Abstract
Although interleukin 13 (IL-13) is an important mediator of asthma and allergic diseases, the molecular mechanisms regulating IL-13 gene expression are not well understood. This study was designed to define the molecular mechanisms governing IL-13 gene expression in T cells. IL-13 expression was examined in human peripheral blood T cells and in the EL-4 T-cell line by enzyme-linked immunosorbent assay and reverse-transcription polymerase chain reaction. An IL-13 promoter deletion analysis was performed using luciferase-based reporter plasmids transiently transfected into EL-4 cells by electroporation. DNA binding factors were investigated using electrophoretic mobility shift assays. In contrast to IL-4 expression, which required concomitant activation of calcium- and protein kinase C- (PKC-) dependent signalling pathways, PKC activation alone was sufficient for IL-13 protein secretion in mitogen-primed (but not resting) peripheral blood T cells, and for IL-13 mRNA expression and promoter activity in EL-4 T cells. Promoter deletion analysis localized a phorbol 12-myristate 13-acetate (PMA) -sensitive element to a proximal promoter region between −109 and −79 base pairs upstream from the IL-13 transcription start site. This promoter region supported the binding of both constitutive and PMA-inducible nuclear factors in gel shift assays.
Keywords: interleukin 13, protein kinase C, T lymphocyte, transcriptional regulation
Introduction
Interleukin-13 (IL-13), a T helper 2 (Th2) cytokine, has recently emerged as a critical mediator of immune and inflammatory responses including allergic asthma. 1–5 It may also promote tumour cell growth6 and inhibit tumour immunosurveillance.7 Thus a detailed understanding of the molecular regulation of IL-13 gene expression is an important goal. The regulation of IL-13 gene expression is co-ordinated with that of other Th2 cytokines, in part as a result of chromatin remodelling at the Th2 locus.8,9 In sharp contrast to IL-4 gene transcription, which is known to be regulated by the binding of numerous enhancing and repressing trans-acting factors to a complex proximal promoter,10–12 relatively few factors have been shown to regulate IL-13 transcription. Dolganov et al. identified a potential nuclear factor of activated T cells (NFAT) site in the human IL-13 promoter which was required for promoter activity in Jurkat cells.13 Kishikawa et al. recently reported that GATA-3 was able to activate the proximal IL-13 promoter, and identified a putative GATA-3 binding site contained therein.14 Calcium signalling and NFAT play particularly important roles in regulating IL-4 gene transcription.15,16 Because calcineurin antagonists can paradoxically enhance IL-13 expression in different experimental settings,17–19 the exact role of this pathway in IL-13 gene regulation remains uncertain.
We recently showed that IL-13 gene expression is regulated at the level of de novo transcription in mitogen-activated mouse splenocytes,20 and wanted to identify the cis-elements and trans-acting factors involved in IL-13 transcriptional regulation. We synthesized a panel of IL-13 promoter reporter constructs and identified the EL-4 mouse T-cell line as a useful model because endogenous IL-13 gene expression and transiently transfected reporter constructs were regulated similarly in these cells. Here we show that a minimal construct containing only 109 base pairs (bp) of the IL-13 promoter is sufficient for PMA-induced IL-13 promoter activity in EL-4 cells. We compared the regulation of IL-13 gene expression in cell lines and primary human T cells, and analysed the nuclear factors binding to this promoter region using the electrophoretic mobility shift assay (EMSA).
Materials and methods
Cells, cell lines and enzyme-linked immunosorbent assay (ELISA)
Peripheral blood T cells were purified from whole blood, taken from healthy volunteers, using the Ficoll cell separation technique and were cultured for 72 hr in the presence of 5 μg/ml phytohaemagglutinin (PHA; Calbiochem, San Diego, CA) and washed prior to restimulation. In some experiments, CD4+ T lymphocytes were obtained by negative selection (RosetteSEP, Stem Cell Technologies, Vancouver, BC, Canada) and analysed without priming with PHA. Volunteer subject recruitment and phlebotomy were carried out using protocols approved by the Johns Hopkins Bayview Institutional Review Board. EL-4 cells (American Type Culture Collection, Manassas, VA) and Jurkat cells (courtesy of Jack Strohminger, Harvard University) were maintained in Iscove's modified Dulbecco's medium (Life Technologies, Gaithersburg, MD) supplemented with gentamycin and 50 μmβ-mercaptoethanol (Life Technologies). Supernatants from 2 × 106 to 5 × 106 cells activated as in Fig. 1 were analysed for cytokine secretion by ELISA using standard kits [those for human IL-4 and IL-13 purchased from BioSource (Camarillo, CA, USA) and Immunotech (Fullerton, CA), respectively; that for mouse IL-13 obtained from R & D Systems (Minneapolis, MN)].
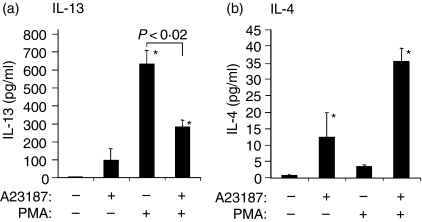
Figure 1.
Signal requirements for IL-13 and IL-4 protein expression in mitogen-primed human peripheral blood T cells; 1 × 106 cells were incubated with or without the Ca2+ ionophore A23187 or PMA as indicated, followed by analysis of cytokine secretion by ELISA (see Materials and methods). Data are the mean ± SEM of n = 5 different subjects. Asterisk indicates P < 0·05 compared to unstimulated cells, which secreted undetectable levels of either cytokine.
Plasmid construction
Genomic DNA was isolated from dispersed splenocytes that had been harvested from A/J mice as described elsewhere.20 IL-13 promoter fragments were amplified from genomic DNA by polymerase chain reaction (PCR). The following 5′ primers were used with a constant 3′ primer annealing at position +33 (numbers refer to nucleotides relative to the putative transcription start site according to ref. 21): −691 to −666: CACTGGCAGAATTAGCATCAGAAGAG; −501 to −478: CCATGCATTGCTTTGGTGATTTAT; −262 to −239: ATTACTGGGGCGGAAGTTAGCTTT; −109 to −80: ATTCAAGATGAGTAAAGATGGGGTTTTCAG; −51 to −30: GTGAGGCGTCATCACTTTGGTT; +33 to +8: AGAGAACCAGGGAGCTGTAGAACTGT.
Primers were ligated in proper orientation into the HindIII and BglII sites of pGL3.Basic (Promega, Madison, WI). All PCR products were sequenced to ensure accurate replication.
Cell lines and transfections
EL-4 cells (12 × 106) were transfected via electroporation with 20 μg reporter plasmids using the Bio-Rad Gene Pulser (Bio-Rad, Hercules, CA; 250 V and 960 μF). Cells were allowed to recover for 18 hr then stimulated with A23187 (0·5 μm; Calbiochem, San Diego, CA) and phorbol 12-myristate 13-acetate (PMA; 20 ng/ml, Calbiochem), either alone or in combination. Eighteen hours later, cells lysates were analysed on a Monolight 3010C luminometer (Analytical Bioluminescence, Gaithersburg, MD) using a luciferase assay kit (PharMingen, San Diego, CA). Multiple different preparations of each reporter plasmid were used. Transfection efficiency under these conditions averaged 15% (determined using a cotransfected green fluorescent protein vector, data not shown). In some experiments, cells were cotransfected with an AP-2 expression vector (kindly provided by Dr Trevor Williams, University of Colorado Health Sciences Center) followed by analysis of reporter gene expression using luminometry.
Reverse transcription (RT) PCR
Total cellular RNA was harvested using the RNeasy RNA isolation kit (Qiagen, Valencia, CA) from 5 × 106 EL-4 cells after a 4-hr stimulation with 0·5 μm A23187 (Calbiochem) and 20 ng/ml PMA (Calbiochem), either alone or in combination. RT-PCR was conducted using Superscript II reverse transcriptase and Taq polymerase (Life Technologies) at an annealing temperature of 60° for 30 cycles using the following primers: mouse IL-13 5′ CAGCATGGTATGGAGTGTGGACCT; mouse IL-13 3′ ACAGCTGAGATGCCCAGGGAT; and mouse β-actin 5′ GTGGGCCGCTCTAGGCACCA; mouse β-actin 3′ TGGCCTTAGGGTGCAGGGGG.
PCR products were analysed by agarose gel electrophoresis and ethidium bromide staining.
Nuclear extracts and EMSA
Nuclear extracts were isolated from Jurkat and EL-4 cells and were analysed by EMSA as described previously,11,12 using the following oligonucleotides and their complements: IL-13 PuB (− 138 to −116) 5′ GCGACACTGGATTTTCCACAAAG 3′; Probe I (− 109 to −79) 5′ ATTCAAGATGAGTAAAGATGTGGTTTTCAGA 3′; Probe II (− 93 to −68) 5′ GATGTGGTTTTCAGATAATGCCCAACAAAG 3′; Probe III (− 78 to −51) 5′ TAATGCCCAACAAAGCAGAGACCAGGG 3′. The nuclear factor-κB (NF-κB) consensus oligonucleotide (PRD-II site) was from the IFN-β gene.22 EMSAs were performed using 5 μg nuclear protein and probes were radiolabelled internally using random priming and the Klenow fragment. Each reaction condition contained 0·8 μg poly dG:dC (Pharmacia, New Jersey, NJ), 84 mm KCl, 34 mm NaCl, 7% glycerol, 20 mm HEPES (pH 7·5), 1 mm dithiothreitol, and 0·1% nonidet-P40 in a final volume of 10 μl. Free probes and protein–DNA complexes were resolved by 5% polyacrylamide gel electrophoresis with 0·5× Tris Borate EDTA (TBE). In competition studies, nuclear extracts were incubated with 50-fold molar excess unlabelled oligonucleotides containing binding sites for: NFAT (human IL-4 promoter P1 sequence 5′ TGAGTTTACATTGGAAATTTTCGTTACACCAGATTG 3′) AP-1, AP-2, CREB, OCT, GRE, and GATA (all from Promega). In pilot experiments, a 50-fold molar excess resulted in optimal competition in these experiments. In antibody assays, extracts were incubated with 1 μg specific antibodies directed against NFATp (Upstate, Waltham, MA), AP-2 (Geneka, Montreal, Canada), and isotype-matched controls for 1 hr at 4° prior to polyacrylamide gel electrophoresis.
Results
Different signalling requirements for expression of IL-4 and IL-13 protein in human peripheral blood T cells
Previous studies have suggested that gene expression of IL-4 and IL-13 is regulated by distinct signals.17 We used mitogen expanded human peripheral blood T cells and compared IL-4 and IL-13 protein secretion using pharmacological stimuli to activate either Ca2+ or protein kinase C (PKC) -dependent signalling pathways. We found that maximal expression of IL-4 protein required costimulation of Ca2+ and PKC-dependent pathways (Fig. 1b), and that there was a slight but significant increase in IL-4 protein when cells were treated with the Ca2+ ionophore A23187 alone. In contrast, the signals necessary for IL-13 expression were strikingly different, requiring only PKC-signalling for maximal activation (Fig. 1a). In fact, costimulation with A23187 inhibited PKC-induced IL-13 secretion in PHA-primed peripheral blood T cells (P < 0·02,Fig. 1a). We next determined if the distinct signal requirements found in mitogen-primed peripheral blood T cells were also observed using resting CD4+ lymphocytes. Peripheral blood CD4+ cells were isolated and stimulated with PMA, with and without A23187, followed by analysis of cytokine secretion by ELISA. In these experiments, PMA alone induced only modest IL-13 secretion; maximal IL-13 secretion required costimulation with A23187. The amounts of secreted IL-13 protein were 45 ± 14 and 634 ± 58 pg/ml from 1 × 106 cells stimulated with PMA alone versus PMA plus A23187, respectively (n = 3, mean ± SEM, both P < 0·05 versus resting cells which produced undetectable levels of IL-13). The signal requirements for maximal IL-13 secretion in mitogen-primed T cells (PKC-dependent, calcium-independent) are therefore different from those in freshly isolated cells (PKC- and calcium-dependent).
PKC pathways alone are sufficient for IL-13 gene expression in EL-4 T cells
We previously found that IL-13 gene expression is modulated at the level of de novo transcription in mouse splenocytes.20 To study the transcriptional regulation of IL-13 gene expression, we used the mouse EL-4 thymoma line because these cells express several transcription factors that are relevant for cytokine gene transcription (such as GATA-3,23 and see below), and that can be reliably transfected by electroporation. We first studied endogenous IL-13 gene expression using RT-PCR and ELISA and found that IL-13 mRNA expression and protein secretion were maximally induced in EL-4 T cells by PKC activation alone (Fig. 2 and data not shown). Interestingly, similar to the results obtained with mitogen-primed human PBT cells, costimulation with A23187 markedly reduced PMA-induced IL-13 mRNA expression (Fig. 2, lane 4).
Figure 2.
Signal requirements for IL-13 mRNA expression. EL-4 T cells were incubated with or without the Ca2+ ionophore A23187 or PMA as indicated for 4 hr, followed by extraction of total RNA and analysis of IL-13 expression by RT-PCR (see Materials and methods). A single transcript of the expected size was detected using both IL-13 and control β-actin primers (arrows). Data are from one experiment and are representative of three. M indicates molecular weight markers.
The proximal mouse IL-13 promoter is sufficient for transcriptional activation
We sequenced the mouse IL-13 promoter and compared this with published human IL-13 promoter sequence data. This analysis revealed that the human and mouse promoter regions are highly conserved, especially immediately upstream of their respective transcription start sites (see Fig. 4 and data not shown). We next performed a detailed deletion analysis using luciferase-based reporter constructs containing varying lengths of the IL-13 promoter transiently transfected into EL-4 cells (see Materials and methods section). Interestingly, a significant increase in PMA-induced promoter activity was observed using a minimal promoter construct containing only 109 bp upstream from the transcription start site (Fig. 3). Deletion of an additional 58 bp drastically reduced both constitutive and inducible transcriptional activation. This suggests that a positive regulatory element resides between −109 and −51 bp upstream from the transcription start site. Importantly, the signal requirements required for IL-13 promoter activation were identical to those for the endogenous IL-13 gene expression (Fig. 2) in that: (1) maximal transcriptional activation was observed using PMA alone, and (2) costimulation with Ca2+ ionophore inhibited promoter activity (Fig. 3). Inhibition by A23187 costimulation was observed using the −109 construct, suggesting that the inhibitory calcium signal is at least partially NFAT-independent. PMA-inducibility increased with progressive promoter deletions indicating that the upstream sequences (e.g. between −501 and −262) contain negative regulatory element(s), an observation that was not pursued further in this report.
Figure 4.
Nuclear factors from Jurkat and EL-4 cells differentially interact with the IL-13 purine box and an NF-kB consensus sequence. Nuclear extracts from 5 × 106 Jurkat (lanes 2–5) or EL-4 cells (lanes 6–9) treated as indicated were analysed by EMSA with (a) oligonucleotides encompassing the IL-13 purine box (IL-13 PuB) or (b) an NF-kB consensus site (PRD-II, see Materials and methods). Aliquots of the same extracts and similar exposure times were used in both panels. (c) Sequence comparison of the human and mouse IL-13 promoters in the PuB region (corresponding to −138 to −116 of the human promoter). Only three nucleotides are different in this region (underlined), and none of these are contained in the indicated core NFAT binding site.
Figure 3.
Deletion analysis of the mouse IL-13 promoter. EL-4 T cells were transfected by electroporation with five IL-13 promoter luciferase-based reporter constructs as well as a promoter-less vector (pGL3 Basic), and allowed to recover for 24 hr. Cells were then activated for 18 hr as indicated followed by analysis of reporter gene expression using luminometry. Data are the mean ± SEM of six experiments; asterisk indicates P < 0·05 versus unstimulated control cells. Numbers underneath refer to nucleotides upstream of the transcription start site. The lower panel shows a cartoon diagram of currently known regulatory elements in the IL-13 promoter including a polymorphic NFAT site (NFAT*, not included in the constructs in this report), as well as the PuB and GATA sites (not drawn to scale).
In a prior analysis of the human IL-13 promoter, Dolganov et al. reported that an element containing the sequence − 137GGAAAA− 142 was required for transcriptional activity in PMA-stimulated and calcium ionophore-stimulated Jurkat cells.13 This purine-rich element (which we term the IL-13 purine-box, or IL-13 PuB) supported the binding of NFATp in EMSA using T-cell nuclear extracts.13 We found that related sequences in the mouse promoter were dispensable for both constitutive and PMA-inducible promoter activity in EL-4 cells (Fig. 3). This apparent discrepancy could be the result of differences in reporter constructs or cell lines used. The nucleotides in this region are highly conserved between mouse and man, with identical purine residues at the core NFAT site (Fig. 4c), suggesting that species-dependent sequence divergence is not the sole contributing factor. Therefore, we next studied the nuclear factors that bound this region using nuclear extracts isolated from EL-4 (mouse) or Jurkat (human) T cells in EMSA (Fig. 4). Interestingly, Jurkat cells express several nuclear factors that interact with an oligonucleotide containing the IL-13 PuB including a predominant calcium-inducible factor (complex I, Fig. 4a). Using specific antisera, we found that complex I contains immunoreactive NFATp (data not shown). In contrast, nuclear extracts from EL-4 T cells do not contain factors that interact with IL-13 PuB (Fig. 4a, lanes 6–9). This is not simply the result of protein degradation or of other technical considerations using EL-4 nuclear extracts because these extracts contain abundant NF-κB (Fig. 4b). Taken together, these results suggest that the molecular regulation of IL-13 gene expression involves different DNA-binding factors in EL-4 and Jurkat T cells.
A PMA-sensitive factor does not bind between −78 and −51
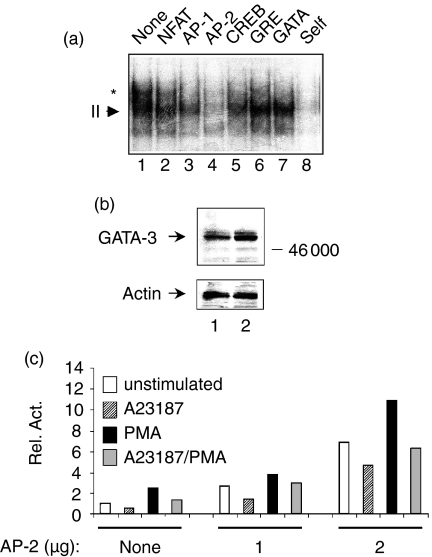
To investigate further the factor(s) involved in regulating PMA-inducible transcription of the proximal IL-13 promoter, we synthesized three overlapping oligonucleotides spanning the region from −109 to −51 and used EL-4 nuclear extracts in EMSA (Fig. 5a). This region contains the three potential binding sites for GATA family members described by Lavenu-Bombled et al.,24 including both consensus and non-consensus GATA-3 sites (Fig. 5). We found that although each oligonucleotide supported the binding of constitutive nuclear factors in EMSA (Fig. 5b), complex formation was enhanced using nuclear extracts from PMA-stimulated cells only using oligo II (complex II, Fig. 5b). The formation of complex II was not altered in extracts from cells stimulated only with calcium ionophore (data not shown). We next performed competition experiments using oligo II with a panel of unlabelled oligonucleotides containing consensus factor binding sites. Complex II contained a sequence-specific factor because its formation was completely prevented by excess self sequences but not by unrelated sequences (Fig. 6a). Interestingly, despite the fact that oligo II contains a potential GATA consensus (5′ CAGATAA 3′), a 100-fold molar excess of a competitor GATA oligonucleotide did not affect the binding of complex II (Fig. 6a, lane 7). In addition, an anti-GATA-3 antibody did not affect complex II formation in additional EMSA (data not shown). This was not because of the lack of nuclear GATA-3 expression in EL-4 cells, which we confirmed by Western immunoblot analysis (Fig. 6b).
Figure 5.
(a) Sequence of the region between −109 to −51, including the three oligonucleotides used in EMSA. Three potential GATA binding sites are underlined. See text for details.(b) Nuclear extracts were prepared from EL-4 T cells treated with and without PMA for 4 hr, and then analysed by EMSA using the three oligonucleotide probes as in (a). Each oligonucleotide supported the binding of one prominent constitutive complex (arrowheads). Complex formation was noticeably enhanced by PMA only using oligo II (complex II, lanes 3 and 4). Results are from one experiment that was representative of three. The exposure time for oligo I was approximately half that of oligo II and oligo III (8 hr versus 18 hr).
Figure 6.
(a) A panel of consensus oligonucleotides was used with nuclear extracts from PMA-treated EL-4 cells and analysed by EMSA using radiolabelled oligo II. Consensus oligonucleotides contained binding sites for NFAT (lane 2), AP-1 (lane 3), AP-2 (lane 4), CREB (lane 5), the GR (lane 6), GATA (lane 7) and Oct (not shown). Only the portion of the gel containing complex II is shown. Arrow indicated complex II, and asterisk indicates a non-specific slower migrating complex.(b) Nuclear extracts (20 μg) from EL-4 cells treated without (lane 1) or with (lane 2) PMA for 4 hr were analysed by Western immunoblot with an anti-GATA-3 antibody. Equal lane loading was ensured by stripping and re-probing for β-actin. (c) Overexpression of AP-2 enhances IL-13 promoter activity in cotransfection assays. EL-4 T cells were transfected with an IL-13 promoter construct containing 109 bp of the proximal promoter (5 μg) together with increasing amounts of an AP-2 expression vector as indicated. After 18 hr, cells were stimulated for an additional 18 hr as indicated and reporter gene expression was analysed by luminometry. Data are from one experiment, representative of n = 3.
Interestingly, an AP-2 consensus oligonucleotide strongly competed for the formation of complex II (Fig. 6a, lane 4). Other consensus sequences including NFAT, AP-1, GRE, CREB and OCT-1, failed to compete, or competed only weakly, for complex II (Fig. 6 and data not shown). AP-2 comprises a family of transcription factors that bind to the sequence 5′ CCCAGGCT 3′.25 Using anti-AP-2 antibodies in EMSA, however, we only observed partial attenuation of complex II without a supershift (data not shown). Taken together with the lack of an obvious AP-2 consensus sequence in oligo II, this suggested that AP-2 or a related factor interacts with low affinity in this region of the IL-13 promoter. To determine if overexpressed AP-2 could enhance IL-13 promoter activity, we cotransfected EL-4 cells with a minimal IL-13 promoter construct and an AP-2 expression vector and analysed reporter gene activity by luminometry.
Discussion
Our study provides several novel observations regarding the regulation of IL-13 gene expression in T cells. We found that the signal requirements for IL-13 gene expression are different in mitogen-primed T cells compared to freshly isolated, unprimed peripheral blood T cells. Activation of the PKC pathway alone was sufficient for IL-13 gene expression and promoter activity in mitogen-primed PBT cells, whereas coactivation of calcium signalling pathways provided an inhibitory signal. We found similar signalling requirements for IL-13 gene expression in the mouse EL-4 thymoma T-cell line, with maximal activation of IL-13 protein secretion, mRNA expression and promoter activity achieved with PKC signalling alone. Further characterization of the promoter revealed a PMA-sensitive element between −109 and −51 bp upstream from the transcription start site. Oligonucleotides from this region supported the binding of a PMA-inducible complex in EMSA which was competed for by an AP-2 consensus oligonucleotide, and overexpression of AP-2 enhanced IL-13 promoter activity in cotransfection assays.
Because PHA is a mitogen that mimics antigen-induced clonal expansion, we interpret our data to mean that the signal requirements for IL-13 expression in clonally expanded T cells (PKC-dependent, calcium-independent) are distinct from otherwise non-expanded lymphocytes (PKC- and calcium-dependent). There are three physiological implications of this conclusion. First, our data suggest that in activated T cells maximal IL-13 expression can be achieved independently of the canonical T-cell receptor activation pathway by cell surface receptors that are capable of activating PKC only. Interestingly, there is growing evidence that signalling by the PKC pathway favours Th2 immune responses.26,27 The identity of these receptors, and whether they could amplify IL-13 gene expression in allergic inflammation, remains to be determined. Potential candidates include G-protein-coupled receptors such as the histamine or lysophosphatidic acid receptor (ref. 28 and Rubenfeld J et al. submitted for publication). Second, altered peptide ligands with reduced T-cell receptor affinity have been shown to induced weak calcium signals and to promote Th2 responses.26,29 Although currently unknown, if these ligands still promote PKC signalling then this could lead to IL-13 secretion in mitogen-primed cells. Third, our results suggest that drugs that antagonize the calcium/calcineurin pathway alone will not be effective in diseases characterized by activated T cells secreting IL-13 (e.g. allergic asthma). Rather, PKC inhibitors may be more effective in this regard.
Our results with mitogen-primed PBT are in keeping with prior studies of IL-13 gene expression in lymphocytes. For example, Luttman et al. reported that PMA alone can induce IL-13 (but not IL-4) expression in PHA-primed peripheral blood mononuclear cells.30 The precise mechanism of this effect requires further study. Furthermore, several groups have reported that calcineurin antagonists enhance IL-13 expression in lymphocytes.17–19 Our results demonstrate that at least part of the inhibitory calcium signal is mediated at the transcriptional level by the proximal IL-13 promoter. Two observations suggest that this is not the result of a direct repressive effect of NFAT on IL-13 transcriptional regulation. First, we found that a proximal promoter construct lacking the NFAT binding site (−109 luc) was still inhibited by coactivation of the pathways for PKC and calcium. Second, overexpression of NFAT can enhance IL-13 promoter activity.31 Therefore, the exact mechanism by which calcium signalling inhibits IL-13 in these models remains to be determined.
In a prior analysis of the human IL-13 promoter, Dolganov et al. reported that the purine-box − 137GGAAAA− 142 which bound NFATp in EMSA, was required for transcriptional activity in PMA-stimulated and calcium ionophore-stimulated Jurkat cells.13 Our results indicate that comparable sequences in the mouse IL-13 promoter are dispensable for promoter inducibility in transiently transfected EL-4 cells (Fig. 3). Interestingly, we also found that although EL-4 cells contain abundant NF-κB, they are devoid of nuclear factors that recognize IL-13 PuB in DNA binding assays (Fig. 4). Taken together these results suggest that IL-13 gene regulation occurs in a distinct fashion in these two cell lines. In a previous study of IL-4 gene regulation, Takemoto et al. found that immunoreactive NFATp bound to two out of four IL-4 promoter P elements using EL-4 nuclear extracts.32 Because each of the P elements readily supports the binding of recombinant NFATp in vitro,33 this suggests that the binding of nuclear NFATp is determined by the promoter context. Therefore, the lack of NFAT binding to IL-13 PuB may indicate that it is a relatively weak NFAT-binding site, and could also reflect relatively low expression of this factor in EL-4 cells. Future experiments will be needed to distinguish between these (and other) possibilities. The deletion analysis and reporter sequences examined in this report are downstream of the recently reported C-T polymorphism at position −1055 of the human IL-13 promoter.34 Interestingly, this polymorphism appears to correlate with both asthma and COPD in different human kindreds.34,35
Our results confirm the recent report by Kishikawa et al. who found that the proximal IL-13 promoter was expressed in a T-cell-dependent manner in transfection assays.14 We also found that a minimal construct containing only 109 bp of the IL-13 promoter was sufficient for inducibility in EL-4 cells (Fig. 3). Lavenu-Bombled et al. recently reported that GATA-3 can activate the mouse IL-13 promoter by binding up to three proximal promoter sequences including − 104AGATGA− 99 (site I), − 94AGATGT− 89 (Site II), and − 82CAGATA− 77 (site III).24 However, in multiple experiments we were unable to demonstrate sequence-specific binding of GATA-3 to this region, despite abundant nuclear expression of this factor (Fig. 5, and data not shown). This may be because the affinity of GATA-3 for individual IL-13 promoter binding sites appears to be relatively low, and DNA binding was only observed under conditions of protein excess.24 However, it remains formally possible that in addition to complex II, GATA-3 (or other factors) could bind in a PMA-dependent manner to additional oligonucleotides spanning the −109 to −52 region. Examining transcription factor binding to the native promoter may answer, at least in part, some of these questions.
In EL-4 cells, we found that PMA alone induced both IL-13 mRNA expression (Fig. 2) and promoter activity (Fig. 3). We localized the PMA response element to a region between nucleotides −109 and −51 upstream from the transcription start site, and found by EMSA that oligonucleotide II (− 93 to −64) supported the binding of both constitutive and PMA-inducible factors. Competition experiments showed that these complexes were effectively competed for by an AP-2 consensus oligonucleotide, and overexpression of AP-2 enhanced the activity of a minimal IL-13 promoter construct. AP-2 comprises a family of developmentally regulated transcription factors that controls the expression of a diverse array of genes including some cytokines.25,36,37 It is well established that AP-2 is a PMA-inducible factor that can mediate phorbol ester-induced gene transcription.38,39 However, the precise link between PMA/PKC-signalling and AP-2 activation remains unclear, and recent studies have highlighted the importance of cofactors40 and post-translational modification41 in regulating the transactivating ability of AP-2. Overexpression of AP-2 resulted in an enhancement of basal promoter activity that was further augmented by stimulation with PMA. This is consistent with a model in which IL-13 promoter activity is regulated by the expression and PMA-induced post-translational modification of AP-2. Our observation that anti-AP-2 antibodies only weakly affected the formation of complex II suggests that this factor binds with low affinity to the proximal IL-13 promoter, possibly in concert with other DNA-bound transcription factors. This is reminiscent of the ability of T-bet to transactivate the interferon-γ promoter with apparently very low affinity DNA-binding that is not detectable by standard gel shift assays.42,43 In summary, we conclude that IL-13 transcriptional regulation involves multiple promoter regulatory elements including a novel PMA-responsive region. A better understanding of the molecular pathways involved in IL-13 gene regulation may lead to the development of novel therapies for asthma and related diseases.
BIND identifiers
One BIND identifier (http://www.bind.ca) is associated with this manuscript: 335751.
Acknowledgments
This work was supported by research grants from the NIH/NHLBI (HLAI61875) and the American Lung Association. S.G. was a recipient of an Allergic Skin Disease Research Award from Fujisawa/AAAAI.
References
- 1.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–61. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 2.Grunig G, Warnock M, Wakil AE, et al. Requirement for IL-13 independently of IL-4 in experimental asthma [see comments] Science. 1998;282:2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graves PE, Kabesch M, Halonen M, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105:506–13. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Z, Homer RJ, Wang Z, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–88. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuperman DA, Huang X, Koth LL, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med. 2002;8:885–9. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 6.Kapp U, Yeh WC, Patterson B, et al. Interleukin 13 is secreted by and stimulates the growth of Hodgkin and Reed-Sternberg cells. J Exp Med. 1999;189:1939–46. doi: 10.1084/jem.189.12.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terabe M, Matsui S, Noben-Trauth N, et al. NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway. Nat Immunol. 2000;1:515–20. doi: 10.1038/82771. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–75. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal S, Viola JP, Rao A. Chromatin-based regulatory mechanisms governing cytokine gene transcription. J Allergy Clin Immunol. 1999;103:990–9. doi: 10.1016/s0091-6749(99)70168-5. [DOI] [PubMed] [Google Scholar]
- 10.Brown MA, Hural J. Functions of IL-4 and control of its expression. Crit Rev Immunol. 1997;17:1–32. doi: 10.1615/critrevimmunol.v17.i1.10. [DOI] [PubMed] [Google Scholar]
- 11.Casolaro V, Keane-Myers AM, Swendeman SL, et al. Identification and characterization of a critical CP2-binding element in the human interleukin-4 promoter. J Biol Chem. 2000;275:36605–11. doi: 10.1074/jbc.M007086200. [DOI] [PubMed] [Google Scholar]
- 12.Guo J, Casolaro V, Seto E, et al. Yin-Yang 1 activates interleukin-4 gene expression in T cells. J Biol Chem. 2001;276:48871–8. doi: 10.1074/jbc.M101592200. [DOI] [PubMed] [Google Scholar]
- 13.Dolganov G, Bort S, Lovett M, et al. Coexpression of the interleukin-13 and interleukin-4 genes correlates with their physical linkage in the cytokine gene cluster on human chromosome 5q23-31. Blood. 1996;87:3316–26. [PubMed] [Google Scholar]
- 14.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by gata-3. J Immunol. 2001;167:4414–20. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 15.Naora H, Altin JG, Young IG. TCR-dependent and -independent signaling mechanisms differentially regulate lymphokine gene expression in the murine T helper clone D10.G4.1. J Immunol. 1994;152:5691–702. [PubMed] [Google Scholar]
- 16.Rao A, Luo C, Hogan P. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–47. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 17.van der Pouw Kraan TC, Boeije LC, Troon JT, Rutschmann SK, Wijdenes J, Aarden LA. Human IL-13 production is negatively influenced by CD3 engagement. Enhancement of IL-13 production by cyclosporin A. J Immunol. 1996;156:1818–23. [PubMed] [Google Scholar]
- 18.Dumont FJ, Staruch MJ, Fischer P, DaSilva C, Camacho R. Inhibition of T cell activation by pharmacologic disruption of the MEK1/ERK MAP kinase or calcineurin signaling pathways results in differential modulation of cytokine production. J Immunol. 1998;160:2579–89. [PubMed] [Google Scholar]
- 19.Pahl A, Zhang M, Kuss H, Szelenyi I, Brune K. Regulation of IL-13 synthesis in human lymphocytes: implications for asthma therapy. Br J Pharmacol. 2002;135:1915–26. doi: 10.1038/sj.bjp.0704656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keen JC, Sholl L, Wills-Karp M, Georas SN. Preferential activation of nuclear factor of activated T cells correlates with mouse strain susceptibility to allergic responses and interleukin-4 gene expression. Am J Respir Cell Mol Biol. 2001;24:58–65. doi: 10.1165/ajrcmb.24.1.3870. [DOI] [PubMed] [Google Scholar]
- 21.McKenzie AN, Li X, Largaespada DA, et al. Structural comparison and chromosomal localization of the human and mouse IL-13 genes. J Immunol. 1993;150:5436–44. [PubMed] [Google Scholar]
- 22.Casolaro V, Georas S, Song Z, Zubkoff I, Abdulkadir S, Thanos D, Ono S. Inhibition of NF-AT-dependent transcription by NF-κB: implications for differential cytokine gene expression. Proc Natl Acad Sci USA. 1995;92:11623–7. doi: 10.1073/pnas.92.25.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegel MD, Zhang DH, Ray P, Ray A. Activation of the interleukin-5 promoter by cAMP in murine EL-4 cells requires the GATA-3 and CLE0 elements. J Biol Chem. 1995;270:24548–55. doi: 10.1074/jbc.270.41.24548. [DOI] [PubMed] [Google Scholar]
- 24.Lavenu-Bombled C, Trainor CD, Makeh I, Romeo PH, Max-Audit I. Interleukin-13 gene expression is regulated by GATA-3 in T cells. Role of a critical association of a GATA and two GATG motifs. J Biol Chem. 2002;277:18313–21. doi: 10.1074/jbc.M110013200. [DOI] [PubMed] [Google Scholar]
- 25.Ye J, Young HA, Zhang X, Castranova V, Vallyathan V, Shi X. Regulation of a cell type-specific silencer in the human interleukin-3 gene promoter by the transcription factor YY1 and an AP2 sequence-recognizing factor. J Biol Chem. 1999;274:26661–7. doi: 10.1074/jbc.274.38.26661. [DOI] [PubMed] [Google Scholar]
- 26.Noble A, Truman JP, Vyas B, Vukmanovic-Stejic M, Hirst WJ, Kemeny DM. The balance of protein kinase C and calcium signaling directs T cell subset development. J Immunol. 2000;164:1807–13. doi: 10.4049/jimmunol.164.4.1807. [DOI] [PubMed] [Google Scholar]
- 27.Marsland BJ, Soos TJ, Spath G, Littman DR, Kopf M. Protein kinase C theta is critical for the development of in vivo T helper (Th) 2 cell but not Th1 cell responses. J Exp Med. 2004;200:181–9. doi: 10.1084/jem.20032229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott KA, Osna NA, Scofield MA, Khan MM. Regulation of IL-13 production by histamine in cloned murine T helper type 2 cells. Int Immunopharmacol. 2001;1:1923–37. doi: 10.1016/s1567-5769(01)00117-5. [DOI] [PubMed] [Google Scholar]
- 29.Tao X, Grant C, Constant S, Bottomly K. Induction of IL-4-producing CD4+ T cells by antigenic peptides altered for TCR binding. J Immunol. 1997;158:4237–44. [PubMed] [Google Scholar]
- 30.Luttmann W, Sengler C, Herzog V, Balkow S, Matthys H, Virchow JC., Jr Differential modulation of interleukin-4 and interleukin-13 secretion from human peripheral blood mononuclear cells. Immunol Lett. 1999;69:225–31. doi: 10.1016/s0165-2478(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 31.Macian F, Garcia-Rodriguez C, Rao A. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. Embo J. 2000;19:4783–95. doi: 10.1093/emboj/19.17.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takemoto N, Koyano-Nakagawa N, Arai N, Arai K, Yokota T. Four P-like elements are required for optimal transcription of the mouse IL-4 gene: involvement of a distinct set of nuclear factors of activated T cells and activator protein-1 family proteins. Int Immunol. 1997;9:1329–38. doi: 10.1093/intimm/9.9.1329. [DOI] [PubMed] [Google Scholar]
- 33.Burke TF, Casolaro V, Georas SN. Characterization of P5, a novel NFAT/AP-1 site in the human IL-4 promoter. Biochem Biophys Res Commun. 2000;270:1016–23. doi: 10.1006/bbrc.2000.2508. [DOI] [PubMed] [Google Scholar]
- 34.Van der Pouw-Krann T, Van Veen A, Boeije LC, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immunity. 1999;1:61–5. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]
- 35.van der Pouw Kraan TC, Kucukaycan M, Bakker AM, et al. Chronic obstructive pulmonary disease is associated with the -1055 IL-13 promoter polymorphism. Genes Immun. 2002;3:436–9. doi: 10.1038/sj.gene.6363896. [DOI] [PubMed] [Google Scholar]
- 36.Fu Y, Ishii KK, Munakata Y, Saitoh T, Kaku M, Sasaki T. Regulation of tumor necrosis factor alpha promoter by human parvovirus B19 NS1 through activation of AP-1 and AP-2. J Virol. 2002;76:5395–403. doi: 10.1128/JVI.76.11.5395-5403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Shin TH, Kudlow JE. Transcription factor AP-2 controls transcription of the human transforming growth factor-alpha gene. J Biol Chem. 1997;272:14244–50. doi: 10.1074/jbc.272.22.14244. [DOI] [PubMed] [Google Scholar]
- 38.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal- transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–60. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 39.Hyman SE, Comb M, Pearlberg J, Goodman HM. An AP-2 element acts synergistically with the cyclic AMP- and phorbol ester-inducible enhancer of the human proenkephalin gene. Mol Cell Biol. 1989;9:321–4. doi: 10.1128/mcb.9.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braganca J, Swingler T, Marques FI, et al. Human CREB-binding protein/p300-interacting transactivator with ED-rich tail (CITED) 4, a new member of the CITED family, functions as a co-activator for transcription factor AP-2. J Biol Chem. 2002;277:8559–65. doi: 10.1074/jbc.M110850200. [DOI] [PubMed] [Google Scholar]
- 41.Eloranta JJ, Hurst HC. Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumoylated in vivo. J Biol Chem. 2002;277:30798–804. doi: 10.1074/jbc.M202780200. [DOI] [PubMed] [Google Scholar]
- 42.Cho JY, Grigura V, Murphy TL, Murphy K. Identification of cooperative monomeric Brachyury sites conferring T-bet responsiveness to the proximal IFN-gamma promoter. Int Immunol. 2003;15:1149–60. doi: 10.1093/intimm/dxg113. [DOI] [PubMed] [Google Scholar]
- 43.Soutto M, Zhang F, Enerson B, Tong Y, Boothby M, Aune TM. A minimal IFN-gamma promoter confers Th1 selective expression. J Immunol. 2002;169:4205–12. doi: 10.4049/jimmunol.169.8.4205. [DOI] [PubMed] [Google Scholar]