google Scholar]
- 18.Alexopoulou L, Thomas V, Schnare M, et al. Hyporesponsiveness to vaccination with Borrelia burgdorferi OspA in humans and in TLR1- and TLR2-deficient mice. Nat Med. 2002;8:878–84. doi: 10.1038/nm732. [DOI] [PubMed] [Google Scholar]
- 19.Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol. 2002;168:554–61. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 20.Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010–7. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hausmann M, Kiessling S, Mestermann S, et al. Toll-like receptors 2 and 4 are up regulated during intestinal inflammation. Gastroenterology. 2002;122:1987–2000. doi: 10.1053/gast.2002.33662. 21. [DOI] [PubMed] [Google Scholar]
- 22.Furrie E, Macfarlane S, Cummings JH, Macfarlane GT. Systemic antibodies towards mucosal bacteria in ulcerative colitis and Crohn's disease differentially activate the innate immune response. Gut. 2004;53:92–9. doi: 10.1136/gut.53.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadkarni MA, Martin FE, Jacques NA. Determination of bacterial load by real time PCR using a broad range (universal) probe and primers set. Microbiology. 2002;148:257–66. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 24.Melmed G, Thomas LS, Lee N, et al. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implications for host–microbial interactions in the gut. J Immunol. 2003;170:1406–15. doi: 10.4049/jimmunol.170.3.1406. [DOI] [PubMed] [Google Scholar]
- 25.Ortega-Cava CF, Ishihara S, Rumi MAK, et al. Strategic compartmentalisation of Toll-like receptor 4 in the mouse gut. J Immunol. 2003;170:3977–85. doi: 10.4049/jimmunol.170.8.3977. [DOI] [PubMed] [Google Scholar]
- 26.Abreu MT, Arnold ET, Thomas LS, Gonsky R, Zhou Y, Hu B, Arditi M. TLR4 and MD-2 expression is regulated by immune-mediated signals in human intestinal epithelial cells. J Biol Chem. 2002;277:20431–7. doi: 10.1074/jbc.M110333200. 10.1074/jbc.M110333200. [DOI] [PubMed] [Google Scholar]
- 27.Boker U, Yerzerskyy O, Feick P, et al. Responsiveness of intestinal epithelial cell lines to lipopolysaccharide is correlated with Toll like receptor 4 but not Toll like receptor 2 or CD14 expression. Int J Colorec Dis. 2003;18:25–32. doi: 10.1007/s00384-002-0415-6. 10.1007/s00384-002-0415-6. [DOI] [PubMed] [Google Scholar]
- 28.Chastre E, Emami S, Rosselin G, Gespach C. Vasoactive intestinal peptide receptor activity and specificity during enterocyte-like differentiation and retrodifferentiation of the human colonic cancerous subclone HT29-18. FEBS Lett. 1985;188:197–204. doi: 10.1016/0014-5793(85)80371-9. [DOI] [PubMed] [Google Scholar]
- 29.Molist A, Romaris M, Lindahl U, Villena J, Touab M, Bassols A. Changes in glycosaminoglycans structure and composition of the main heparin sulphate proteoglycan from human colon carcinoma cells (perlecan) during cell differentiation. Eur J Biochem. 1998;25:4371–7. doi: 10.1046/j.1432-1327.1998.2540371.x. [DOI] [PubMed] [Google Scholar]
- 30.Bertrand V, Couturier-Turpin MH, Louvel A, Panis Y, Couturier D. Relation between cytogenetic characteristics of two human colonic adenocarcinoma cell lines and their ability to grow locally or metastasize or both: an experimental study in the nude mouse. Cancer Genet Cytogenet. 1999;113:36–44. doi: 10.1016/s0165-4608(98)00194-0. [DOI] [PubMed] [Google Scholar]
- 31.Warhurst AC, Hopkins SJ, Warhurst G. Interferon gamma induces differential upregulation of alpha and beta chemokine secretion in colonic epithelial cell lines. Gut. 1998;42:208–13. doi: 10.1136/gut.42.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuntz S, Wenzel U, Daniel H. Comparative analysis of the effects of flavonoids on proliferation, cytotoxicity, and apoptosis in human colon cancer cell lines. Eur J Nutr. 1999;38:133–42. doi: 10.1007/s003940050054. 10.1007/s003940050054. [DOI] [PubMed] [Google Scholar]
- 33.Rousset M. The human colon carcinoma cell lines HT29 and CACO-2: two in vitro models for the study of intestinal differentiation. Biochimie. 1986;68:1035–40. doi: 10.1016/s0300-9084(86)80177-8. [DOI] [PubMed] [Google Scholar]
- 34.Jacobs BL, Langland JO. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219:339–49. doi: 10.1006/viro.1996.0259. 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 35.Doyle SE, Vaidya SA, O'Connell R. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–63. doi: 10.1016/s1074-7613(02)00390-4. 10.1016/S1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 36.Doyle SE, O'Connell R, Vaidya SA, Chow EK, Yee K, Cheng G. Toll-like receptor 3 mediates a more potent antiviral response than Toll-like receptor 4. J Immunol. 2003;170:3565–71. doi: 10.4049/jimmunol.170.7.3565. [DOI] [PubMed] [Google Scholar]
- 37.Oshiumi H, Matsumoto M, Funamim K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–7. doi: 10.1038/ni886. 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 38.Harte MT, Haga IR, Maloney G, et al. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J Exp Med. 2003;197:343–51. doi: 10.1084/jem.20021652. 10.1084/jem.20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinz S, Haehnel V, Karaghiosoff M, Schwarzfischer L, Muller M, Krause SW, Rehli M. Species-specific regulation of Toll-like receptor 3 genes in men and mice. J Biol Chem. 2003;278:21502–9. doi: 10.1074/jbc.M301476200. 10.1074/jbc.M301476200. [DOI] [PubMed] [Google Scholar]
- 40.Christensen HR, Frokiaer H, Pestka JJ. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol. 2002;168:171–8. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 41.Dai D, Walker WA. Protective nutrients and bacterial colonization in the immature human gut. Adv Pediatr. 1999;46:353–82. [PubMed] [Google Scholar]
- 42.Montalto M, Maggiano N, Ricci R, et al. Lactobacillus acidophilus protects tight junctions from aspirin damage in HT-29 cells. Digestion. 2004;69:225–8. doi: 10.1159/000079152. 10.1159/000079152. [DOI] [PubMed] [Google Scholar]
- 43.Baumgart DC, Dignass AU. Intestinal barrier function. Curr Opin Clin Nutr Metab Care. 2002;5:685–94. doi: 10.1097/00075197-200211000-00012. [DOI] [PubMed] [Google Scholar]
- 44.Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G613–26. doi: 10.1152/ajpgi.00341.2003. 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 45.Hertz CJ, Wu Q, Porter EM, et al. Activation of Toll-like receptor 2 on human tracheobronchial epithelial cells induces the antimicrobial peptide human beta defensin-2. J Immunol. 2003;171:6820–6. doi: 10.4049/jimmunol.171.12.6820. [DOI] [PubMed] [Google Scholar]
- 46.Rackley RR, Bandyopadhyay SK, Fazeli-Matin S, Shin MS, Appell R. Immunoregulatory potential of urothelium: characterization of NF-kappaB signal transduction. J Urol. 1999;162:1812–6. doi: 10.1016/s0022-5347(05)68243-2. 10.1097/00005392-199911000-00075. [DOI] [PubMed] [Google Scholar]
- 47.Shuto T, Imasato A, Jono H, et al. Glucocorticoids synergistically enhance nontypeable Haemophilus influenzae-induced Toll-like receptor 2 expression via a negative cross-talk with p38 MAP kinase. J Biol Chem. 2002;277:17263–70. doi: 10.1074/jbc.M112190200. 10.1074/jbc.M112190200. [DOI] [PubMed] [Google Scholar]
- 48.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Inan MS, Tolmacheva V, Wang QS, Rosenberg DW, Giardina C. Transcription factor NF-kappaB participates in regulation of epithelial cell turnover in the colon. Am J Physiol Gastrointest Liver Physiol. 2000;279:G1282–91. doi: 10.1152/ajpgi.2000.279.6.G1282. [DOI] [PubMed] [Google Scholar]
- 50.Bailey M, Plunkett FJ, Rothkotter HJ, Vega-Lopez MA, Haverson K, Stokes CR. Regulation of mucosal immune responses in effector sites. Proc Nutr Soc. 2001;60:427–35. doi: 10.1079/pns2001118. [DOI] [PubMed] [Google Scholar]
- 51.Maggio-Price L, Shows D, Waggie K, Burich A, Zeng W, Escobar S, Morrissey P, Viney JL. Helicobacter bilis infection accelerates and H. hepaticus infection delays the development of colitis in multiple drug resistance-deficient (mdr1a−/−) mice. Am J Pathol. 2002;160:739–51. doi: 10.1016/S0002-9440(10)64894-8. [DOI] [PMC free article] [PubMed] [Google Scholar]