Abstract
Cytotoxic T-lymphocyte antigen-4 (CTLA-4; CD152) is a member of the immunoglobulin gene superfamily with strong homology to the receptor CD28 with which it shares the ligands CD80 and CD86. Unlike CD28, a potent costimulator of T-cell responses, CTLA-4 is transiently expressed on the cell surface of activated T cells and appears to operate predominantly as a negative regulator of T-cell proliferation. Signal transduction mechanisms utilized by CTLA-4 remain unclear although several mechanisms have been implicated. In this study, we show that the cytoplasmic domain of CTLA-4, but not of CD28, binds to STAT5 in yeast two-hybrid assay and in coimmunoprecipitation assays. Mutations of Tyr165 and Tyr182 in CTLA-4 did not abrogate the interaction of STAT5 with CTLA-4. Finally, the overexpression of CTLA-4 in Jurkat T cells inhibits STAT-mediated activation of STAT5 responsive elements. These results suggest that CTLA-4 and STAT5 interact in T cells and that this interaction is important for CTLA-4 signalling.
Keywords: CTLA-4 (CD154), STAT5, T-cell activation
Introduction
Cytotoxic T-lymphocyte antigen-4 (CTLA-4; CD152) is a glycoprotein member of the immunoglobulin superfamily. It shares 31% amino acid identity with CD28,1 including an MYPPY motif involved in binding to B7.1 (CD80) and B7.2 (CD86) on antigen-presenting cells.2–4 In contrast to CD28, CTLA-4 is not present on resting T cells, but its synthesis is induced following T-cell activation.5,6 While the CD28 signal is required to sustain and enhance T-cell activation, the interaction of the B7 isoforms with CTLA-4 apparently elicits an inhibitory signal required for down-regulation of the T-cell response.7 In vitro, CTLA-4 ligation in the context of T-cell activation blocks cytokine production and cell cycle progression.5,8,9 In mouse studies, anti-CTLA-4 monoclonal antibodies (mAb) augment immunity and aggravate autoimmunity in multiple models.10–12 Finally, CTLA-4-deficient mice suffer from a rampant, massive T-cell proliferation to which they succumb rapidly within a few weeks of birth.13,14
There is 100% phylogenetic conservation of the CTLA-4 intracytoplasmic domain, suggesting an important signalling function for T cells.1 The mechanisms by which CTLA-4 down-regulates T-cell activation are complex and remain incompletely elucidated. Two tyrosine phosphorylation sites are present in the cytoplasmic tail of CTLA-4, the phosphorylation of which is not required for the function of CTLA-4.15 Yet the phosphorylation of tyr165 is important for cell surface retention, in that it regulates interactions of CTLA-4 with the clathrin adaptor complexes activator proteins 1 and 2.16–18 Because of its relatively higher affinity for CD80 and CD86, CTLA-4 is able to compete effectively, even at low surface expression, with CD28.19,20 In addition, several ligand-dependent negative signalling mechanisms have been described. Following T-cell activation, CTLA-4 becomes colocalized within the immunological synapse in a CD80/CD86-dependent manner, where it can inhibit T-cell receptor-ζ chain phosphorylation, presumably by recruitment of the SHP-2 phosphatase that binds the CTLA-4 cytoplasmic tail.21,22 In addition, the CTLA-4 cytoplasmic tail also binds the serine/threonine phosphatase PP2A, which can act as a repressor of the inhibitory function of CTLA-4.23,24 The role of transforming growth factor-β in CTLA-4-induced down-regulation of immune responses remains somewhat controversial.25,26 Finally, recent data also indicate that CTLA-4 can exert its negative regulatory role in the absence of ligand binding.27 However, the molecular mechanisms underlying this ligand-independent immunoregulatory role have not been defined.
In the present study, we screened the human lymphocyte Matchmaker cDNA library for proteins interacting with the CTLA-4 cytoplasmic tail, and identified STAT5 as a new candidate CTLA-4-interacting protein. This interaction was also confirmed in transfected 293T cells. Finally, overexpression of CTLA-4 in transfected Jurkat T cells inhibits STAT5-mediated activation of a STAT5 responsive element.
Materials and methods
Reagents and plasmids
Anti-haemagglutinin (HA) mAb was a gift from Innogenetics S.A. (Zwijnaarde, Belgium) and anti-STAT5a and anti-STAT5b polyclonal antibody was purchased from PharMingen International (San Diego, CA). Anti-CTLA-4 mAb was produced as described previously.28 CTLA-4 and CD28 plasmids were kindly provided by Paul Sörensen (AB Biotech, Lund, Sweden). The pCDNA3 HA-STAT5a was a gift from Nancy E. Hynes (Friedrich Miescher Institute, Basel, Switzerland) and the vector pACTIIA P50 was a gift from Ellen Chuang (University of Chicago, Chicago, IL).
Two-hybrid screening
Two-hybrid screening in yeast was carried out with the Matchmarker II kit (Clontech, Erembodegen, Belgium) using the cytoplasmic tail of CTLA-4 as bait and the human lymphocyte Matchmaker cDNA library (Clontech) was used as the source of prey vectors. Transformations of the yeast strain Y190 were performed according to the method of Gietz et al.29 The screening was performed on a selective medium containing 30 mm 3-amino-1,2,3-triazol. A result was scored positive when growth occurred in the absence of histidine and when β-galactosidase activity was observed.
Cell lines and transient transfections
Human embryonic kidney (HEK) 293T cells were maintained in Dulbecco's modified Eagle's medium containing 4·5 g glucose/l and 10% fetal bovine serum. Jurkat T-cell lines stably expressing wild-type CTLA-4 were provided by J. Madrenas (John P. Robarts Research Institute, London, ON, Canada). They were maintained in RPMI-1640 containing 10% fetal bovine serum and hygromycin (Life Technologies, Gaithersburg, MD). CTLA-4 mRNA expression in these transfectants was induced by incubation with doxycyclin at 100 ng/ml (Sigma, St Louis, MO).
Transient transfection for coimmunoprecipitation analysis was performed with Fugene 6 (Roche Molecular Biochemicals, Vilvoorde, Belgium), using 2 μl Fugene per μg plasmid DNA. The 293T cells (1·2 × 106) were transfected with 2 µg of each expression vector.
Transfections of the Jurkat T-cell line for luciferase assays were carried out with diethylaminoethyl (DEAE) –dextran;30 10 μg of the reporter construct and 1 μg of cytomegalovirus–β-gal plasmid were prepared in a transfection mixture consisting of 500 μl Tris-buffered-saline–dextrose (pH 7·4), 40 μl DEAE–dextran (50 mg/ml) and 9·4 ml RPMI-1640. The cells were added to the mix and incubated for 20 min at 37° and 5% CO2, after which the suspension was washed in RPMI-1640 supplemented with 15 U heparin. The cells were kept overnight in complete medium at 37°.
Flow cytometry
Cells were suspended in 100 μl of phosphate-buffered saline containing 1% w/w bovine serum albumin (Sigma Chemicals). The cells were incubated for 30 min at 4° with phycoerythrin (PE)-conjugated anti-CTLA-4 mAb (BNI3, purchased from Becton Dickinson (Erembodegem, Belgium). After two washes, the cells were fixed with 1% w/v paraformaldehyde, and acquisition was performed on a FACSCAN flow cytometer (Becton Dickinson). Data were analysed with cellquest software (Becton Dickinson).
Co-immunoprecipitation and Western blotting
Cells were harvested 24–48 hr after transfection in 300 μl lysis buffer (50 mm Tris–HCl, pH 7·4, 200 mm NaCl, 10% glycerol, 0·2% Nonidet P-40, 50 mm NaF, 1 mm Na4P2O7, 5 mm Na3VO4, 1 mm phenylmethylsulphonyl fluoride, 3 μg aprotinine/ml). Cells were then lysed by incubation for 20 min on ice or by passing five times through a 22-gauge needle. Cellular debris and nuclei were removed by centrifugation (Eppendorf 4517R, 17 500 g at 4° for 10 min). Then, 5 μg antibody was added to the lysate and the mixture was incubated for 3 hr at 4°. Subsequently, 20 μl of a 50% slurry of protein-G Sepharose (Amersham Pharmacia Biotech, Roosendaal, The Netherlands) was added to the samples, and incubation was continued for 1 hr. Next, the Sepharose was washed four times in 750 μl of the lysis buffer for 10 min at 4°. Finally, the beads were mixed with 20 μl sample buffer, and the samples were analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). To verify the expression levels of different proteins, 0·1% of the cytoplasmic extract was analysed on Western blot. Proteins were separated on 10% Tris–Tricine gels and transferred onto polyvinyldifluoride (PVDF) membranes (NEN Life Science Products, Zaventem, Belgium) using a semidry blotting apparatus (Sigma). For Western blot analysis, the membrane was blocked in 3% skimmed milk in TBS-T (10 mm Tris–HCl, pH 7·4, 150 mm NaCl, 0·2% Tween-20). After sequential incubations with primary antibody and horseradish peroxidase-conjugated secondary antibody (Jackson Laboratories, West Grove, PA) for 1 hr at 24°, proteins were visualized with the ECC chemiluminescence detection system (NEN Life Science Products).
Results and discussion
A yeast two-hybrid screen was undertaken to identify putative proteins interacting with the cytoplasmic domain of CTLA-4. The cDNA encoding the cytoplasmic region of CTLA-4 (amino acids K156 to N190) was cloned into the pActII vector and used as bait. A cDNA library obtained from the HTLV-1-transformed T-cell line SLB-I was available in the prey vector. After transformation of bait and prey vectors into yeast cells, approximately 2 million transformants were screened on selection media without Trp and Leu, and about 200 colonies were found to express both the HIS3 marker and the LacZ reporter. The insert of 10 of these positive prey plasmids encoded a 635 amino acid polypeptide corresponding to the N-terminal region of STAT5a.
As confirmation and additional control, cDNAs encoding the 635 amino acid region of STAT5a, the protein AP50 (the medium chain of the clathrin-associated coated pit adaptor protein complex AP2) and Xenopus SMAD1, respectively, were cloned into the prey vector and used to retransform yeast cells along with the CTLA-4 cytoplasmic tail bait vector. This confirmed interaction of the CTLA-4 cytoplasmic tail with STAT5a, and showed interaction with AP50, as predicted, but not with the negative control Xenopus-SMAD1.16,31 As shown in Fig. 1, no interaction between the cytoplasmic tail of CD28 and STAT5a could be demonstrated in this yeast two-hybrid assay. Thus, in this assay, STAT5a interacts specifically with the cytoplasmic tail of CTLA-4, but not with CD28. This interaction was not affected by a Tyr → Phe substitution at position 165 or position 182. In contrast, substitution of Y165 and Y182, respectively, abolished or greatly reduced the interaction of AP50, with the CTLA-4 cytoplasmic tail, which is consistent with published data16,18,31,32 (Fig. 1).
Figure 1.
Yeast two-hybrid assay. The cytoplasmic domain of human CTLA-4 was used to screen a cDNA library generated from a HTLV-1-transformed T-cell line SLB-I, using a yeast two-hybrid system. Positive interaction was confirmed by expression of both the HIS3 and LacZ gene, thereby conferring ability on the positive clones to grow on media lacking His and to turn blue on media containing the chromogenic substrate X-Gal. Ten of the isolated clone inserts encoded 635 amino acids of STAT5a. To establish the specificity of this interaction, the plasmid encoding the 635 amino acid long segment of STAT5a was re-isolated from the library plasmid pACT2. This was then retransformed in yeast along with a vector encoding the cytoplasmic domain of CTLA-4 (CTLA-4 wt), CTLA-4 with a mutation at position 165 (CTLA-4 ms) or position 185 (CTLA-4 md) and CD28, respectively. Vectors encoding AP50 or Xenopus SMAD1 were used as positive and negative controls, respectively. Positive interaction was scored by the expression of HIS or LacZ. The relative β-galactosidase activity is indicated (blue colonies shown in grey). Two representative colonies are shown for each interaction.
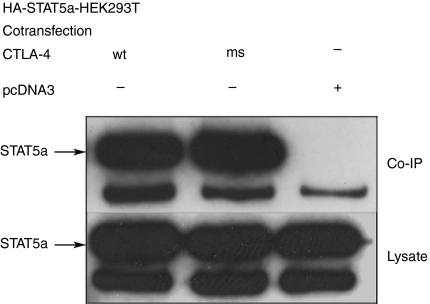
We further sought to demonstrate that full-length CTLA-4 and STAT5a/b can interact with one another in mammalian cells. To this end, we cotransfected expression vectors encoding full-length CTLA-4 and HA-tagged STAT5a or STAT5b (not HA-tagged) into HEK293T cells. CTLA-4 was immunoprecipitated using anti-human CTLA-4 mAb, followed by SDS–PAGE and immunoblotting with an anti-HA-antibody or with a polyclonal anti-STAT5b antibody. As shown in Fig. 2(a,b), both STAT5a and STAT5b were specifically coimmunoprecipitated by anti-CTLA-4. As it has been shown previously that the signalling function of CTLA-4 does not require the presence of a tyrosine at position 165,15 we also cotransfected the STAT5a vector with a plasmid encoding a mutant CTLA-4 cytoplasmic tail with phenylalanine in position 165. When immunoprecipitated with anti-CTLA-4, this mutant CTLA-4 tail was also shown to coimmunoprecipitate STAT5a (Fig. 3), confirming our yeast two-hybrid assay result. Therefore, the association of STAT5a with the CTLA-4 cytoplasmic tail in mammalian cells does not require the presence of Tyr165.
Figure 2.
Coimmunoprecipitation of STAT5a and STAT5b with CTLA-4. Two micrograms HA-tagged STAT5a in pcDNA3 (a) or STAT5b in pcDNA3 (b)were transfected in HEK293T cells together with 2 μg CTLA-4-pcDNA3 or with the pcDNA3 empty vector, as indicated (+ or −). Between 24 and 36 hr after transfection, cells where harvested and lysed. Lysates were immunoprecipitated with anti-CTLA-4 mAbs, the immunoprecipitate was subjected to SDS–PAGE, blotted onto PVDF and immunostained with anti-HA mAbs (a) or anti-STAT5b polyclonal antibodies (b) (upper panel). Total lysates were separated by SDS–PAGE, blotted onto PVDF immunostained with anti-HA mAbs (a) or anti-STAT5b polyclonal antibodies (b) showing comparable amount of STATS5a/b in each lane.
Figure 3.
Coimmunoprecipitation of STAT5a with CTLA-4 mutants. Two micrograms STAT5a-pcDNA3 was transfected in HEK293T cells together with 2 μg CTLA-4-pcDNA3, 2 μg CTLAms-4-pcDNA3 or with the pcDNA3 empty vector as indicated. 24–36 hr after transfection cells where harvested and lysed. Upper panel: lysates were immunoprecipitated with anti-CTLA-4 mAbs, the immunoprecipitate was subjected to SDS–PAGE, blotted onto PVDF and immunostained with anti-HA mAbs. Lower panel: total lysates were separated by SDS–PAGE, blotted onto PVDF immunostained with anti-HA mAb.
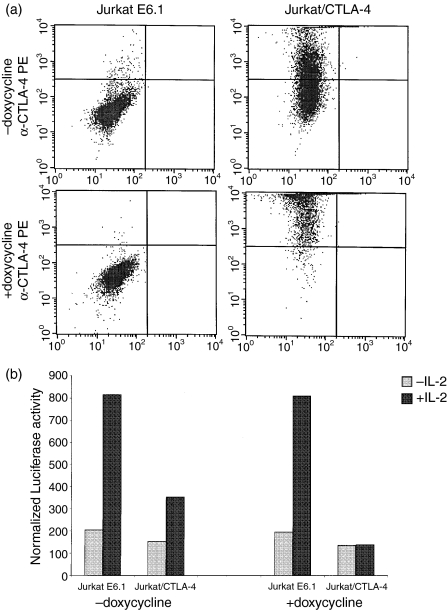
The T-cell leukaemia cell line Jurkat E6.1 expresses STAT5 at low levels, migrating as a 95 000 molecular weight band after immunoprecipitation (data not shown). However, Jurkat E6.1 cells do not express CTLA-4 protein or CTLA-4 mRNA (ref. 6 and data not shown). In an attempt to demonstrate an association between STAT5 and the CTLA-4 cytoplasmic tail in a mammalian cell system representative of T cells, we used stably transformed Jurkat cells with doxycyclin-inducible CTLA-4 expression. However, before or after induction with doxycyclin, we were unable to coimmunoprecipitate STAT5a using human CTLA-4 antibodies, not even from 20 × 106 CTLA-4-expressing Jurkat cells. Yet, it is possible that the low endogenous levels of STAT5 in Jurkat cells preclude detection in this coimmunoprecipitation assay of a potential physical association between STAT5 and the CTLA-4 cytoplasmic tail. Therefore, we tried to obtain functional evidence supporting the hypothesis that the CTLA-4 cytoplasmic tail associates with STAT5a. STAT5a/b belong to the stat (signal transducer and activator of transcription) family of proteins, comprising several related proteins that mediate intracellular signal transduction for growth factors such as CNTF (Ciliary Neurotropic Factor), interleukin-2 (IL-2) and IL-4. Upon binding of cytokines to their cognate receptors, specific STAT family members will become phosphorylated, will dimerize and will translocate to the nucleus where they directly regulate transcription of target genes.33 STAT5a and STAT5b are of pivotal importance for signal transduction through the IL-2 receptor, as is shown by the grossly impaired T-cell response to IL-2 and the immunodeficiency state in mice deficient in STAT5a or STAT5b.34,35 We hypothesized that the association of STAT5a with CTLA-4 might reduce its intracellular availability and thus antagonize signal transduction through the IL-2 receptor. To test this, a STAT5-responsive luciferase reporter construct was transiently transfected into Jurkat E6.1 cells, and into Jurkat cells expressing CTLA-4 in a doxycyclin-inducible manner.36 Of note, the CTLA-4-transfected Jurkat cells have a leaky phenotype in that they express CTLA-4 in the absence of doxycyclin, albeit in lower amounts than after doxycyclin induction (Fig. 4a). Exposure of Jurkat E6.1 cells to IL-2 in a concentration of 200 U/ml for 24–36 hr led to a five-fold up-regulation of luciferase activity. A smaller increase was observed in the Jurkat cells stably transformed with a doxycyclin-sensitive CTLA-4 transgene (Fig. 4b). In the presence of doxycyclin, the IL-2-induced up-regulation of luciferase activity was completely abolished in the CTLA-4-transfected cells, while doxycyclin exposure did not affect the up-regulation in wild-type Jurkat cells. Therefore, these results are consistent with the hypothesis that expression of CTLA-4 expression negatively interferes with STAT5-regulated transcriptional events. Moreover, the inhibition of IL-2-induced luciferase activity seems to correlate with surface expression levels of CTLA-4 (Fig. 4a,b).
Figure 4.
CTLA-4 expression in Jurkat cells and its effect on a STAT5-responsive reporter. (a)Wild Jurkat E6.1 and CTLA-4-transfected Jurkat cells were incubated without or with doxycyclin. Following immunostaining with PE-conjugated CTLA-4 mAb, surface levels of CTLA-4 were analysed by flow cytometry. Dot plots of reactivity with PE-conjugated anti-CTLA-4 (y-axis) are shown. (b)Jurkat E6.1 or Jurkat cells stably transfected to express CTLA-4 were transfected with a GAS-luciferase reporter plasmid. Each transfection mixture was split into two different wells, one with and one without doxycyclin. 24 hr after transfection, samples were left unstimulated (– IL-2) or were stimulated with IL-2 (200 U/ml) (+ IL-2). The results are representative of four independent transfections.
In this report, we thus show that the cytoplasmic domain of CTLA-4 binds to STAT5 in yeast two-hybrid and in coimmunoprecipitation assays using CTLA-4 and STAT5 overexpressing 293T cells. Mutations of Tyr165 and Tyr182 in CTLA-4, which do not influence the inhibitory function of CTLA-4,15 did not abrogate the interaction with STAT5. Finally, the expression of CTLA-4 in Jurkat T cells down-regulates IL-2-induced STAT5-mediated activation of target gene expression in an expression-dependent way and in the absence of ligation of CTLA-4 by antibody or CD80/CD86 ligand. These results suggest that CTLA-4 may compete for intracellular STAT5 and that this competition may be important for the function of CTLA-4. We admit that we were not able to demonstrate a physical interaction between STAT5 and CTLA-4 in primary T cells nor in Jurkat cells; probably because of low endogenous STAT5 levels in these cell types. Given the difficulties of investigating the effect of CTLA-4 expression on STAT5-driven transcriptional events in primary T cells, comparative studies in wild-type and CTLA-4-deficient mice will be instrumental in confirming the physiological relevance of our findings.
However, our findings are novel and important in that they suggest a link between CTLA-4 signal transduction and STAT5-mediated transcriptional events of the IL-2 pathway. CTLA-4 can also inhibit other transcription factors such as stat6, nuclear factor-κB and AP1.37,38 Our findings could fit with the recently proposed model of ligand-independent signalling of CTLA-4, that affects downstream signalling in T cells independently of the proximal TCR-mediated signalling events.27 Further investigations in mouse models are required to demonstrate the physiological relevance of the interaction between CTLA-4 and STAT5 in CTLA-4 signal transduction and T-cell immunity.
Acknowledgments
We thank J. Madrenas for providing Jurkat T-cell lines stably expressing wild-type CTLA-4. Many thanks also go to D. Huylebroeck for critical reading of the manuscript and discussions. This research was supported by grants 1.5.994.95 and G.0156.96 of Fonds voor Wetenschappelijk Onderzoek (FWO)-Vlaanderen to P.V. P.V. is a Clinical Investigator of FWO-Vlaanderen; S.P. was supported by a research fellowship of the Flemish Institute for the Promotion of Scientific/Technological Research in Industry (IWT), Brussels. J.E.R. was supported by VIB, and L.A.v.G. was a postdoctoral fellow of FWO-Vlaanderen. M.S. is recipient of a stipend from the Sophea-CRUH foundation.
References
- 1.Harper K, Balzano C, Rouvier E, Mattei MG, Luciani MF, Golstein P. CTLA-4 and CD28 activated lymphocyte molecules are closely related in both mouse and human as to sequence, message expression, gene structure, and chromosomal location. J Immunol. 1991;147:1037–44. [PubMed] [Google Scholar]
- 2.Freeman GJ, Gribben JG, Boussiotis VA, Ng JW, Restivo VA, Jr, Lombard LA, Gray GS, Nadler LM. Cloning of B7-2: a CTLA-4 counter-receptor that costimulates human T cell proliferation. Science. 1993;262:909–11. doi: 10.1126/science.7694363. [DOI] [PubMed] [Google Scholar]
- 3.Freeman GJ, Borriello F, Hodes RJ, et al. Uncovering of functional alternative CTLA-4 counter-receptor in B7-deficient mice. Science. 1993;262:907–9. doi: 10.1126/science.7694362. [DOI] [PubMed] [Google Scholar]
- 4.Lenschow DJ, Su GH, Zuckerman LA, Nabavi N, Jellis CL, Gray GS, Miller J, Bluestone JA. Expression and functional significance of an additional ligand for CTLA-4. Proc Natl Acad Sci USA. 1993;90:11054–8. doi: 10.1073/pnas.90.23.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, Thompson CB, Bluestone JA. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1:405–13. doi: 10.1016/1074-7613(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 6.Freeman GJ, Lombard DB, Gimmi CD, et al. CTLA-4 and CD28 mRNA are coexpressed in most T cells after activation. Expression of CTLA-4 and CD28 mRNA does not correlate with the pattern of lymphokine production. J Immunol. 1992;149:3795–801. [PubMed] [Google Scholar]
- 7.Riley JL, June CH. The CD28 family. A T-cell rheostat for therapeutic control of T-cell activation. Blood. 2005;105:13–21. doi: 10.1182/blood-2004-04-1596. [DOI] [PubMed] [Google Scholar]
- 8.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182:459–65. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med. 1996;183:2533–40. doi: 10.1084/jem.183.6.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karandikar NJ, Vanderlugt CL, Walunas TL, Miller SD, Bluestone JA. CTLA-4: a negative regulator of autoimmune disease. J Exp Med. 1996;184:783–8. doi: 10.1084/jem.184.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eagar TN, Karandikar NJ, Bluestone JA, Miller SD. The role of CTLA-4 in induction and maintenance of peripheral T cell tolerance. Eur J Immunol. 2002;32:972–81. doi: 10.1002/1521-4141(200204)32:4<972::AID-IMMU972>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Luhder F, Chambers C, Allison JP, Benoist C, Mathis D. Pinpointing when T cell costimulatory receptor CTLA-4 must be engaged to dampen diabetogenic T cells. Proc Natl Acad Sci USA. 2000;97:12204–9. doi: 10.1073/pnas.200348397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers CA, Sullivan TJ, Allison JP. Lymphoproliferation in CTLA-4-deficient mice is mediated by costimulation-dependent activation of CD4+ T cells. Immunity. 1997;7:885–95. doi: 10.1016/s1074-7613(00)80406-9. [DOI] [PubMed] [Google Scholar]
- 14.Waterhouse P, Penninger JM, Timms E, et al. Lymphoproliferative disorders with early lethality in mice deficient in CTLA-4. Science. 1995;270:985–8. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 15.Baroja ML, Luxenberg D, Chau T, Ling V, Strathdee CA, Carreno BM, Madrenas J. The inhibitory function of CTLA-4 does not require its tyrosine phosphorylation. J Immunol. 2000;164:49–55. doi: 10.4049/jimmunol.164.1.49. [DOI] [PubMed] [Google Scholar]
- 16.Shiratori T, Miyatake S, Ohno H, Nakaseko C, Isono K, Bonifacino JS, Saito T. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity. 1997;6:583–9. doi: 10.1016/s1074-7613(00)80346-5. [DOI] [PubMed] [Google Scholar]
- 17.Schneider H, Martin M, Agarraberes FA, Yin L, Rapoport I, Kirchhausen T, Rudd CE. Cytolytic T lymphocyte-associated antigen-4 and the TCR zeta/CD3 complex, but not CD28, interact with clathrin adaptor complexes AP-1 and AP-2. J Immunol. 1999;163:1868–79. [PubMed] [Google Scholar]
- 18.Chuang E, Alegre ML, Duckett CS, Noel PJ, Vander Heiden MG, Thompson CB. Interaction of CTLA-4 with the clathrin-associated protein AP50 results in ligand-independent endocytosis that limits cell surface expression. J Immunol. 1997;159:144–51. [PubMed] [Google Scholar]
- 19.Collins AV, Brodie DW, Gilbert RJ, et al. The interaction properties of costimulatory molecules revisited. Immunity. 2002;17:201–10. doi: 10.1016/s1074-7613(02)00362-x. [DOI] [PubMed] [Google Scholar]
- 20.van der Merwe PA, Bodian DL, Daenke S, Linsley P, Davis SJ. CD80 (B7-1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J Exp Med. 1997;185:393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee KM, Chuang E, Griffin M, et al. Molecular basis of T cell inactivation by CTLA-4. Science. 1998;282:2263–6. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 22.Marengere LE, Waterhouse P, Duncan GS, Mittrucker HW, Feng GS, Mak TW. Regulation of T cell receptor signaling by tyrosine phosphatase SYP association with CTLA-4. Science. 1996;272:1170–3. doi: 10.1126/science.272.5265.1170. [DOI] [PubMed] [Google Scholar]
- 23.Chuang E, Fisher TS, Morgan RW, et al. The CD28 and CTLA-4 receptors associate with the serine/threonine phosphatase PP2A. Immunity. 2000;13:313–22. doi: 10.1016/s1074-7613(00)00031-5. [DOI] [PubMed] [Google Scholar]
- 24.Baroja ML, Vijayakrishnan L, Bettelli E, et al. Inhibition of CTLA-4 function by the regulatory subunit of serine/threonine phosphatase 2A. J Immunol. 2002;168:5070–8. doi: 10.4049/jimmunol.168.10.5070. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan TJ, Letterio JJ, van Elsas A, et al. Lack of a role for transforming growth factor-beta in cytotoxic T lymphocyte antigen-4-mediated inhibition of T cell activation. Proc Natl Acad Sci USA. 2001;98:2587–92. doi: 10.1073/pnas.051632398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Jin W, Wahl SM. Engagement of cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) induces transforming growth factor beta (TGF-beta) production by murine CD4(+) T cells. J Exp Med. 1998;188:1849–57. doi: 10.1084/jem.188.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chikuma S, Abbas AK, Bluestone JA. B7-independent inhibition of T cells by CTLA-4. J Immunol. 2005;175:177–81. doi: 10.4049/jimmunol.175.1.177. [DOI] [PubMed] [Google Scholar]
- 28.Vandenborre K, Delabie J, Boogaerts MA, De Vos R, Lorre K, De Wolf-Peeters C, Vandenberghe P. Human CTLA-4 is expressed in situ on T lymphocytes in germinal centers, in cutaneous graft-versus-host disease, and in Hodgkin's disease. Am J Pathol. 1998;152:963–73. [PMC free article] [PubMed] [Google Scholar]
- 29.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucl Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. Embo J. 1995;14:2876–83. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradshaw JD, Lu P, Leytze G, Rodgers J, Schieven GL, Bennett KL, Linsley PS, Kurtz SE. Interaction of the cytoplasmic tail of CTLA-4 (CD152) with a clathrin-associated protein is negatively regulated by tyrosine phosphorylation. Biochemistry. 1997;36:15975–82. doi: 10.1021/bi971762i. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Allison JP. Interaction of CTLA-4 with AP50, a clathrin-coated pit adaptor protein. Proc Natl Acad Sci USA. 1997;94:9273–8. doi: 10.1073/pnas.94.17.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivashkiv LB, Hu X. Signaling by STATs. Arthritis Res Ther. 2004;6:159–68. doi: 10.1186/ar1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakajima H, Liu XW, Wynshaw-Boris A, Rosenthal LA, Imada K, Finbloom DS, Hennighausen L, Leonard WJ. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- 35.Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, Leonard WJ. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188:2067–74. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carreno BM, Bennett F, Chau TA, Ling V, Luxenberg D, Jussif J, Baroja ML, Madrenas J. CTLA-4 (CD152) can inhibit T cell activation by two different mechanisms depending on its level of cell surface expression. J Immunol. 2000;165:1352–6. doi: 10.4049/jimmunol.165.3.1352. [DOI] [PubMed] [Google Scholar]
- 37.Olsson C, Riesbeck K, Dohlsten M, Michaelsson E. CTLA-4 ligation suppresses CD28-induced NF-kappaB and AP-1 activity in mouse T cell blasts. J Biol Chem. 1999;274:14400–5. doi: 10.1074/jbc.274.20.14400. [DOI] [PubMed] [Google Scholar]
- 38.Pioli C, Gatta L, Ubaldi V, Doria G. Inhibition of IgG1 and IgE production by stimulation of the B cell CTLA-4 receptor. J Immunol. 2000;165:5530–6. doi: 10.4049/jimmunol.165.10.5530. [DOI] [PubMed] [Google Scholar]