Abstract
Bifidobacteria comprise a dominant microbial population group in the human intestinal tract with purported beneficial health effects on the host. In this study, we characterized the molecular mechanisms for the initial interaction of probiotic Bifidobacterium lactis strain BB12 with native and intestinal epithelial cell (IEC) lines. We showed that B. lactis-monoassociated Fisher F344 rats transiently induce phosphorylation/activation of the NF-κB transcriptionally active subunit RelA and the mitogen-activated protein kinase (MAPK) p38 in native IEC at day 5 after initial bacterial colonization. In addition, Interleukin 6 (IL-6) gene expression was significantly increased at day 5, demonstrating the physiological relevance of transient transcription factor activation in IEC. In contrast, Bacteroides vulgatus-monoassociated Fisher rats revealed RelA but not p38 MAPK phosphorylation and failed to trigger significant IL-6 gene expression in native IEC. Moreover, we demonstrated that B. lactis triggers NF-κB RelA and p38 MAPK phosphorylation in IEC lines. Adenoviral delivery of mutant IKK-β (Ad5dnIKKβ) and inhibition of the p38 MAPK pathway through the pharmacological inhibitor SB203580 significantly blocked B. lactis-induced IL-6 gene expression in IEC, suggesting that B. lactis triggers NF-κB and MAPK signaling to induce gene expression in the intestinal epithelium. Regarding the mechanisms of bacteria epithelial cell cross-talk, B. lactis-induced IL-6 gene expression was completely inhibited in TLR2 deficient mouse embryogenic fibroblasts (MEF TLR2−/−) as well as TLR2ΔTIR transfected Mode-K cells. In conclusion, we demonstrated that probiotic bacteria transiently trigger innate signal transduction and pro-inflammatory gene expression in the intestinal epithelium at early stages of bacterial colonization.
Keywords: intestinal epithelial cells, Toll-like receptor 2, probiotic bacteria, nuclear factor (NF)-κB, gnotobiology
Introduction
The mucosal surfaces and cavities of the gastrointestinal (GI) tract in humans and animals are populated by a complex mixture of more than 400 microbial species with spatial differences in population size and relative species predominance.1–4 Studies in gnotobiotic animals have shown that association of germ-free rodents with single bacterial species has a profound impact on the anatomical, physiological, and immunological development of the host, including effects on epithelial cell functions and the gut-associated lymphoid tissue.5–7 The complex homeostasis between non-pathogenic intestinal micro-organisms and the host is an intriguing immunological paradox as the normal mucosal immune system acquires tolerance (hyporesponsiveness) to the enteric microbiota, while protective cell-mediated and humoral immune responses to enteropathogens are maintained. Although the pattern of bacterial colonization in the premature neonatal or infant gut is different from the adult intestine, bifidobacteria are a dominant microbial population group in the human intestine.8–10 Clinical and animal studies provide evidence that certain strains of Bifidobacterium animalis (lactis), B. longum, B. infantis and B. breve may be effective in the prevention and/or treatment of gastroenteritis, necrotizing enterocolitis and chronic intestinal inflammation.11,12 The molecular mechanisms underlying these protective effects of probiotic bifidobacteria in the gut are completely unknown.
Intestinal epithelial cells (IEC), which make up the actual barrier that separates the host from the gut luminal environment, constitutively express or can be induced to express Toll-like receptors (TLR), costimulatory molecules, components of the human major histocompatibility complex (MHC) and a wide range of inflammatory and chemoattractant cytokines when activated by enteric pathogens or inflammatory products.13–16 Most of these molecules are in part transcriptionally regulated by the transcription factor nuclear factor (NF)-κB.17 Although bacteria trigger host responses by multiple mechanisms, the cornerstone of innate signalling is mediated by TLRs, a set of 10 well conserved pattern recognition receptors (PRR).18–20 TLRs are transmembrane proteins characterized by an extracellular domain containing leucine-rich repeats and an intracellular domain homologous to the interleukin (IL)-1R, or Toll/IL-1R (TIR). Ligand-specific binding to TLR promotes interaction of the cytoplasmic TIR domain with adaptor proteins followed by the recruitment of multiple kinases and activation of downstream target effector systems, including the mitogen-activated kinases (MAPK) as well as the IκB/NF-κB transcriptional system.20,21 We previously demonstrated that non-pathogenic Gram negative Bacteroides vulgatus induce RelA (NF-κB p65 subunit) phosphorylation and NF-κB activation in IEC through a TLR4-dependent manner.22,23 Monoassociation of germ-free Fisher rats with Bacteroides vulgatus induced transient nuclear localization of phosphorylated-RelA in the intestinal epithelium, demonstrating the physiological relevance for bacteria–epithelial cell interaction at the mucosal surface of the gut.
In this study, we characterized the molecular mechanisms for the cellular interaction of the probiotic bacterial strain B. lactis BB12 with the intestinal epithelium in native epithelium and cell lines. We showed that B. lactis BB12 transiently triggered NF-κB RelA and p38 MAPK phosphorylation as well as IL-6 gene expression in native epithelium after the initial monoassociation of Fisher F344 rats. Similarly, B. lactis BB12 induced RelA and p38 phosphorylation in the IEC line Mode-K. In contrast, B. vulgatus-monoassociated Fisher rats revealed RelA but not p38 MAPK phosphorylation and failed to trigger significant IL-6 gene expression in IEC. The inhibition of the NF-κB and p38 MAPK pathways using the adenoviral vector Ad5dnIKKβ and the pharmacological inhibitor SB203580 significantly inhibited IL-6 protein secretion in B. lactis BB12-stimulated Mode-K cells. Interestingly, the mRNA expression level of A20 which is a negative regulator of the TLR-mediated NF-κB signal transduction cascade was significantly up-regulated in primary and IEC lines.24–26In vitro, the induction of IL-6 gene expression was completely blocked in TLR2 gene deficient mouse embryogenic fibroblast cells (TLR2–/– MEF) and in TLR2ΔTIR transfected Mode-K cells, suggesting an important role of this pattern recognition receptor to mediate the initial interaction of B. lactis BB12 with the intestinal epithelium.
Materials and methods
Animals and bacterial monoassociation
Germ-free Fisher F344 rats in the Gnotobiotic Animal Facility at the German Institute of Human Nutrition (DIFE Potsdam-Rehbrücke, Germany). They were obtained from the germ-free breeding colony of the department. Twice a month the germ-free status of the animals was monitored. The animals were maintained in positive-pressure isolators (Metall & Plastik, Radolfzell, Germany) and housed in polycarbonate cages on irradiated wood chips at 22 ± 2°, 55 ± 5% relative humidity on a 12 hr light–dark cycle. They had free access to irradiated diet (Altromin fortified type 1314; Altromin, Lage, Germany) and autoclaved distilled water. Rats were monoassociated at 12–16 weeks of age with B. lactis BB12 as well as B. vulgatus (a generous gift from Dr R. Balfour Sartor, University of North Carolina at Chapel Hill, NC) and maintained under gnotobiotic conditions. This B. vulgatus strain was used to induce chronic intestinal inflammation in the HLA-B27 transgenic rat model of experimental colitis.27 Bacterial monoassociation and absence of contamination by other bacterial species were confirmed by culturing samples from the small and large intestine at necropsy and culturing serial faecal samples. The protocol for the animal experiment was approved by the Ministry of Nutrition, Agriculture and Forestry, Brandenburg, Germany. Rats were killed 3, 5, and 7 days after initial bacterial colonization. Germ-free rats were used as controls. Histological analysis of paraffin embedded colonic tissue sections assessed the degree of lamina propria mononuclear cell infiltration, crypt hyperplasia, goblet cell depletion and architectural distortion27 and revealed no signs of inflammation in B. lactis BB12 monoassociated rats.
Isolation of primary rat intestinal epithelial cells
Gnotobiotic and germ-free rats were killed, and the cecum as well the as colon were removed and placed in Dulbecco's modified Eagle's minimal essential medium (DMEM; Invitrogen Life Technologies, Karslruhe, Germany) containing 5% fetal calf serum (FCS). Caecum and colon were cut longitudinally, washed three times in calcium/magnesium-free Hank's balanced salt solution (Gibco BRL, Invitrogen, Karlsruhe, Germany), cut into pieces 0·5 cm long and incubated at 37° in 40 ml DMEM containing 5% FCS and 1 mm dithiothreitol (DTT) for 30 min in an orbital shaker. The supernatant was filtered, centrifuged for 5 min at 400 g and the cell pellet was resuspended in DMEM containing 5% FCS. The remaining tissue was incubated in 30 ml phosphate-buffered saline (PBS) (1×) containing 1·5 mm ethylenediaminetetra-acetic acid for an additional 10 min. The supernatant was filtered, centrifuged for 5 min at 400 g and the cell pellet was resuspended in DMEM containing 5% FCS. Finally, primary IEC were collected by centrifugation through a 25/40% discontinuous Percoll gradient at 600 g for 30 min. Primary rat IEC from caecum and colon were combined and collected in sample buffer for subsequent RNA isolation and Western blot analysis.
Cell culture and bacterial stimulation
The mouse IEC line Mode-K (passage 10–30; a generous gift from Dr Ingo B. Autenrieth, University of Tübingen, Germany) was grown in a humidified 5% CO2 atmosphere at 37° to confluency in six-well tissue culture plates (Cell Star, Greiner bio-one, Frickenhausen, Germany) as previously described.23 Mode-K cells that were generated from C3H/He mice are lipopolysaccharide unresponsive.28 Reconstitution of Mode-K cells with the TLR4/MD2 complex conferred LPS responsiveness (data not shown). We used Mode-K cells to selectively characterize Gram-positive bacteria-induced activation of epithelial cells. In addition, Mode-K cells were transfected with pZERO vector (InvivoGen, San Diego, CA) for murine TLR2ΔTIR lacking the cytoplasmic TIR domain. Stable transfected TLR2ΔTIR Mode-K cells were established through serial plating in Puromycin-containing medium. Wild type (WT) and TLR2–/– MEF cells were a generous gift from Dr Carsten J. Kirschning (Technical University of Munich, Germany).
B. lactis BB12 was anaerobically grown at 37° in MRS broth (Sigma Aldrich, Taufkirchen, Germany) containing 1% Tween-80. Bacteria were harvested by centrifugation (3000 g, 15 min) at stationary growth phase, washed in phosphate-buffered saline (1× PBS pH 7·4) and diluted in DMEM (Invitrogen). Confluent epithelial cell monolayers were infected with B. lactis BB12 at a bacterium-to-epithelial cell ratio (multiplicity of infection (m.o.i.)) of 3–30 at various time points. Based on the average number of Mode-K cells per six-well (1·5 × 106), we used 4·5 × 106 (m.o.i. 3) or 4·5 107 (m.o.i. 30) total bacteria per 2 ml of medium. Where indicated, Mode-K cells were pretreated with the p38 MAPK inhibitor SB203580 (Merck Bioscience, Schwalboch, Germany) and the adenoviral vector Ad5dnIKKβ (a generous gift from Dr Christian Jobin, University of North Carolina at Chapel Hill, NC) as previously described.
RNA isolation and real-time reverse transcription–polymerase chain reaction (PCR)
RNA from IEC was extracted using Trizol Reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. Extracted RNA was dissolved in 20 µl water containing 0·1% diethyl-pyrocarbonate. For reverse transcription, 1 µg total RNA was added to 30 µl of reaction buffer containing 8 µl 5× first-strand buffer, 4 µl DTT (100 mm), 6 µl desoxyribonucleoside triphosphate mixture (300 µm) (all reagents from Invitrogen Life Technologies) and incubated for 5 min at 65°. After adding 10 µl of a solution containing 0·2 µg random hexamers, 40 U RNase Out and 200 U murine Moloney leukaemia virus reverse transcriptase (all reagents from Invitrogen Life Technologies), the total mixture was incubated for an additional 60 min at 37° followed by a final 1 min heating step at 99°.
Real time PCR was performed in glass capillaries using a Light CyclerTM system (Roche Diagnostics, Mannheim, Germany). Primer sequences and amplicon sizes were as follows: rat IL-6-F 5′-ccactgccttccctac-3′, rat IL-6-R 5′-gtgcatcatcgctgtt-3′ (amplicon size 183 bp), rat A20-F 5′-catctgcccttcctgg-3′, rat A20-R 5′-tcttcttggttcttaggct-3′ (amplicon size 274 bp), rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-F 5′-ccaaggagtaagaaaccc-3′; rat GAPDH-R 5′-ggtgcagcgaactttatt-3′ (amplicon size 209 bp), mouse A20-F 5′-gaacaatgtcccgtgc-3′, mouse A20-R-5′-acctactcgttggctt-3′ (amplicon size 277 bp). For real time PCR, 1 µl reverse transcribed cDNA was added to a total volume of 10 µl PCR reaction buffer containing 1× LC-FastStart DNA Master Mix (Roche), MgCl2 (4 µm), forward and reverse primers (20 µm). The PCR programme was one cycle of denaturation at 95° for 10 min followed by 50 cycles of 95° for 15 s, annealing at 60° for 10 s and extension at 72° for 20 s. The amplified product was detected by the fluorescent dye SYBR green. Melting curve analysis and gel electrophoresis were used to document the amplicon specificity. Calibration curves were generated by measuring serial dilutions of stock cDNA to calculate the amplification efficiency (E). The crossing point (Cp) of the log-linear portion of the amplification curve was determined. The relative induction of gene mRNA expression was calculated using the following equation EΔCp (control samples − treated samples) and normalized for the expression of GAPDH mRNA.29 Triplicate samples were measured in duplicates and blotted as fold increase between treated and untreated control samples.
Western blot analysis
Purified primary IEC or Mode-K cells were lysed in 1× Laemmli buffer, and 20–50 µg of protein was subjected to electrophoresis on 10% sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis (PAGE) gels. Where indicated IEC cells were pretreated for 1 hr with 20 µm of the proteasome inhibitor MG132 (BioMol, Plymouth Meeting, PA). Anti-phospho-RelA (Ser536), RelA, antiphospho-p38 (Thr180/Tyr182), p38, acetylated-phosphorylated-histone 3 (Ac-P-H3), histone 3 (all antibodies from Cell Signaling (Beverly, MA)), CD45 (Santa Cruz Biotechnology, Santa Cruz, CA) and β-actin (ICN, Costa Mesa, CA) were used to detect immunoreactive phospho-RelA, RelA, phospho-p38, p38, phospho-acetyl-histone 3, histone 3, CD45 and β-actin, respectively, using an enhanced chemiluminescence light-detecting kit (Amersham, Arlington Heights, IL) as previously described23.
Enzyme-linked immunosorbent assay (ELISA) analysis
Protein concentrations were determined in spent culture supernatants of IEC cultures using an ELISA technique. IL-6 protein production was determined by mouse specific ELISA assay kits according to the manufacturer's instructions (R & D Systems, Heidelberg, Germany).
Statistical analysis
Data are expressed as the means ± S.D. of triplicates. Statistical analysis was performed by the two-tailed Student's t-test for paired data and considered significant if P-values were <0·05 (*) or <0·01 (**).
Results
NF-κB RelA and p38 phosphorylation as well as IL-6 gene expression in native IEC after bacterial colonization of germ-free rats with B. lactis BB12 and B. vulgatus
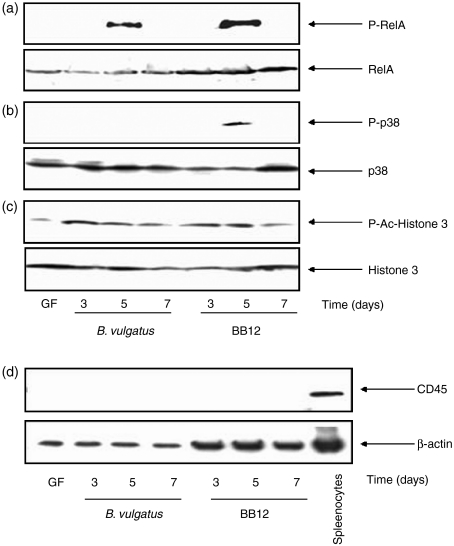
To investigate molecular mechanisms for the interaction of the Gram-positive probiotic bacterial strain B. lactis BB12 with the host after the initial colonization of the gut, we first associated germ-free Fisher F344 rats with B. lactis BB12 for 1 week. In parallel, we monoassociated germ-free rats with our reference strain B. vulgatus.23 Monoassociated rats were killed after 3, 5 and 7 days after bacterial colonization. Primary IEC from the large intestine (caecum + colon) were isolated and pooled samples from two independent experiments were subjected to Western blot analysis. As shown in Fig. 1 B. lactis BB12 induced RelA (Fig. 1a) and p38 (Fig. 1b) phosphorylation in native IEC at day 5 but not at day 3 and day 7 after the association of the rats. In contrast, B. vulgatus triggered RelA but not p38 phosphorylation after 5 days of bacterial colonization. Total RelA and p38 protein expression was confirmed in IEC for germ-free control and monoassociated rats. Of note, germ-free control rats revealed no phospho-RelA and phospho-p38 expression in native IEC, suggesting that B. lactis BB12 transiently triggered activation of the NF-κB and p38 MAPK transcription factor systems in native epithelium. Histone acetylation and phosphorylation are post-transcriptional mechanisms leading to chromatin unfolding in order to provide access of transcrition factors to gene promoter binding sites and to achieve coordinate gene expression. As shown in Fig. 1(c) B. lactis BB12 induced histone 3 acetylation/phosphorylation in IEC on days 3, 5 and 7 after the initial bacterial colonization. The slightly elevated level of P-Histone 3 in germ-free IEC may reflect basic activation of gene expression. In addition, the absence of lymphocyte contaminations in the purified epithelial cell preparations was confirmed by determining the lymphocyte marker CD45 using Western blot analysis. As shown in Fig. 1(d), CD45 protein expression was absent in isolated epithelial cells but was clearly detectable in control samples from spleenocytes.
Figure 1.
(a–c) NF-κB RelA and p38 MAPK phosphorylation in native IEC from B. vulgatus and B. lactis BB12-monoassociated Fisher rats. Germ-free Fisher F344 rats were monoassociated with B. vulgatus and B. lactis BB12 at 12–16 weeks of age. Rats (n = 2) were killed at day 3, 5, and 7 after initial bacterial colonization and native IEC from large intestine (caecum + colon) were isolated as described in Materials and Methods. IEC from germ-free rats were used as controls. Total protein was extracted and 50 µg of protein was subjected to SDS–PAGE followed by (a) phospho-RelA and RelA (b) phospho-p38 and p38 (c) actetylated-phosphorylated H3 and total H3 as well as (d) CD45 and b-actin immunoblotting using the ECL technique. Gels represent the combined protein samples from each group.
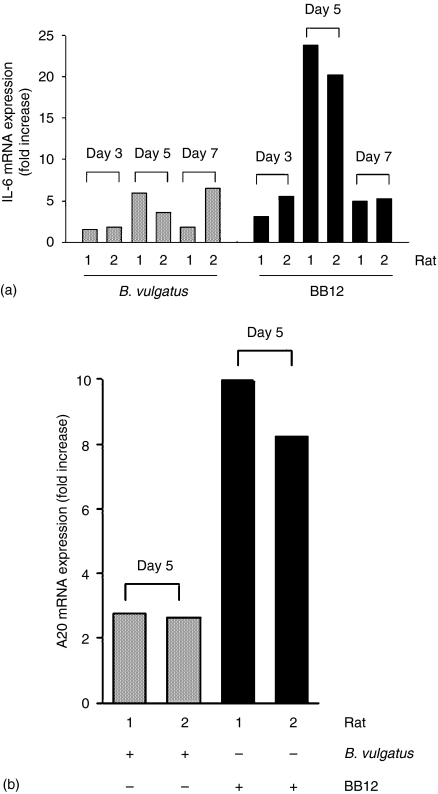
We next sought to investigate IL-6 gene expression in native IEC from gnotobiotic Fisher rats compared to germ-free controls using real-time quantitative Light Cycler reverse transcription–PCR. As shown in Fig. 2(a), the rats monoassociated with B. lactis triggered a significantly higher IL-6 mRNA expression on day 5 (20–23-fold increase) than on days 3 (three- to fivefold increase) and 7 (four- to fivefold increase) in both gnotobiotic experiments, confirming the transient activation of the NF-κB and MAPK signalling cascades in IEC. Interestingly, IL-6 gene expression in IEC was significantly lower in B. vulgatus-monoassociated rats.
Figure 2.
IL-6 gene and A20 gene expression in native IEC from B. vulgatus and B. lactis BB12-monoassociated Fisher rats. Germ-free Fisher F344 rats were monoassociated with B. lactis BB12 at 12–16 weeks of age. Rats (n = 2) were killed at day 3, 5, and 7 after initial bacterial colonization and native IEC from large intestine (caecum + colon) were isolated as described in Materials and methods. IEC from germ-free rats were used as controls. Total RNA was extracted, reverse transcribed and real-time PCR was performed using the Light Cycler system with specific primers for IL-6, A20 and GAPDH. The induction of IL-6 (a) and A20 (b) mRNA was calculated relative to germ-free controls (mean fold increase ± SD) using the crossing point of the log-linear portion of the amplification curve after normalization with GAPDH.
A20 is an inducible molecule implicated in the negative regulation of NF-κB-dependent gene expression.24 Parallel to the induction of RelA phosphorylation and IL-6 gene expression at day 5 after bacterial colonization, A20 mRNA expression was significantly higher in B. lactis-monoassociated rats (10- and eightfold increase) compared with B. vulgatus-colonized rats (two- and twofold increase) (Fig. 2b).
B. lactis BB12 triggers NF-κB RelA and p38 phosphorylation as well as IL-6 and A20 expression in Mode-K cells
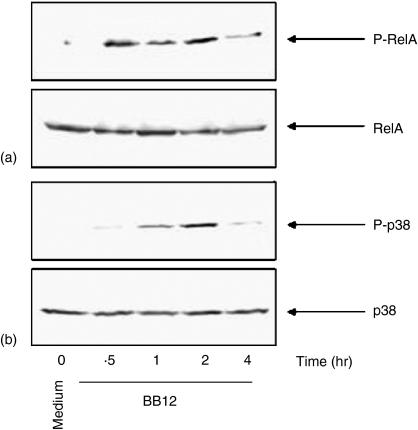
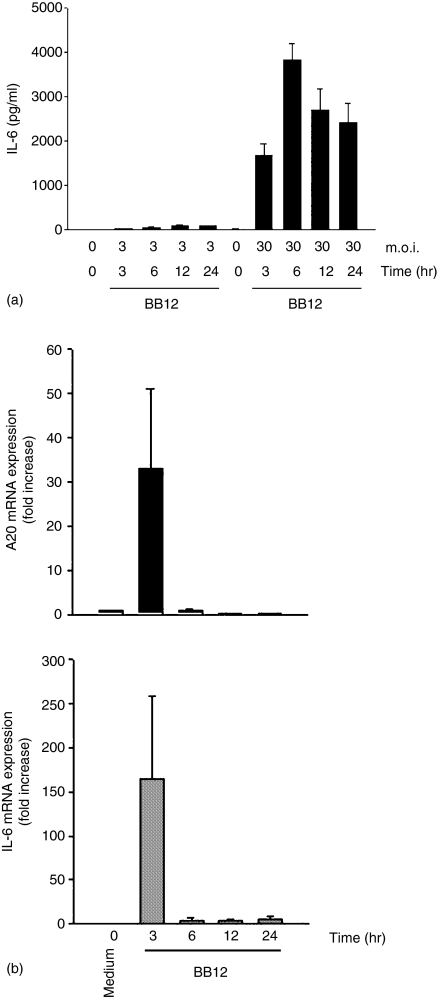
Because B. vulgatus triggers RelA phosphorylation and IL-6 gene expression in IEC through the TLR4 signalling cascade, we next asked the question whether B. lactis BB12 can directly induce RelA and p38 phosphorylation in IEC. We first stimulated Mode-K cells with B. lactis BB12 at m.o.i. 30 for 0–4 hr. Interestingly, B. lactis BB12 transiently induced phospho-RelA (Fig. 3a) as well as phospho-p38 (Fig. 3b), suggesting that B. lactis directly triggers these signalling pathways in native epithelium (Fig. 1). We next measured IL-6 protein secretion in the culture supernatant of stimulated Mode-K cells using ELISA technique. As shown in Fig. 4(a).B. lactis BB12 induced IL-6 protein secretion in Mode-K cells in a dose- and time-dependent manner reaching maximal stimulation at m.o.i. 30 after 6 hr of stimulation.
Figure 3.
B. lactis BB12 triggers transient RelA and p38 phosphorylation in Mode-K epithelial cells. Mode-K cells were stimulated with B. lactis BB12 for various times at m.o.i. 30. Total protein was extracted and 20 µg of protein was subjected to SDS-PAGE followed by immunoblotting using the ECL technique with antibodies for (a) phospho-RelA and RelA as well as (b) phospho-p38 and p38phospho-RelA. Representative gels from at least three different experiments are shown.
Figure 4.
B. lactis BB12 triggers IL-6 and A20 gene expression in Mode-K epithelial cells. Mode-K cells were stimulated with B. lactis BB12 for various times at m.o.i. 3 and 30. (a) IL-6 protein was measured in the spent culture supernatant from stimulated Mode-K cells using ELISA. (b) IL-6 and A20 mRNA expression was measured in B. lactis BB12 stimulation (m.o.i. 30). Total RNA was extracted, reverse transcribed and real-time PCR was performed using the Light Cycler system with specific primers for IL-6, A20 and GAPDH. The bars represent the combined mean value (± SD) of three experiments.
At the level of mRNA expression, IL-6 was strongly induced after 3 hr of bacterial stimulation of Mode-K cells followed by a complete down-regulation after 6 hr to 24 hr of bacterial stimulation (Fig. 4b). Interestingly and in accordance with our in vivo data, the induction of A20 mRNA expression in Mode-K cells was associated with the induction of IL-6 gene expression. These data may suggest that A20 gene expression could be involved in the negative regulation of NF-κB activity and NF-κB-dependent gene expression in IEC.
B. lactis BB12 induce TLR2 signalling to trigger IL-6 gene expression in IEC through the NF-κB and p38 MAPK pathways
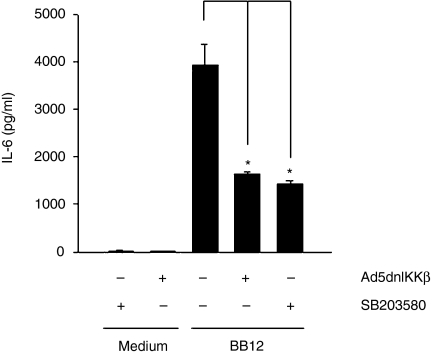
To further dissect the role of NF-κB and p38 MAPK signalling in B. lactis-induced IEC activation, we measured IL-6 secretion in Mode-K cells in the absence or presence of Ad5dnIKKβ as well as the pharmacological p38 inhibitor SB203580. Figure 5 shows that B. lactis-induced IL-6 production was significantly inhibited in the presence of dominant negative IKKβ as well as SB203580, suggesting an important role for both signalling pathways in mediating B. lactis-induced IL-6 gene expression.
Figure 5.
B. lactis BB12 triggers IL-6 secretion in Mode-K epithelial cells through the induction of NF-κB and p38 MAPK signalling. Mode-K cells were stimulated with B. lactis BB12 for 24 hr m.o.i. 30. Where indicated Mode-K cells were infected with adenoviral dominant negative (dn) IKKβ (Ad5dnIKKβ) and the pharmacological p38 MAPK inhibitor SB203580 (20 µm). IL-6 protein was measured in the spent culture supernatant from stimulated Mode-K cells using ELISA methods. The bars represent the combined mean value (± SD) of three experiments. *P < 0·05.
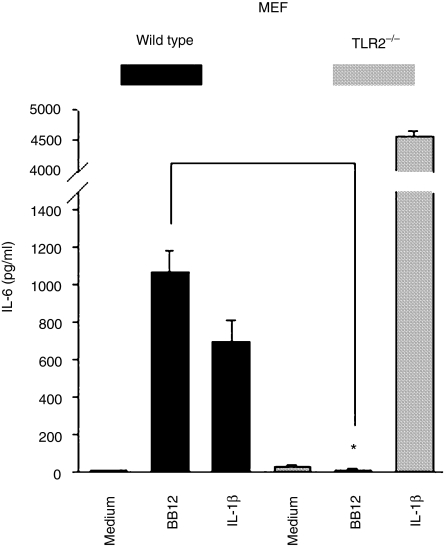
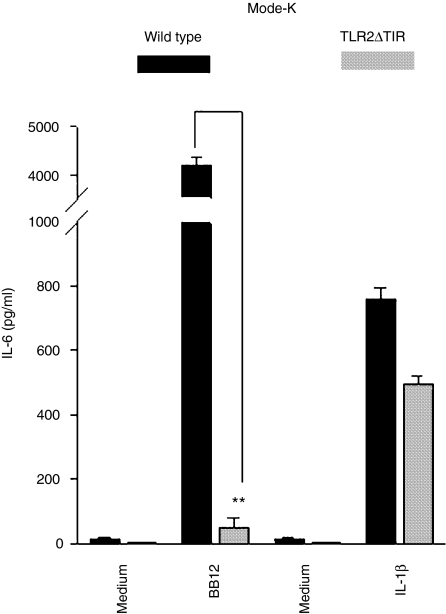
Gram-positive bacterial products have been shown to trigger cell activation through the pattern recognition receptor TLR2. We first used wild type and TLR2–/– MEF cells to evaluate the role of TLR2 in triggering IL-6 production. As shown in Fig. 6, B. lactis BB12 induced significant IL-6 expression in wild type cells but completey failed to trigger IL-6 secretion in TLR2–/– MEF. In contrast, IL-1β-induced IL-6 secretion was intact in both cell types, demonstrating the specificity for the inhibition of B. lactis BB12-mediated IL-6 production in TLR2–/– MEF. To further confirm the capability of B. lactis to signal through the pattern recognition receptor TLR2 in IEC, we established stable transfected TLR2ΔTIR Mode-K cells as described in Materials and methods. As shown in Fig. 7, B. lactis BB12-induced IL-6 protein secretion in Mode-K cells was completely blocked in TLR2ΔTIR stable transfected cells. In contrast, IL-1β-induced IL-6 production was intact confirming the responsiveness of these cells.
Figure 6.
B. lactis BB12 signals through the pattern recognition receptor TLR2 cascade to induce IL-6 gene expression in MEF cells. Wild type and TLR2–/– MEF cells were stimulated with IL-1β (20 ng/ml) and B. lactis BB12 at m.o.i. 30 for 12 hr. IL-6 protein was measured in the spent culture supernatant from stimulated Mode-K cells using ELISA methods. The bars represent the combined mean value (± SD) of three experiments. *P < 0·05.
Figure 7.
B. lactis BB12 signals through the pattern recognition receptor TLR2 cascade to induce IL-6 gene expression in IEC. Mode-K and stable transfected TLR2ΔTIR Mode-K cells were stimulated with IL-1β (20 ng/ml) and B. lactis BB12 at m.o.i. 30 for 6 hr. IL-6 protein was measured in the spent culture supernatant from stimulated Mode-K cells using ELISA methods. The bars represent the combined mean value (± SD) of three experiments. **P < 0·01.
In conclusion, these results demonstrate that the Gram-positive probiotic strain B. lactis BB12 signals through the TLR2 cascade to induce IL-6 gene expression. Of note, B. lactis BB12-induced IL-6 gene expression in IEC was mediated through the NF-κB and p38 MAPK pathways. In addition, we demonstrated that B. lactis BB12 transiently triggered NF-κB and p38 MAPK signalling as well as IL-6 gene expression in native IEC from monoassociated rats, demonstrating the physiological relevance of these mechanisms for bacteria–epithelial cell interaction in vivo.
Discussion
The molecular mechanisms of activating epithelial cell signal transduction in intestinal epithelium cells by probiotic bacteria may be relevant for initiating and maintaining intestinal homeostasis. We showed that the probiotic bacterial strain B. lactis BB12 signals through the NF-κB and p38 MAPK pathways to trigger IL-6 gene expression in both native and IEC lines. Most importantly, the monoassociation of germ-free Fisher F344 rats with B. lactis BB12 induced NF-κB RelA and p38 MAPK phosphorylation as well as IL-6 gene expression in native IEC. In contrast, B. vulgatus-monoassociated Fisher rats revealed RelA but not p38 MAPK phosphorylation and failed to trigger significant IL-6 gene expression in IEC. Of note, B. vulgatus triggers chronic intestinal inflammation in gnotobiotic HLA-B27 transgenic but not normal rats.27 Although B. lactis triggered pro-inflammatory signal transduction and gene expression in IEC, histological signs of inflammation were absent in B. lactis-monoassociated rats, suggesting that the normal host developed feedback mechanisms to control the mucosal immune responses to the constant challenge by commensal bacteria. We have previously shown that Gram-positive colitogenic Enterococcus faecalis initially triggered transient NF-κB signal transduction and pro-inflammatory gene expression in IEC from bacteria-monoassociated wild type and IL-10–/– mice. Interestingly and parallel to the development of chronic intestinal inflammation, NF-κB activity and pro-inflammatory gene expression was sustained in IEC IL-10–/– mice.30 Considering these findings it seems important to evaluate and compare the physiological consequences of B. lactis and B. vulgatus signalling to the epithelium on the development of chronic intestinal inflammation in animal models for experimental colitis including HLA-B27 transgenic rats and IL-10–/– mice.
Miettinen et al. showed that the probiotic Gram-positive strain Lactobacillus rhamnosus GG induces NF-κB activation and IL-6 gene expression in freshly isolated human leucocyte cultures.31 Here, we provide evidence that the Gram-positive B. lactis strain BB12 triggers IL-6 gene expression through TLR2-mediated activation of the NF-κB and p38 MAPK signalling cascades. Adenoviral delivery of dominant negative IKK-β and the addition of p38 MAPK inhibitor SB203580 significantly inhibited B. lactis-induced IL-6 gene expression in IEC, suggesting that both NF-κB and p38 signalling pathways contribute to the TLR2-meditaed activation of gene expression. Although B. vulgatus triggers TLR4-mediated RelA phosphorylation and IL-6 gene expression in epithelial cell lines23 but failed to trigger p38 phosphorylation in native epithelium, the physiological consequences of p38 MAPK signalling in the epithelium for bacteria-mediated host responses remains to be elucidated. We previously characterized molecular mechanisms for the inhibition of B. vulgatus and E. faecalis-mediated NF-κB activity and IL-6 gene expression in IEC, including effects that involve pattern recognition receptor stability, chromatin remodelling and modulation of phosphatase activity.32 It appears from several studies that transforming growth factor-β (TGF-β) and 15-deoxy-Δ12,14-prostaglandin J2 are important mediators in the negative regulation of acute and chronic inflammatory in the gut. Indeed, we showed that TGF-β-activated Smad signalling induced TLR2 degradation30 and inhibited C basic protein/p300-mediated histone phosphorylation in IEC.22 In addition, 15-deoxy-Δ12,14-prostaglandin J2-induced ERK signalling triggered phosphoprotein phosphatase (PP2A) activity, which directly dephosphorylated RelA and inhibited IL-6 gene expression in IEC.33 In addition, we currently characterize the protective role of IL-10-induced p38 signalling in the intestinal epithelium (Ruiz and Haller, unpublished observation). Induction of the zinc finger protein A20 is also involved in the negative regulation of TLR-mediated activation of the NF-κB signal transduction cascade likely through the inhibition of IKK activity by binding to the TRAF/RIP proteins.25,26,34,35 We showed that the induction of IL-6 mRNA expression by B. lactis was associated with the up-regulation of A20 mRNA expression in native and IEC lines suggesting a possible mechanism for the termination of NF-κB signalling and IL-6 gene expression.
Clinical and animal studies with probiotic micro-organisms provide some evidence that certain bifidobacterial strains are effective in the modulation of mucosal immune responses including the prevention and/or treatment of acute and chronic inflammation. For example, B. lactis BB12 antagonized Helicobacter pylori growth and/or metabolism in vitro as well as in vivo.36 In addition, B. lactis BB12 was effective at least partially to maintain oral tolerance to β-lactoglobulin in mice.37 Moreover, B. lactis HN019 and B. longum BB4638 were shown to increase resistence to experimental salmonellosis in mice. B. lactis HN019 also revealed immune stimulatory functions in elderly people including effects on natural killer cell and granulocyte activation.39 In addition, B. infantis strain 35624 attenuated experimental chronic colitis in IL-10–/– mice40 and dextran sodium sulphate-induced acute colitis in Sprague–Dawley rats.41 Furthermore, the probiotic mixture VSL#3 with eight different lactic acid bacterial species including B. longum, B. infantis and B. breve were effective in the treatment and/or treatment of experimental colitis in IL-10–/– mice42 as well as inflammatory bowel disease patients with pouchitis.43,44 The underlying cellular and molecular mechanisms for these beneficial health effects of bifidobacteria including their capability to activate and/or inhibit immune functions in the gut remain unclear.
Several studies now point to the fact that certain protective effects of probiotic bacteria may be mediated through the improvement of the gut epithelial cell barrier function. Indeed, ligand-specific activation of the TLR2 system greatly enhance transepithelial resistance in IEC lines associated with apical tightening and sealing of tight junction-associated ZO-1.45 In addition, Yang and Polk suggested that L. rhamnosus GG inhibits tumour necrosis factor (TNF)-mediated pro-apoptotic mechanisms in IEC YAMC cultures through the blockade of TNF-induced p38 MAPK activation46. Interestingly and consistent with our data, one mechanism to trigger antiapoptotic signalling pathways includes the induction of the NF-κB signalling cascade.47 Accordingly, the selective inhibition of NF-κB activity in enterocytes of IKKβ gene deficient mice sensitized these animals to ischaemia–reperfusion-induced apoptosis in IEC, which was associated with a loss of mucosal integrity.48 It appears from these results that although the pro-inflammatory NF-κB transcription factor system plays an important role in perpetuating the late/chronic state of certain inflammatory processes, the maintenance of normal mucosal homeostasis and epithelial cell integrity may require a fine-tuned balance between anti- and proapoptotic signals mediated at least in part by the presence of different bacterial species in the luminal gut microbiota.
In conclusion, we identified innate mechanisms for the probiotic B. lactis strain BB12 to activate transient pro-inflammatory host responses in the intestinal epithelium. We have shown that B. lactis BB12 triggers transient IL-6 gene expression in native epithelium and in cell lines. The initial and transient induction of TLR-mediated activation of pro-inflammatory transcription factor systems may play an important role in initiating epithelial cell homeostasis at early stages of bacterial colonization, while the persistent induction of NF-κB signalling associated with the lack of control mechanisms may lead to intestinal pathology including the development of chronic intestinal inflammation in the genetically susceptible host.
Acknowledgments
Grant support: This work was supported by Die Deutsche Forschungsgemeinschaft grant HA 3148/1–2 to D. Haller.
Abbreviations
- IEC
intestinal epithelial cells
- NF-κB
nuclear factor κB
- MAPK
mitogen-activated protein kinase
- IL-6
interleukin-6
- TLR2
Toll-like receptor 2
- MEF
mouse embryogenic fibroblasts
References
- 1.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 2.Moore WE, Holdeman LV. Human fecal flora. The normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med. 1968;127:67. doi: 10.1084/jem.127.1.67. 10.1084/jem.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon JI, Hooper LV, McNevin MS, Wong M, Bry L. Epithelial cell growth and differentiation. III. Promoting diversity in the intestine. conversations between the microflora, epithelium, and diffuse GALT. Am J Physiol. 1997;273:G565. doi: 10.1152/ajpgi.1997.273.3.G565. [DOI] [PubMed] [Google Scholar]
- 5.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 6.Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem. what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards CA, Parrett AM. Intestinal flora during the first months of life: new perspectives. Br J Nutr. 2002;88:S11. doi: 10.1079/BJN2002625. 10.1079/BJN2002625. [DOI] [PubMed] [Google Scholar]
- 9.Mitsuoka T. Intestinal flora and aging. Nutr Rev. 1992;50:438. doi: 10.1111/j.1753-4887.1992.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 10.Mountzouris KC, McCartney AL, Gibson GR. Intestinal microflora of human infants and current trends for its nutritional modulation. Br J Nutr. 2002;87:405. doi: 10.1079/BJNBJN2002563. [DOI] [PubMed] [Google Scholar]
- 11.Matto J, Malinen E, Suihko ML, Alander M, Palva A, Saarela M. Genetic heterogeneity and functional properties of intestinal bifidobacteria. J Appl Microbiol. 2004;97:459. doi: 10.1111/j.1365-2672.2004.02340.x. 10.1111/j.1365-2672.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- 12.Reid G, Jass J, Sebulsky MT, McCormick JK. Potential uses of probiotics in clinical practice. Clin Microbiol Rev. 2003;16:658. doi: 10.1128/CMR.16.4.658-672.2003. 10.1128/CMR.16.4.658-672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jobin C, Sartor RB. The I kappa B/NF-kappa B system. a key determinant of mucosal inflammation and protection. Am J Physiol Cell Physiol. 2000;278:C451. doi: 10.1152/ajpcell.2000.278.3.C451. [DOI] [PubMed] [Google Scholar]
- 14.Hershberg RM, Framson PE, Cho DH, Lee LY, Kovats S, Beitz J, Blum JS, Nepom GT. Intestinal epithelial cells use two distinct pathways for HLA class II antigen processing. J Clin Invest. 1997;100:204. doi: 10.1172/JCI119514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kagnoff MF, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:6. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumberg RS, Terhorst C, Bleicher P, McDermott FV, Allan CH, Landau SB, Trier JS, Balk SP. Expression of a nonpolymorphic MHC class I-like molecule, CD1D, by human intestinal epithelial cells. J Immunol. 1991;147:2518. [PubMed] [Google Scholar]
- 17.Pahl HL. Activators and target genes of Rel/NF-kB transcription factors. Oncogene. 1999;18:5853. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 18.Backhed F, Hornef M. Toll-like receptor 4-mediated signaling by epithelial surfaces: necessity or threat? Microbes Infect. 2003;5:951. doi: 10.1016/s1286-4579(03)00189-8. [DOI] [PubMed] [Google Scholar]
- 19.Dunne A, O'Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003 doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 20.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335. doi: 10.1146/annurev.immunol.21.120601.141126. 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 21.Zhang G, Ghosh S. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J Clin Invest. 2001;107:13. doi: 10.1172/JCI11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haller D, Holt L, Kim SC, Schwabe RF, Sartor RB, Jobin C. Transforming growth factor-{beta}1 inhibits non-pathogenic Gram-negative bacteria-induced NF-{kappa}B recruitment to the interleukin-6 gene promoter in intestinal epithelial cells through modulation of histone acetylation. J Biol Chem. 2003;278:23851. doi: 10.1074/jbc.M300075200. 10.1074/jbc.M300075200. [DOI] [PubMed] [Google Scholar]
- 23.Haller D, Russo MP, Sartor RB, Jobin C. IKK beta and phosphatidylinositol 3-kinase/Akt participate in non-pathogenic Gram-negative enteric bacteria-induced RelA phosphorylation and NF-kappa B activation in both primary and intestinal epithelial cell lines. J Biol Chem. 2002;277:38168. doi: 10.1074/jbc.M205737200. 10.1074/jbc.M205737200. [DOI] [PubMed] [Google Scholar]
- 24.Beyaert R, Heyninck K, Van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor- kappa B-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60:1143. doi: 10.1016/s0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 25.Boone DL, Lee E, Chai S, et al. A20 is required for the negative regulation of LPS-induced NF-kappab in macrophages. Gastroenterology. 2003;124:A45. [Google Scholar]
- 26.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350. doi: 10.1126/science.289.5488.2350. 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rath HC, Herfarth HH, Ikeda JS, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest. 1996;98:945. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vidal K, Grosjean I, Revillard JP, Gespach C, Kaiserlian D. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. Phenotypic and immune characterization of the MODE-K cell line. J Immunol Methods. 1993;166:63. doi: 10.1016/0022-1759(93)90329-6. 10.1016/0022-1759(93)90329-6. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene deficient mice lack TGF-beta/Smad signaling and fail to inhibit pro-inflammatory gene expression in intestinal epithelial cells after the colonization with colitogenic Enterococcus faecalis. J Immunol. 2005;174:2990. doi: 10.4049/jimmunol.174.5.2990. [DOI] [PubMed] [Google Scholar]
- 31.Miettinen M, Lehtonen A, Julkunen I, Matikainen S. Lactobacilli and streptococci activate NF-kappa B and STAT signaling pathways in human macrophages. J Immunol. 2000;164:3733. doi: 10.4049/jimmunol.164.7.3733. [DOI] [PubMed] [Google Scholar]
- 32.Haller D, Jobin C. Interaction between resident luminal bacteria and the host: can a healthy relationship turn sour? J Pediatr Gastroenterol Nutr. 2004;38:123. doi: 10.1097/00005176-200402000-00004. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz PA, Kim SC, Balfour Sartor R, Haller D. 15-deoxy-Delta{12,14}-prostaglandin J2-mediated ERK signaling inhibits gram negative bacteria-induced RelA phosphorylation and IL-6 gene expression in intestinal epithelial cells through modulation of protein phosphatase 2A activity. J Biol Chem. 2004;21:21. doi: 10.1074/jbc.M405032200. [DOI] [PubMed] [Google Scholar]
- 34.Zhang SQ, Kovalenko A, Cantarella G, Wallach D. Recruitment of the IKK signalosome to the p55 TNF receptor. RIP and A20 bind to NEMO (IKKgamma) upon receptor stimulation. Immunity. 2000;12:301. doi: 10.1016/s1074-7613(00)80183-1. 10.1016/S1074-7613(00)80183-1. [DOI] [PubMed] [Google Scholar]
- 35.Hu X, Yee E, Harlan JM, Wong F, Karsan A. Lipopolysaccharide induces the antiapoptotic molecules, A1 and A20, in microvascular endothelial cells. Blood. 1998;92:2759. [PubMed] [Google Scholar]
- 36.Wang KY, Li SN, Liu CS, et al. Effects of ingesting Lactobacillus- and Bifidobacterium-containing yogurt in subjects with colonized Helicobacter pylori. Am J Clin Nutr. 2004;80:737. doi: 10.1093/ajcn/80.3.737. [DOI] [PubMed] [Google Scholar]
- 37.Prioult G, Fliss I, Pecquet S. Effect of probiotic bacteria on induction and maintenance of oral tolerance to beta-lactoglobulin in gnotobiotic mice. Clin Diagn Lab Immunol. 2003;10:787. doi: 10.1128/CDLI.10.5.787-792.2003. 10.1128/CDLI.10.5.787-792.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva AM, Barbosa FH, Duarte R, Vieira LQ, Arantes RM, Nicoli JR. Effect of Bifidobacterium longum ingestion on experimental salmonellosis in mice. J Appl Microbiol. 2004;97:29. doi: 10.1111/j.1365-2672.2004.02265.x. 10.1111/j.1365-2672.2004.02265.x. [DOI] [PubMed] [Google Scholar]
- 39.Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am J Clin Nutr. 2001;74:833. doi: 10.1093/ajcn/74.6.833. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy J, O'Mahony L, O'Callaghan L, et al. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975. doi: 10.1136/gut.52.7.975. 10.1136/gut.52.7.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osman N, Adawi D, Ahrne S, Jeppsson B, Molin G. Modulation of the effect of dextran sulfate sodium-induced acute colitis by the administration of different probiotic strains of Lactobacillus and Bifidobacterium. Dig Dis Sci. 2004;49:320. doi: 10.1023/b:ddas.0000017459.59088.43. 10.1023/B:DDAS.0000017459.59088.43. [DOI] [PubMed] [Google Scholar]
- 42.Madsen K, Cornish A, Soper P, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 43.Gionchetti P, Rizzello F, Helwig U, et al. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202. doi: 10.1016/s0016-5085(03)00171-9. 10.1016/S0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 44.Gionchetti P, Rizzello F, Venturi A, et al. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology. 2000;119:305. doi: 10.1053/gast.2000.9370. [DOI] [PubMed] [Google Scholar]
- 45.Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 enhances ZO-1-associated intestinal epithelial barrier integrity via protein kinase C. Gastroenterology. 2004;127:224. doi: 10.1053/j.gastro.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002;277:50959. doi: 10.1074/jbc.M207050200. 10.1074/jbc.M207050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheikh MS, Huang Y. Death receptor activation complexes: it takes two to activate TNF receptor 1. Cell Cycle. 2003;2:550. [PubMed] [Google Scholar]
- 48.Chen LW, Egan L, Li ZW, Greten FR, Kagnoff MF, Karin M. The two faces of IKK and NF-kappaB inhibition: prevention of systemic inflammation but increased local injury following intestinal ischemia-reperfusion. Nat Med. 2003;9:575. doi: 10.1038/nm849. [DOI] [PubMed] [Google Scholar]