Abstract
Antibodies are adaptor molecules that neutralize pathogens and link humoral and cellular defence mechanisms. Immunoglobulin D (IgD), one of the five antibody classes present in mammals, is expressed as an antigen receptor on naïve B cells. The functional role that IgD plays in the immune response is still poorly understood, but the recent characterization of immunoglobulin heavy constant delta genes (IGHD) in a variety of species challenges the view that IgD is of minor importance and is not present in many animals. On the basis of serological studies, IgD appears to be expressed in the majority of mammalian species examined. To confirm, at the molecular level, that IgD is present in different species, we cloned and sequenced IGHD cDNA from dogs and five non-human primate species (chimpanzee, rhesus macaque, cynomolgus macaque, baboon and sooty mangabey). Our results show that in all six species, IgD heavy chains possess three immunoglobulin domains and a long hinge region encoded by two exons. Only the hinge region of non-human primates is similar to the human hinge region, with conservation of O-glycosylation sites and multiple charged residues at opposing ends. The preservation of IgD in primates, dogs and previously characterized species suggests an important functional role for IgD, possibly involving binding to a receptor. The high degree of similarity existing between the structural features of human and non-human primate IgD suggests that non-human primates are suitable for in vivo studies designed to define the role that IgD plays in the immune response.
Keywords: immunoglobulin heavy constant delta, IgD, non-human primates, dog, B-cell receptor
Introduction
Immunoglobulin D (IgD) is the least understood of the five antibody classes found in mammals, both from a functional and evolutionary perspective. On the surface of naïve B cells, IgD functions as an antigen receptor in apparent redundancy with IgM. Indeed, IgM heavy chain gene (IGHM) knockout mice appear to be healthy, with only a slight reduction of their B-cell compartment.1 IgD is also present in a secreted form, with sera concentrations (40 µg/ml in adults) considerably less than those of IgG, IgA and IgM.2 Attempts to identify the function of the secreted form have languished in part because of the difficulty of obtaining purified IgD.2 Initially described in 1965 as a human myeloma protein3 IgD was subsequently characterized in humans, mice and rats at both the protein and genetic levels.3–8 The presence of IgD in non-human primates was firmly established soon after that of human IgD.9 Putative IgD molecules were also identified in other species including chicken, rabbit, dog and tortoise.10–15 Additional studies failed to identify IgD either at the protein or the genetic level in other species (swine, cows, sheep, duck and African clawed frog) as well as in the same species (chicken and rabbit).16–20 The finding that mouse and human IgD are structurally different argued against a conserved IgD function.5 These combined observations contributed to the speculation that IgD evolved recently, was repeatedly deleted in different species, and lacks a major function. However, results from recent studies indicate that IgD plays an important role in the immune system.2 IgD expression is differentially regulated from that of IgM21 and antigen-binding properties differ for IgM and IgD because of differences in their hinge regions.22 Antigen cross-linking of IgD on B cells leads to a stronger and more prolonged signal than that of IgM23 and in IgD-deficient mice affinity maturation is slower than that of normal mice.24
Analysis of newly described immunoglobulin heavy constant delta genes (IGHD) has greatly altered and expanded the understanding of IgD biology and evolution. Until the late 1990s, only human and mouse DNA sequences were available in addition to a partial rat sequence. Even with this paucity of genetic information, it is apparent that IGHD properties are quite divergent between species. The human IGHD consists of eight exons: one encoding the first immunoglobulin domain (CH1), two encoding 58 amino acids of an extended hinge region (H1 and H2), two encoding the second and third immunoglobulin domains (CH2 and CH3), one encoding a hydrophilic secretory tail (CH-S), and two encoding the membrane tail (M1 and M2).5 By contrast mouse and rat IGHD have only 6 exons (CH1, a single hinge exon, CH3, CH-S, M1 and M2).6–8 The human IgD hinge region is characterized by a highly O-glycosylated N-terminal end encoded by H1 and a highly charged C-terminal end encoded by H2. The rodent IgD hinge region is shorter and appears to be structurally unrelated to the human hinge.7 In 1997, Wilson et al.25 described an IGHD in channel catfish, which contains seven tandem immunoglobulin exons and lacks any hinge exon. IGHD has since been identified in Atlantic cod, Japanese flounder, carp, Atlantic salmon, Atlantic halibut, rainbow trout, fugu and zebra fish. These genes also encode IgD heavy chains without hinge regions and consist of various numbers of tandem immunoglobulin domain encoding exons, which for some species are repeated in clusters.26–35 The fish IgD heavy chains are characterized by the fusion of their N-terminal end with the CH1 of IgM, which results in unique chimeric molecules. Recently, IGHD of cow, sheep, pig and horse have been sequenced. Their exon configuration is similar to that found in humans.36–38 With the exception of the pig IgD, which has the H2 exon spliced out,37 in ungulates the IgD hinge regions are all encoded by two exons. However, their hinge regions are dissimilar to those of humans and rodents in sequence. Interestingly, pig IGHD transcripts can have the IgM CH1 fused to their 5′ end as described in fish.37 Together, these data demonstrate that IGHD has an ancient origin, is distributed widely across vertebrate taxa, and is structurally diverse particularly within the hinge region.
Despite the early recognition of the presence of IgD both on B cells and as a secreted protein in non-human primates9,39–42 IgD has not been studied in these animals at the genetic level. In pioneering studies of IgD function, injection of anti-IgD antiserum into rhesus macaques was shown to enhance antibody responses to antigen in an adjuvant-like manner and lead to hypergammaglobulinaemia, indicating a role for IgD in regulation of humoral responses.43,44 Because of their similarities to humans, non-human primates are commonly used as models to understand pathogenesis for a variety of human diseases and to develop therapeutic and preventive approaches.45,46 Although IgD in non-human primates appears well conserved with human IgD based on serology42 more detailed studies are required to determine the extent to which this is so, particularly in light of the divergence of IgD seen in other species. A dog immunoglobulin with IgD-like properties, including B-cell surface expression and lack of cross-reactivity with antibodies against the other dog isotypes, has been identified.14 As pointed out by Naessens16 conclusive evidence that this immunoglobulin is indeed canine IgD, such as cross-reactivity with known anti-IgD antibodies, is still lacking. Hence, the presence of IgD in dogs remains to be established, particularly as it has previously been proposed that IgD may have been deleted from many mammals.16,26
The human IgD hinge region has structural features in common with the hinge region of IgA, including a repetitive sequence. It has been suggested that this repetition is possibly responsible for the genetic instability and diversification of the IgA hinge region.47 If this is correct, the hinge regions of IgD from species closely related to humans might be expected to be highly divergent and polymorphic. On the other hand, the O-glycans of the human hinge are responsible for forming interactions with an IgD receptor expressed on human CD4+ and CD8+ T cells.48,49 If the human IgD receptor has orthologues in non-human primates and it is important for survival, then the hinge region of non-human primates may be well conserved with that of human IgD. Therefore, we have sequenced IGHD from chimpanzee, rhesus macaque, cynomolgus macaque, baboon and sooty mangabey. We have also sequenced dog IGHD, thus confirming the presence of IgD in Carnivora, and expanding the growing body of evidence that IgD is present in most mammals and likely to be functionally important.
Materials and methods
Blood samples and RNA extraction
Total RNA was extracted from heparinized whole blood of two rhesus macaques (Macaca mulatta), three cynomolgus macaques (Macaca fascicularis), two baboons (Papio hamadryas anubis) and one sooty mangabey (Cercocebus torquatus), whereas total RNA was extracted from isolated peripheral blood mononuclear cells (PBMC) of two chimpanzees (Pan troglodytes) and one dog (Canis familiaris) using the QIAamp RNA Blood Mini Kit (Qiagen Inc., Valencia, CA). PBMC were isolated from whole blood by Histopaque®-1077 (Sigma-Aldrich, St. Louis, MO) centrifugation. All animals used were healthy. Macaques and baboons samples were from animals housed at the Southwest National Primate Research Center (San Antonio, TX). The sooty mangabey and the chimpanzees, Tika and Manuel, were housed at the Yerkes National Primate Research Center (Emory University, Atlanta, GA). Whole dog blood was purchased from Harlan Bioproducts for Science, Inc. (Madison, WI).
Amplification, cloning and sequencing of IgD heavy chain cDNAs
RNA was reverse transcribed into cDNA using oligo d(T)17 primers, followed by primer extension with AMV reverse transcriptase (Roche Molecular Biochemicals, Indianapolis, IN). Polymerase chain reaction (PCR) amplification of cDNA was performed with Expand High Fidelity polymerase (Roche Molecular Biochemicals). Primers for amplification of primate IGHD were designed on the basis of the human sequence assuming conserved homology between primates. The forward primer IgD7 (5′-CGGATGTGTTCCCCATCATATCAG-3′) is located in the 5′ end of the human CH1 exon (16–39nt). Two reverse primers were used with IgD7, IgD3 (5′-ACCCAGAAGTGTTCACCTCACG-3′) located in the center of the CH3 exon (135–156nt) and IgD13 (5′-AGCTGACTTCTAGGCTCCGGCT-3′) located at the 3′ end of CH3 exon (303–324nt). Canine primers were designed from DNA sequences of a contig (GenBank accession number NW_140211) predicted to encode an IgD heavy chain-like protein. The canine forward primer K9IGHDF1 (5′-ATCGTCACTTCTGCTCCCCTTG-3′) is located in the canine IgD CH1 exon (12–33nt). The canine reverse primer K9IGHDB6 (5′-AGCAAAAAGGCAAGGGGCTG-3′) is located in a region upstream of a polyadenylation signal and downstream of the M2 exon. After initial denaturation at 95° for 10 min, cDNAs was amplified for 40 cycles, with each cycle consisting of 94° for 1 min, 56° for 1 min and 72° for 1 min 30 s. A final step at 72° for 10 min was used to ensure complete extension. Primers IgD7 and IgD13 yield a human product of 1110 bp and primers IgD7 and IgD3 yield one of 942 bp. All reactions were performed in at least two independent reverse transcription PCRs to verify product sequences. At least 10 clones were sequenced for each animal of the primate species and six clones were sequenced for dog. An additional primer set was used to amplify the baboon IgD hinge region exons and the immediate surround nucleotides (BGDF 5′-AGTACAAATGCACCGTCAAGCAT-3′ and BGDR 5′-CGAAGCAGGTGAAGGTGACTTTG-3′).
Cloning of the amplified gene sequences
For cloning, 100 µl of a reverse transcription PCR was run on a 1% agarose gel. The specific band of interest was excised from the gel and purified using a QIAquick® Gel Extraction Kit (QIAgen). The cDNA was ligated into TopoTA vector and transformed into Top10 Esherichia coli (Invitrogen, Carlsbad, CA). Plasmid DNA was purified using a FastPlasmid® Mini kit (Hamburg, Germany) and screened on a 1% agarose gel after digestion with EcoRI to confirm the correct size of the DNA fragments. All DNA sequences were determined using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on an ABI 3100 Genetic Analyzer (Perkin Elmer, Wellesley, MA). The forward and reverse M13 primers were used for the sequencing. For the canine DNA fragments, an additional primer, K9IGHDB2 (5′-TGATCCAGGTGAGGAGGATGTCAG-3′), located in the CH3 exon, was used.
Analysis of DNA sequences
Overlapping regions were identified and sequences were edited using the MacVector software program (Accelrys Inc., San Diego, CA). Sequences were aligned with each other and other known IGHD using the CLUSTAL function of the MEGALIGN part of the LASERGENE software package (DNASTAR Inc., Madison, WI). ImMunoGeneTics (IMGT) standardized nomenclature and numbering has been used to show and discuss data based on human reference sequences.50 The GenBank accession numbers for the IGHD sequences of the various species used for analysis which were previously available are human K02875–K02883, horse AY631942, cow AF411240, pig AF411239 and AY228508, sheep AF411238, rat AY148494 and AY148495, and mouse V00786-V00788 and J00450. The GenBank accession numbers for the IGHD sequences described in this study are: chimpanzee DQ297173–DQ297174; baboon DQ297175–DQ297176; cynomolgus macaque DQ297177–DQ297178; rhesus macaque DQ297179–DQ297181; sooty mangabey DQ297182–DQ297184; and dog DQ297185.
Results
Chimpanzee IGHD
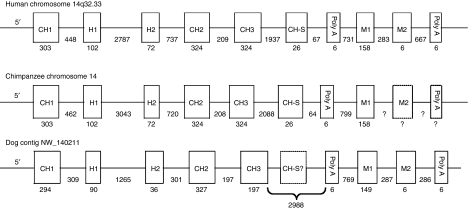
We cloned and sequenced IGHD using total RNA of PBMC from two different chimpanzees. Tika and Manuel's IGHD were sequenced using the primers IgD7 and IgD3, which amplify a 942 bp product with a sequence encoding amino acids from the CH1 position 7 to the CH3 position 52. Tika's IGHD was also amplified using the primer pair IgD7 and IgD13 allowing for sequencing through the 3′ terminus of the CH3 and yielding a 1110 bp product. Attempts to sequence Manuel's IGHD with this latter primer pair were unsuccessful. The region amplified for both animals had identical sequence except for a single G/C difference in the CH1 codon 14 at position 3, which is silent. Through screening of GenBank for matches with the chimpanzee mRNA sequences IGHD, the genomic sequence was identified on a contig from the chimpanzee chromosome 14 (NW_115908). The nucleotide sequence of the contig has 99·4% identity to that of the cDNA, with a G in the third position of CH1 codon 14. In the contig sequence three nucleotide differences from the cDNA sequences are present: an insertion of C at CH2 codon 16 between positions 2 and 3, which results in a reading frame shift; a silent A to C substitution of CH2 codon 24 position 3; and a missense A to C substitution of CH3 codon 106 at position 3 resulting in a coding change from glutamate to aspartate. All exons had the anticipated boundaries and were of equivalent sizes to the human counterparts. Besides the CH1-3 identified by reverse transcription PCR, we identified an exon for the secretory tail and the first exon for the transmembrane tail from the contig (Figs 1 and 2).
Figure 1.
Chain diagram of IGHD of human, chimpanzee and dog. Numbers indicate nucleotides in exons or introns. Domains which have not been identified are indicated by dashed lines.
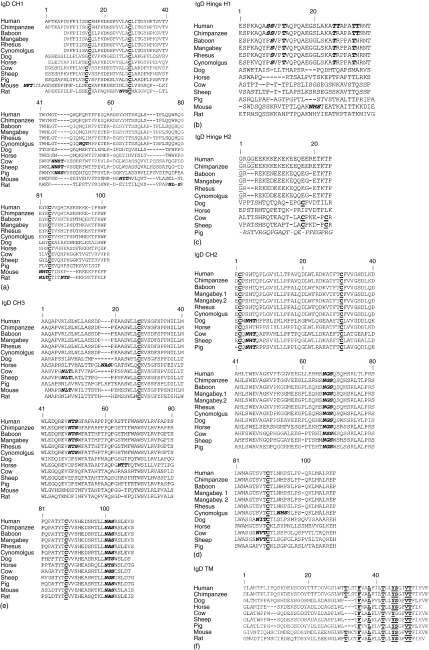
Figure 2.
Alignment of IgD heavy chain deduced amino acid sequences for each domain: CH1 (a), hinge H1 (b) and H2 (c), CH2 (d), CH3 (e) and transmembrane (f). Mouse and rat hinge is encoded by a single exon. Pig H2 is spliced out of the mature mRNA and its deduced sequence is denoted in italics to reflect this.37 (b) Glycines in the middle of the hinge, potentially contributing to flexibility, are underlined. O-glycan sites in human H1 and the corresponding conserved residues in non-human primates are bolded and italicized. (a, c, d, and e) N-glycosylation motifs (NXS or NXT where X is not proline) are bolded and italicized. Cysteines that form disulphide bonds within immunoglobulin domains and between immunoglobulin chains are bolded and underlined. (f) Amino acid of the conserved antigen receptor transmembrane motif (CART) are underlined and bolded. Mangabey.1 and mangabey.2 in D are two sequence variants. Numbering is based on the IMGT numbering for human IgD heavy chain and disregards insertions and deletions found in other species. GenBank accession numbers are given in the methods sections.
The chimpanzee IGHD deduced amino acid sequence, which is 98·1% identical to that of human IGHD, is shown aligned with IGHD of other species in Fig. 2. All cysteines responsible for inter- and intrachain disulphide bonds found in human IGHD are conserved in the chimpanzee (CH1 positions 15, 28 and 84; CH2 positions 2, 31 and 90; CH3 positions 27 and 88). Similarly, the three N-glycosylation motifs (CH2 asparagine 66 and CH3 asparagines 49 and 100) are conserved between humans and chimpanzees. The human IgD N-terminal portion of the hinge is highly O-glycosylated.4 Identified sites of O-glycosylation in human IgD hinge include H1 encoded residues S8, S9, T12, T25, T26, T30 and T31. All of these O-glycosylation sites are present in chimpanzee IgD with the exception of T25, which is replaced by an arginine in chimpanzee.
IGHD mRNA transcripts in Cercopithecoidea
With the same strategy used to clone and sequence chimpanzee IGHD, we identified IGHD transcripts for four old world monkey species commonly used in biological research. IGHD were sequenced for two rhesus macaques, three cynomolgus macaques, two baboons and one sooty mangabey. Amplification of monkey IGHD cDNA with the primer pair IgD7 and IgD13 resulted in an 1107 bp product for all four species. Because IGHA in macaques and baboons is highly polymorphic51,52 multiple animals were used for some of the species to allow for identification of potentially high levels of IGHD polymorphisms in these species. A single IGHD sequence was present for each species, except sooty mangabey, which had two sequences in one animal that varied at five nucleotides resulting in two amino acid substitutions. As discussed below, polymorphisms of IGHD appear to be less extensive than those of IGHA in non-human primates.
The deduced amino acid sequences for each species are shown in Fig. 2. The percentage identities of the old world monkey sequences with those of the human and chimpanzee (in parentheses if different) sequences are: 72% baboon; 77% (78%) sooty mangabey; 77% rhesus macaque; and 76% cynomolgus macaque. The percentage identities between old world monkeys are higher: 96% baboon–sooty mangabey; 97% baboon–rhesus macaque; 94% baboon–cynomolgus macaque; 98% sooty mangabey–rhesus macaque; 96% sooty mangabey–cynomolgus macaque; and 97% rhesus macaque–cynomolgus macaque (Table 1). The transcripts encoded all expected domains: CH1, hinge, CH2 and CH3. In the old world monkeys, CH1 is two amino acids longer than the hominoid CH1 (excluding any differences in the first six amino acid which are encoded by nucleotides prior to where our forward PCR primer annealed), while the old world monkey hinge is three amino acids shorter than the hominoid IgD hinge. These differences result in old world monkey IgD heavy chains being one amino acid shorter in their described CH1–CH3 portion than is found in chimpanzee and human IgD heavy chain. Comparing hominoids to old world monkeys, CH1 of old world monkeys lacks arginine 60, has an insertion of proline and threonine between S71 and T72, and an insertion of lysine between E96 and I97. Within the carboxy-terminal half of the hinge region, three amino acids present in human and chimpanzee IGHD are absent in the old world monkeys: G3, G4 and R19. All N-glycosylation motifs and cysteines involving inter- and intrachain disulphide bonds in humans are conserved in old world monkey IgD. Cynomolgus macaques have additional potential N-glycosylation sites at CH1 N50 and CH2 N93. These position are not N-glycosylated in any of the other known mammalian IgD. CH1 N50 is also present in pig and the other old world primates and CH2 N93 is present in all the primates examined. However, in these species, neither is followed by the N-glycosylation consensus sequence. Human O-glycosylation sites in the first half of the hinge are partially conserved in the old world monkeys; H1 encoded residues S9, T12, T25 and T30 are conserved, but S8, T26, T31 are proline, arginine and asparagine, respectively, in old world monkeys.
Table 1.
Range of percent identities for deduced amino acid sequences between the IgD heavy chain domains of different mammalian phyla
| CH1 | Hinge | CH2 | CH3 | CH1–3 | |
|---|---|---|---|---|---|
| Human–chimpanzee | 98·9 | 96·6 | 97·2 | 99·1 | 98·1 |
| Hominoids–Cercopithecoidea | 57·7–58·8 | 61·8–65·5 | 84·3–89·8 | 88·9–90·7 | 71·5–77·7 |
| Within Cercopithecoidea | 91·8–95·9 | 96·4–100 | 93·5–99·1 | 99·1–100 | 93·8–98·4 |
| Primates–Laurasiatheria | 21·6–32·0 | 13·2–19·6 | 48·6–55·6 | 51·4–60·2 | 27·3–42·4 |
| Primates–rodents | 15·5–18·7 | 23·5–31·4 | N/A* | 47·2–51·9 | 12·9–32·3 |
Groups used are based on representative species for which IGHD has been identified and include: hominoids (human and chimpanzee); Cercopithecoidea (baboon, mangabey, rhesus macaque and cynomolgus macaque); primates (combined Cercopitheocoidea and hominoids); laurasiatheria (dog, horse, pig, sheep and cow); rodents (mouse and rat).
For the described species of rodents, IgD does not encode a CH2 domain.
Unexpectedly, in addition to the 1107 bp inserts from the reverse transcription PCR clones, 1044 bp clones were isolated from all four old world primate species. These clones have the same sequence as the larger product but, inferring a similar intron–exon arrangements and boundaries as found in humans and chimpanzees, with an absent H2 exon. This H2 deletion was present in clones from PCR products of both IgD3 and IgD13, and IgD7 and IgD13 primer sets. In two of the three cynomolgus macaques examined, all of the clones sequenced had the H2 deletions. Conversely, the clones of the third cynomolgus macaque included the H2 exon. A single clone for each of the other species (baboon, rhesus macaque and sooty mangabey) was sequenced that lacked the H2. In addition to clones that were verified by sequencing, clones for each species with inserts of size corresponding to the H2 deletion product were observed on 1% agarose gels. No clones missing the H2 exon were observed for chimpanzee clones or human clones when the same primer sets were employed. Besides the two primer pairs described, we also designed a primer set (BGDF and BGDR) flanking the hinge to amplify small PCR fragments. These primers were tested with cDNA of two different baboons than those reported here and produced primarily fragments of the size expected for the IgDΔH2 product when visualized as bands on an agarose gel. The bands for the product with a complete hinge were faint. These experiments likely under represent the full-length hinge product, because PCR amplification would favour the smaller IgDΔH2 product, which was nearly half the size of the product with H2.
Dog IGHD
We performed reverse transcription PCR on dog PBMC total RNA using primers K9IGHDF1 and K9IGHDB6 derived from sequences within a contig (NW_140211) that had a high percent identity with IGHD of other species. The resulting 1468nt product consisted of seven exons (CH1, H1, H2, CH2, CH3, M1 and M2) encoding a 416 amino acids polypeptide (Fig. 2), and a portion of the 3′ untranslated region. Comparison with the genomic sequence shows the dog IGHD spans 8·25kb and all the introns follow the GT…AG rule with the possible exception of the H2/CH3 intron, which does not begin with GT in the contig sequence. The cDNA sequences agree with the dog contig for all but two nucleotides. Residues corresponding to human IgD CH1 positions 56 and 63 are methionine and glutamate in the contig, but are lysine and aspartate, respectively, in our clones. All cysteines that form intra- and interchain disulphide bonds in human IgD heavy chains are conserved in the dog IgD heavy chain. An additional cysteine is present in the dog H2 at position 16. The predicted molecular weight of the unglycosylated transmembrane protein is 45 409 MW, and without the transmembrane domain is 39 836 MW. There are four potential N-glycosylation sites in the dog IgD heavy chain. N-glycosylation sites at CH2 N66 and CH3 N100 are sites conserved with other mammalian IgDs. The N-glycosylation site at CH2 N4 is shared with artiodactyls and the N-glycosylation site at CH2 N87 is also present in cow and sheep. Dog IgD heavy chain amino acid identities with horse, pig, cow and sheep are similar to those seen between these species. CH1 and the hinge are least conserved, while CH2, CH3 and transmembrane domains are more conserved. Residues of the conserved antigen receptor transmembrane (CART) motif are present in the dog IgD transmembrane domain with the exception of T31, which is also absent in the other species of the Laurasiatheria group, i.e. pig, cow, sheep and horse.53 Dog IgD is most similar to horse IgD for CH1, hinge, CH2 and CH3 with percentage identities of 40·2%, 37·3%, 57·8% and 67·3%, respectively. Dog IgD TM is most similar to that of pig and sheep with 62% identity to each. The percentage identity of dog IgD CH1–CH3 with that of primates is 39% (human 39·3%, cynomolgus 38·7%, mangabey 38·6%, rhesus 38·7% and baboon 38·5%).
Phylogenetic analysis and comparative analysis of IgD heavy chains
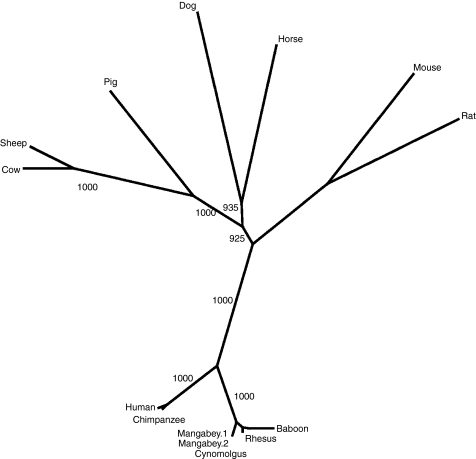
To gain further insight into the evolution of IGHD, additional analysis of IGHD in mammals was performed. A phylogenetic tree was constructed from IgD CH1–CH3 of different mammals (Fig. 3). The relationship between taxa corresponds to accepted phylogeny, with the exception of that of the old world monkeys.
Figure 3.
Neighbour-joining phylogenetic tree constructed from the deduced amino acid sequences of mammalian IgD heavy chains for domain CH1–CH3 created using the CLUSTAL X method. Mangabey.1 and mangabey.2 represent two sequence variants. Accepted phylogenetic relationships between the different species are present in the tree except for old world monkeys, which likely result from the high conservation of IgD within these species. The values shown represent the number of occurrences of branches over 1000 bootstrap resampling of the data sets.
The human IgD H1 may have originated from IgM CH2.4 Therefore, we compared the amino acid sequences from IgD H1 and IgM CH2 of different species. Human H1 is 35·3% identical to a portion of human IgM CH2 (R46-T79). The H1 of the chimpanzees and old world monkeys have 32·4% identity with the same portion of IgM CH2, but the percent identity was lower when dog or mouse IgM CH2 was used for comparison. IgD H1 S8 and T26 in humans in this scenario would derive from IgM CH2 S53 and T71, respectively. However, with the exception of humans, IgD H1 S8 is only conserved in chimpanzees and IgD H1 T26 is replaced by an arginine in all the examined non-human primate species. IgD H1 A22 of hominoids is substituted with S22 in all old world monkeys, which aligns with the conserved IgM CH2 S67. IgD H1 of species other than primates have less than 17% amino acid identities with IgM CH2.
In previously characterized IGHD sequences, different exons are conserved at different degrees. Table 1 summarizes the overall trends in deduced amino acid conservation observed between groups of mammals for the CH1–CH3 exons. As reported by others38 the CH1 and the hinge exons are the least conserved. CH3 is the most conserved exon. In rodents, IgD CH2 is absent, but otherwise CH2 is conserved nearly as well as CH3 for other mammals. IGHD is well conserved in primates, both within the old world monkey and hominoid groups, and between the two groups. Comparing old world monkeys with hominoids, the IgD domain conservation is CH3 > CH2 > hinge > CH1. IgD CH3 is conserved 100% between baboon, mangabey and cynomolgus monkeys. The IgD hinge region of rhesus and mangabey is identical. The secretory tail (CH-S) of IgDs is poorly conserved between species. Of all the IgD domains, CH-S has the lowest percentage identity between chimpanzees and humans (77·8%; chimpanzee CH-S: YVTDRGPVK versus human CH-S: YVTDHGPMK).
Discussion
Here we have sequenced IGHD cDNA from five non-human primate species and from dog. Through comparison of these sequences and previously available IGHD sequences, we have examined issues related to the evolution of IgD and gained insight into structural IgD features that are most likely involved in functional properties. In agreement with what was previously known about IgD, CH3 is the most conserved domain among the different species (Table 1). In contrast, the hinge region has diversified extensively between different mammalian groups, but is well conserved within primates. Hence, the hinge region may have evolved unique functional roles in primates.
Despite the importance of non-human primates in research, the immunoglobulin heavy chain constant regions of these species are only partially characterized. Sequences of genes are currently available for only a few antibody classes, mainly IgG and IgA, and are limited to a few species.51,52,54–56 Furthermore, corresponding immunoglobulin Fc receptors, which are responsible for initiating cellular immune responses to antibody bound antigens, generally have not been characterized for non-human primates beyond identification by cross-reactive antibodies. The biology of humans and mice with regards to Fc receptors is often quite different.57 Therefore, non-human primates may provide useful alternative models to study antibody/Fc receptor interactions. Mice lack a homologue of the human IgA Fc receptor I57 but homologues are present in macaques.58 In non-human primates, IgA is highly polymorphic and sequence differences from the human counterpart may result in modifications of their functional properties.51,52 Overall, few studies have been performed to evaluate the functional properties of non-human primate antibodies, in part because of their incomplete molecular characterization.
Understanding of IgD has lagged compared to other immunoglobulin classes, but evidence has mounted over the past years for distinct roles played by IgD in the immune response. When immunized with a model antigen, the antibody repertoire of IgD+ IgM– mice differs from that of the IgD+ IgM+ mice by VH gene usage, degree of affinity maturation, and reduced isotype switching to the IgG2a subclass.59 Both secretory and membrane bound IgD can bind to IgD receptors found on T cells of humans and mice.49,60 In contrast to Fc receptors for IgE, IgG and IgA, these IgD receptors, identified over a decade ago, have remained poorly characterized, making it difficult to assess their true function. In mice, antigen-specific responses involving the cognate interactions of T and B cells are enhanced by the combined presence of IgD on B cells and up-regulation of the IgD receptor on CD4+ T cells.61 IgD can activate the alternate complement pathway.62 Despite the low concentration of IgD in normal sera, IgD serum concentrations are elevated under some circumstances.2 For example, increased serum IgD levels appear early and persist in human immunodeficiency virus infections.63 IgD levels also increase in humans with atopy.64 Pathogen-specific IgD can be produced in response to infection.2 IgD makes up 3–10% of immunoglobulins in nasal, lacrimal and parotid glands, and in IgA-deficient individuals IgD increases to 34–57% of the total immunoglobulins at these locations.65 Elevated levels of serum IgD are characteristic of an autosomal recessive disorder, hyper-IgD and periodic fever syndrome (HIDS), which is caused by mutations in the mevalonate kinase gene.66 Although symptomatic attacks in HIDS patients do not correlate to IgD serum levels, attacks are marked by high levels of interleukin-1, interleukin-2 and tumour necrosis factor-α, the same cytokines induced by incubation of normal PBMC with IgD, suggesting a possible link between IgD and HIDS pathology.67
Our findings show IgD structure of non-human primates and dog is similar to that found in human, horse and artiodactyls (cow and sheep). Three CH domains and a long hinge are present in all the examined primates and in dog. By contrast, mouse and rat IgD have no CH2, although a mouse pseudo-exon related to CH2 has been described.6,7,68 Our results cannot establish whether or not the dog immunoglobulin identified by Yang et al.14 corresponds to the one that would be produced from dog IGHD; however, our data is compatible with this possibility. The calculated molecular weight of dog IgD CH1–CH3 domains is 40 000 MW. A complete IgD heavy chain including glycans at its four N-glycosylation sites, the heavy chain variable domain and a secretory tail (which we did not identify) would be expected to be of a greater molecular weight. This would be consistent with the reported molecular weight of 55 000 MW of the putative dog IgD heavy chain found in sera14.
N-glycosylation is an important feature of all immunoglobulin molecules including IgD. All the N-glycosylation sites found in humans are conserved in non-human primates. In humans, the N-glycosylation site at CH2 N66 is necessary for the association of IgD heavy chains to form a complete antibody and for secretion from the endoplasmic reticulum.69 The N-glycosylation site at CH3 N49 is characteristic of all the examined primates and not found in IgD of the other species. Cynomolgus macaque IgD has additional N-glycosylation motifs present at CH1 N50 and CH2 N93. Mice possess an IgD receptor different from that of humans, which is expressed on CD4+ T cells and is specific for N-linked glycans on murine IgD.60,70 It is unknown if similar IgD receptors are present in other species, but possible N-glycan sites are present in dog IgD that might provide points of interaction with such an IgD receptor.
The structure of the immunoglobulin hinge regions is critical for their function; it gives antibodies the flexibility needed to bind antigen and provide sites of interaction with Fc receptors and complement. The IgD hinge is quite diverse in structure between species. Rodents have a shorter IgD hinge encoded by a single exon. The hinge regions of dog and chimpanzee are encoded by two exons. Old world monkey IgD hinge regions, similarly, have two distinct segments that are highly conserved with those of hominoids, including the large number of lysine and glutamate residues of the C-terminal portion. These charged residues may favour the formation of an α-helix structure and act to separate the two hinge segments by repulsion.71 The IgD hinge region is the longest of that found in all human antibody isotypes and, besides the IgG3 hinge, is the only other immunoglobulin hinge encoded by multiple exons.50 The dog IgD hinge region is structurally distinct from the corresponding primate regions and is related more closely to that of ungulates; it does not have a highly charged second domain. The second half of the dog hinge region contains a cysteine which is at a conserved position with cysteines found in the hinge regions of sheep and cow. In the latter two species a second cysteine is also present in the IgD H2. It is possible that these cysteines in the IgD hinge regions of dogs, sheep, and cows are involved in forming inter-heavy chain disulphide bonds, as described for the cysteines of IgG hinge regions.
Structurally, IgD and IgA1 hinges of humans share common features. IgA1 is the only other immunoglobulin with O-glycosylation in its upper hinge. As previously demonstrated for IgA, the flexible arms of IgD Fab can be separated by a wide angle thus resulting in an average antibody conformation similar to that of a T-shape in contrast to the more typical immunoglobulin Y-shape.22,71 Comparable to the hinge of IgD, IgA of different mammals is highly variable in length and amino acid sequence. IgD H2 is high repetitive. It has been suggested that the repetitive genetic structure of the IgA hinge region has led to its evolutionary instability.47 Indeed, the hinge of two mouse species has diverged 25% in the mere 4–8 million years since the two species have separated.72 Old world monkeys, unlike humans, have only a single IGHA that encodes a short hinge more like the human IgA2 hinge without multiple O-glycosylation sites.52 The IgA hinge within rhesus macaques is highly polymorphic.52 By contrast, non-human primate IgD amino acid substitutions were found only in a sooty mangabey at two positions in CH2. More importantly, the hinge is highly conserved between primates; rhesus macaque and sooty mangabey IgD hinge regions are identical. Therefore, the hinge region of IgD appears to be less variable and evolutionarily more stable than the hinge region of IgA in non-human primates.
In humans, three glycine residues encoded at the start of H2 contribute to IgD hinge segmental flexibility.71 Though conserved in chimpanzee, the glycines at H2 positions 3 and 4 are deleted in old world monkeys. Loss of these glycines may be compensated for by a glycine dyad created by a glycine substitution next to a conserved glycine in IgD H1 and by another glycine substitution in the middle of IgD H2. Besides being important for IgD receptor interactions49 the O-glycans of the hinge contribute to structural rigidity.71 We predict that IgD H1 O-glycosylation is reduced in non-human primates. At equivalent positions of the human O-glycan sites, IgD of chimpanzee has one substitution and IgD of old world monkeys have three substitutions. In old world monkeys, only IgD H1 S22 offers a potential O-glycosylation site not found in human IgD H1. Dog IgD hinge is not well conserved with that of primates. Hence, if present, dog IgD O-glycosylation is quite different.
In mangabey, baboon, rhesus macaque and cynomolgus macaque, we identified IGHD clones in which the hinge region that corresponds to the hominoid H2 exon was deleted. These transcripts (IgDΔH2) may be the result of (1) an artefact of the reverse transcription PCR, (2) a polymorphism encoded by a second IGHD gene or allele or (3) alternative splicing events. Several lines of evidence argue against the first possibility. Under the same PCR conditions, and using the same primer sets, IgDΔH2 transcripts were not found in any of the chimpanzee clones, nor in control experiments using human RNA. The percentage of clones for each transcript varied between animals. Multiple primer pairs produced IgDΔH2 clones. The deletion was always exact so that it maintained the reading frame. Finally, alignment of the hinge regions and CH3 were compared and no unusual sequences with high identity were found that might be conducive to PCR jumping. The latter two possible explanations remain to be tested and are not necessarily exclusive. For example, an allelic variant in the intron between H1 and H2 could alter splicing. Pig IGHD has an H2 exon that is spliced out because of a branchpoint mutation in the H1–H2 hinge intron.37 When a single T nucleotide is introduced into this sequence, pig IGHD transcripts include H2. Zhao et al.37 speculated that some transcripts with H2 might be produced normally in pigs, although none of their clones included the H2 exon. Future studies will be needed to test whether or not these primates express IgD without the H2 portion of the molecule, as such a difference may have profound effects on antigen binding properties.
The origin of the hinge in IgD is still an open question. Fish IgD have no hinge.25–35 Putnam et al.4 have made the case that the human IgD N-terminal half of the hinge (H1) may have originated from a duplication of the IGHM CH2 domain, as human IgD H1 residues have a significant percentage identity with those conserved between IgM CH2 of different species. Complementing this hypothesis is evidence that IGHM and IGHD are closely related and have exchanged genetic material in the past. Such evidence includes the probable origination of IGHD from an ancient duplication of IGHM8 and the demonstration that a duplicated IGHM CH1 replaced the original IGHD CH1 in artiodactyls.36 IgD H1 of primate appears to have a common origin, and non-human primate IgD H1 has a 32·4% identity with the same section of human IgM CH2 that has a 35·3% identity to human IgD H1. Such supports the hypothesis for a possible IgM CH2 origin of the primate IgD H1. This hypothesis does not fit as well for species other than primates, which possess IgD hinges with less than 17% amino acid identities with IgM CH2. If the hypothesis is correct, then it would seem likely that IgD hinge regions arose in evolution three or more times, once for rodent hinge, once for primate hinge, and once for Laurasiatheria hinge. Additionally, the H2 of primates appears to be unique and may have yet another origin.
As anticipated on the basis of cross-reactivity obtained with antihuman antibodies, IgD deduced amino acid sequences of non-human primates are well conserved among the examined primates and humans. By contrast, dog IgD is not as well conserved, and it is not surprising that the available anti-IgD raised against IgD of other species do not cross-react with dog IgD. IgD CH1 and CH2 domains compared to CH3 and transmembrane domains are less conserved at the amino acid levels (Table 1). This would seem to indicate that the IgD CH1 and CH2 domains are less critical for functional properties of IgD that are conserved across species, although these domains may have important species-specific functions. For example, in human IgD, these domains contain cysteines necessary for integrity of the quaternary structure. CH1 C15 forms bonds with the immunoglobulin light chains and CH2 C2 forms the only covalent bond between heavy chains. These cysteines are conserved in non-human primates and in dog.
IgD CH1 of artiodactyls shares a high degree of conservation with CH1 of IgM as a result of the IGHM CH1 having replaced the original IGHD CH1.8,36 Our data are in agreement with the hypothesis that the IgD CH1 replacement with IgM CH1 in artiodactyls was recent. Dog IgD CH1 is not highly conserved with the published dog IgM CH1 protein sequence (14·3% identity).73 Horse IgD CH1 is not highly conserved with IgM CH1.38 Therefore, the genetic event leading to the replacement of the original IgD CH1 in artiodactyls occurred after the evolution of Carnivora, Perissodactyla and Cetartiodactyla as distinct phyla.
The IGHD M1 and M2 exons encoding the transmembrane domain are highly conserved between all species examined reflecting the importance of the transmembrane domain that they encode in establishing interactions with the B-cell receptor-associated signalling chains. As expected, dog IGHD maintains the M1 and M2 exon arrangement seen in other species. Presumably, this is also the case for chimpanzee IGHD, for which we could not identify the M2 (because of the incomplete resolution of the chimpanzee contig sequence). Importantly, the dog and chimpanzee transmembranes domains contain the CART motif found in antigen receptors described by Campbell et al.53 This motif is involved in forming interactions with the B-cell receptor signalling polypeptides.53
Knowledge at the genetic level of IgD in species other than human and rodents has only begun to accumulate recently, and has demonstrated that IgD is widespread throughout vertebrates and is extremely diverse. Despite this diversity, IgD is surprisingly well conserved between non-human primates. In contrast to the view that IgD function as a BCR is redundant with IgM and that secreted IgD is unimportant, this high degree of conservation indicates that IgD may have valuable biological roles as yet unappreciated. Future studies should include the molecular characterization of IgD receptors and the determination of whether or not IgD receptors are a common feature of mammals, thus leading to a clear definition of the roles that IgD plays in the immune response. The IgD sequences identified here would be valuable in such endeavours.
Acknowledgments
The authors thank and fondly remember the late Dr Harold McClure (Yerkes National Primate Research Center) for providing chimpanzee, rhesus macaque and sooty mangabey blood samples, and Drs Jerilyn Pecotte and Kathy Brasky (Southwest National Primate Research Center) for providing cynomolgus macaque and baboon blood samples. This work was supported in part by NIH grants RR10755 and RR00165, by the Research Program Enhancement from the GSU Office of Research and Sponsored Programs and by the Georgia Research Alliance. Support for Kenneth A. Rogers was provided by the Molecular Basis of Disease program at Georgia State University.
Abbreviations
- CART
conserved antigen receptor transmembrane
- IGHD
immunoglobulin heavy constant delta
- IGHM
immunoglobulin heavy constant mu
- PBMC
peripheral blood mononuclear cells
References
- 1.Lutz C, Ledermann B, Kosco-Vilbois MH, Ochsenbein AF, Zinkernagel RM, Köhler G, Brombacher F. IgD can largely substitute for loss of IgM function in B cells. Nature. 1998;393:797–801. doi: 10.1038/31716. [DOI] [PubMed] [Google Scholar]
- 2.Preud'homme JL, Petit I, Barra A, Morel F, Lecron JC, Lelièvre E. Structural and functional properties of membrane and secreted IgD. Mol Immunol. 2000;37:871–87. doi: 10.1016/s0161-5890(01)00006-2. [DOI] [PubMed] [Google Scholar]
- 3.Rowe DS, Fahey JL. A new class of human immunoglobulins. J Exp Med. 1965;121:171–99. doi: 10.1084/jem.121.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Putnam FW, Takahashi N, Tetaert D, Debuire B, Lin L. Amino acid sequence of the first constant region domain and the hinge region of the δ heavy chain of human IgD. Proc Natl Acad Sci USA. 1981;78:6168–72. doi: 10.1073/pnas.78.10.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White MB, Shen AL, Word CJ, Tucker PW, Blattner FR. Human immunoglobulin D. Genomic sequence of the delta heavy chain. Science. 1985;228:733–7. doi: 10.1126/science.3922054. [DOI] [PubMed] [Google Scholar]
- 6.Mushinski JF, Blattner FR, Owens JD, Finkelman FD, Kessler SW, Fitzmaurice L, Potter M, Tucker PW. Mouse immunoglobulin D. Construction and characterization of a cloned δ chain cDNA. Proc Natl Acad Sci USA. 1980;77:7405–9. doi: 10.1073/pnas.77.12.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sire JA, Auffray C, Jordan BR. Rat immunoglobulin delta heavy chain gene: nucleotide sequence derived from cloned cDNA. Gene. 1982;20:377–86. doi: 10.1016/0378-1119(82)90206-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, Hammarström L. Cloning of the complete rat immunoglobulin δ gene: evolutionary implications. Immunology. 2003;108:288–95. doi: 10.1046/j.1365-2567.2003.01610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin LN, Leslie GA, Hindes R. Lymphocyte surface IgD and IgM in non-human primates. Int Arch Allergy Appl Immunol. 1976;51:320–9. doi: 10.1159/000231606. [DOI] [PubMed] [Google Scholar]
- 10.Chen CL, Lehmeyer JE, Cooper MD. Evidence for an IgD homologue on chicken lymphocytes. J Immunol. 1982;129:2580–5. [PubMed] [Google Scholar]
- 11.Sire J, Collé A, Bourgois A. Identification of an IgD like surface Ig on rabbit lymphocytes. Eur J Immunol. 1977;9:13–6. doi: 10.1002/eji.1830090104. [DOI] [PubMed] [Google Scholar]
- 12.Wilder RL, Yuen CC, Coyle SA, Mage RG. Demonstration of a rabbit cell surface Ig that bears light chain and VH9, but lacks mu-, alpha-, and gamma-allotypes-rabbit IgD? J Immunol. 1979;122:464–8. [PubMed] [Google Scholar]
- 13.Eskinazi DP, Bessinger BA, McNicholas JM, Leary LA, Knight KL. Expression of an unidentified Ig isotype on rabbit Ig-bearing lymphocytes. J Immunol. 1977;122:469–74. [PubMed] [Google Scholar]
- 14.Yang M, Becker AB, Simons FER, Peng Z. Identification of a dog IgD-like molecule by a monoclonal antibody. Vet Immunol Immunopathol. 1995;47:215–24. doi: 10.1016/0165-2427(94)05401-d. [DOI] [PubMed] [Google Scholar]
- 15.Fiebig H, Ambrosius H. Cell surface Ig of lymphocytes in lower vertebrates. In: Wright RK, Cooper EL, editors. Phylogeny of Thymus and Bone Marrow-Bursa Cells. North Holland, Amsterdam: Elsevier; 1976. p. 195. [Google Scholar]
- 16.Naessens J. Surface Ig on B lymphocytes from cattle and sheep. Int Immunol. 1996;9:349–54. doi: 10.1093/intimm/9.3.349. [DOI] [PubMed] [Google Scholar]
- 17.Butler JE, Sun J, Navarro P. The swine Ig heavy chain locus has a single JH and no identifiable IgD. Int Immunol. 1996;8:1897–904. doi: 10.1093/intimm/8.12.1897. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Rabbani H, Shimizu A, Hammarström L. Mapping of the chicken immunoglobulin heavy-chain constant region gene locus reveals and inverted alpha gene upstream of a condensed epsilon gene. Immunol. 2000;101:348–53. doi: 10.1046/j.1365-2567.2000.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundqvist ML, Middleton DL, Hazard S, Warr GW. The immunoglobulin heavy chain locus of the duck. Genomic organization and expression of D, J, and C region genes. J Biol Chem. 2001;276:46729–36. doi: 10.1074/jbc.M106221200. [DOI] [PubMed] [Google Scholar]
- 20.Knight KL, Tunyaplin C. Immunoglobulin heavy chain genes of rabbit. In: Honjo T, Alt FW, editors. Immunoglobulin Genes. New York: Academic Press; 1995. pp. 289–314. [Google Scholar]
- 21.Loder F, Mutschler B, Ray RJ, Paige CJ, Sideras P, Torres R, Lamers MC, Carsetti R. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J Exp Med. 1999;190:75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Løset GÅ, Roux KH, Zhu P, Michaelsen TE, Sandlie I. Differential segmental flexibility and reach dictate the antigen binding mode of chimeric IgD and IgM: Implications for the function of the B cell receptor. J Immunol. 2004;172:2925–34. doi: 10.4049/jimmunol.172.5.2925. [DOI] [PubMed] [Google Scholar]
- 23.Kim KM, Reth M. The B cell antigen receptor of class IgD induces a stronger and more prolonged protein tyrosine phosphorylation than that of class IgM. J Exp Med. 1995;181:1005–14. doi: 10.1084/jem.181.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roes J, Rajewsky K. Immunoglobulin D (IgD)–deficient mice reveal an auxiliary receptor function for IgD in antigen-mediated recruitment of B cells. J Exp Med. 1993;177:45–55. doi: 10.1084/jem.177.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson M, Bengtén E, Miller NW, Clem LW, Pasquier LD, Warr GW. A novel chimeric Ig heavy chain from a teleost fish shares similarities to IgD. Proc Natl Acad Sci USA. 1997;94:4593–7. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stenvik J, Jørgensen TØ. Immunoglobulin D (IgD) of Atlantic cod has a unique structure. Immunogenetics. 2000;51:452–61. doi: 10.1007/s002510050644. [DOI] [PubMed] [Google Scholar]
- 27.Hirono I, Nam B, Enomoto J, Uchino K, Aoki T. Cloninig and characterisation of a cDNA encoding Japanese flounder Paralichthys olivaceus IgD. Fish Shellfish Immunol. 2003;15:63–70. doi: 10.1016/s1050-4648(02)00139-0. [DOI] [PubMed] [Google Scholar]
- 28.Srisapoome P, Ohira T, Hirono I, Aoki T. Genes of the constant regions of functional immunoglobulin heavy chain of Japanese flounder. Paralichthys olivaceus. Immunogenetics. 2004;56:292–300. doi: 10.1007/s00251-004-0689-7. [DOI] [PubMed] [Google Scholar]
- 29.Savan R, Aman A, Nakao M, Watanuki H, Sakai M. Discovery of a novel immunoglobulin heavy chain gene chimera from common carp (Cyprinus carpio L.) Immunogentics. 2005;57:458–63. doi: 10.1007/s00251-005-0015-z. [DOI] [PubMed] [Google Scholar]
- 30.Saha NR, Suetake H, Kikuchi K, Suzuki Y. Fugu immunoglobulin D. a highly unusual gene with unprecedented duplications in its constant region. Immunogenetics. 2004;56:438–47. doi: 10.1007/s00251-004-0693-y. [DOI] [PubMed] [Google Scholar]
- 31.Hordvik I. Identification of a novel immunoglobulin – transcript and comparative analysis of the genes encoding IgD in Atlantic salmon and Atlantic halibut. Mol Immunol. 2002;39:85–91. doi: 10.1016/s0161-5890(02)00043-3. [DOI] [PubMed] [Google Scholar]
- 32.Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebra fish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6:295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- 33.Bengtén ES, Quiniou M-A, Stuge TB, Katagiri T, Miller NW, Clem LW, Warr GW, Wilson M. The IgH locus of the Channel Catfish, Ictalurus punctatus, contains multiple constant region gene sequences: different genes encode heavy chains of membrane and secreted IgD. J Immunol. 2002;169:2488–97. doi: 10.4049/jimmunol.169.5.2488. [DOI] [PubMed] [Google Scholar]
- 34.Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isoype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci USA. 2005;102:6919–24. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hordvik O, Thevarajan J, Samdal I, Bastani Krossøy B. Molecular cloning and phylogenetic analysis of the Atlantic salmon immunoglobulin D gene. Scand J Immunol. 1999;50:202–10. doi: 10.1046/j.1365-3083.1999.00583.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Kacskovics I, Pan Q, Liberles DA, Geli J, Davis SK, Rabbani H, Hammarström L. Artiodacyl IgD: the missing link. J Immunol. 2002;169:4408–16. doi: 10.4049/jimmunol.169.8.4408. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Pan-Hammarström Q, Kacskovics I, Hammarström L. The porcine Igδ gene: Unique chimeric splicing of the first constant region domain in its heavy chain transcripts. J Immunol. 2003;171:1312–8. doi: 10.4049/jimmunol.171.3.1312. [DOI] [PubMed] [Google Scholar]
- 38.Wagner B, Miller DC, Lear TL, Antczak DF. The complete map of the Ig Heavy chain constant gene region reveals evidence for seven IgG isotypes and for IgD in the horse. J Immunol. 2004;173:3230–42. doi: 10.4049/jimmunol.173.5.3230. [DOI] [PubMed] [Google Scholar]
- 39.Voormolen-Kalova M, van den Berg P, Rádl J. Immunoglobulin levels as related to age in nonhuman primates in captivity. I. Chimpanzees. J Med Prim. 1974;3:335–42. doi: 10.1159/000460104. [DOI] [PubMed] [Google Scholar]
- 40.Voormolen-Kalova M, van den Berg P, Rádl J. Immunoglobulin levels as related to age in nonhuman primates in captivity. II. Rhesus monkeys. J Med Prim. 1974;3:343–50. doi: 10.1159/000460105. [DOI] [PubMed] [Google Scholar]
- 41.Finkelman FD, Scher I. Rhesus monkey B lymphocyte surface immunoglobulin: analysis with a fluorescence-activated cell sorter. J Immunol. 1979;122:1757–62. [PubMed] [Google Scholar]
- 42.Black CM, McDougal JS, Holman RC, Evatt BL, Reimer CB. Cross-reactivity of 75 monoclonal antibodies to human immunoglobulin with sera of non-human primates. Immunol Lett. 1993;37:207–13. doi: 10.1016/0165-2478(93)90032-w. [DOI] [PubMed] [Google Scholar]
- 43.Pernis B. The effect of anti-IgD serum on antibody production in rhesus monkeys. In: Seligman M, Preud'homme JL, Kourilsky FM, editors. Membrane Receptors of Lymphocytes. New York: Elsevier Inc.; 1975. pp. 25–6. [Google Scholar]
- 44.Martin LN, Leslie GA. In vivo effects of antiserum to IgD on surface immunoglobulins, serum immunoglobulins and lymphocyte blastogenesis in rhesus monkeys. Immunology. 1979;37:253–62. [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy RC, Shearer MH, Hildebrand W. Nonhuman primate models to evaluate vaccine safety and immunogenicity. Vaccine. 1997;15:903–8. doi: 10.1016/s0264-410x(96)00277-0. [DOI] [PubMed] [Google Scholar]
- 46.Carlsson HE, Schapiro SJ, Farah I, Hau J. Use of primates in research: a global overview. Am J Prim. 2004;63:225–37. doi: 10.1002/ajp.20054. [DOI] [PubMed] [Google Scholar]
- 47.Flanagan JG, Lefrance MP, Rabbitts TH. Mechanisms of divergence and convergence of the human immunoglobulin alpha 1 and alpha 2 constant region gene sequences. Cell. 1984;36:681–8. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- 48.Tamma SML, Coico RF. IgD-receptor-positive human T lymphocytes. II. Identification and partial characterization of human IgD-binding factor. J Immunol. 1992;148:2050–7. [PubMed] [Google Scholar]
- 49.Rudd P, Fortune F, Lehner T, et al. Lectin–carbohydrate interactions in disease. T-cell recognition of IgA and IgD; mannose binding protein recognition of IgG0. Adv Exp Med Biol. 1995;376:147–52. [PubMed] [Google Scholar]
- 50.Lefranc M-P, Lefranc G. The Immunoglobulin Factsbook. San Diego: Academic Press; 2001. [Google Scholar]
- 51.Scinicariello F, Attanasio R. Intraspecies heterogeneity of immunoglobulin alpha-chain constant region genes in rhesus macaques. Immunology. 2001;103:441–8. doi: 10.1046/j.1365-2567.2001.01251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scinicariello F, Engleman CN, Jayashankar L, McClure HM, Attanasio R. Rhesus macaque antibody molecules: sequence and heterogeneity of alpha and gamma constant regions. Immunology. 2004;111:66–74. doi: 10.1111/j.1365-2567.2003.01767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell KS, Bäckström BT, Tiefenthaler G, Palmer ECART. a conserved antigen receptor transmembrane motif. Semin Immunol. 1994;6:393–410. doi: 10.1006/smim.1994.1049. [DOI] [PubMed] [Google Scholar]
- 54.Ueda S, Kawamura S. Immunoglobulin CH gene family in hominoids and its evolutionary history. Genomics. 1992;13:194–200. doi: 10.1016/0888-7543(92)90220-m. [DOI] [PubMed] [Google Scholar]
- 55.Calvas P, Apoil P, Fortenfant F, Roubinet F, Andris J, Capra D, Blancher A. Characterization of the three immunoglobulin G subclasses of macaques. Scand J Immunol. 1999;49:595–610. doi: 10.1046/j.1365-3083.1999.00540.x. [DOI] [PubMed] [Google Scholar]
- 56.Attanasio R, Jayashankar L, Engleman CN, Scinicariello F. Baboon immunoglobulin constant region heavy chains: identification of four IGHG genes. Immunogenetics. 2002;54:556–61. doi: 10.1007/s00251-002-0505-1. [DOI] [PubMed] [Google Scholar]
- 57.Takai T. Fc Receptors and theirs role in immune regulation and autoimmunity. J Clin Immunol. 2005;25:1–17. doi: 10.1007/s10875-005-0353-8. [DOI] [PubMed] [Google Scholar]
- 58.Rogers KA, Scinicariello F, Attanasio R. Identification and characterization of macaque CD89 (immunoglobulin A Fc receptor) Immunol. 2004;113:178–86. doi: 10.1111/j.1365-2567.2004.01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han S, Zhang X, Xu R, Finkelman FD, Brombacher F, Zheng B. IgD+ IgM– B cells mount immune responses that exhibit altered antibody repertoire. Eur J Immunol. 2004;34:661–8. doi: 10.1002/eji.200324493. [DOI] [PubMed] [Google Scholar]
- 60.Amin AR, Tamma SML, Oppenheim JD, Finkelman FD, Kieda C, Coico RF, Thorbecke GJ. Specificity of the murine IgD receptor on T cells is for N-linked glycans on IgD molecules. Proc Natl Acad Sci USA. 1991;88:9238–42. doi: 10.1073/pnas.88.20.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y, Tamma SML, Lima V, Coico R. Facilitated antigen presentation by B cells expressing IgD when responding T cells express IgD-receptors. Cell Immunol. 1999;192:194–202. doi: 10.1006/cimm.1998.1453. [DOI] [PubMed] [Google Scholar]
- 62.Spiegelberg HL. Biological role of different antibody classes. Int Arch Allergy Appl Immunol. 1989;90(1):22–7. doi: 10.1159/000235071. [DOI] [PubMed] [Google Scholar]
- 63.Raiteri R, Albonico M, Deiana R, Marietti G, Sinicco A. Serum IgD behaviour in HIV-1 infected patients. Allergol Immunopathol. 1991;19:6–10. [PubMed] [Google Scholar]
- 64.Peng Z, Fisher R, Adkinson NF., Jr Total serum IgD is increased in atopic subjects. Allergy. 1991;46:436–44. doi: 10.1111/j.1398-9995.1991.tb04222.x. [DOI] [PubMed] [Google Scholar]
- 65.Brandzaeg P, Farstad IN, Johansen FE, Morton HC, Norderhaug IN, Yamanaka T. The B-cell system of human mucosae and exocrine glands. Immunol Rev. 1999;171:45–87. doi: 10.1111/j.1600-065X.1999.tb01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Centola M, Aksentijevich I, Kastner DL. The hereditary periodic fever syndromes: molecular analysis of a new family of inflammatory diseases. Human Mol Genet. 1998;7:1581–8. doi: 10.1093/hmg/7.10.1581. [DOI] [PubMed] [Google Scholar]
- 67.Drenth JP, Goertz J, Daha MR, vander Meer JW. Immunoglobulin D enhances the release of tumor necrosis factor-alpha, and interleukin-1 beta as well as interleukin-1 receptor antagonist from human mononuclear cells. Immunology. 1996;88:355–62. doi: 10.1046/j.1365-2567.1996.d01-672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richards JE, Gilliam AC, Shen A, Tucher PW, Blattner FR. Unusual sequences in the murine immunoglobulin µ-δ heavy-chain region. Nature. 1983;306:483–7. doi: 10.1038/306483a0. [DOI] [PubMed] [Google Scholar]
- 69.Gala FA, Morrison SL. The role of constant region carbohydrate in the assembly and secretion of human IgD and IgA. J Biol Chem. 2002;277:29005–11. doi: 10.1074/jbc.M203258200. [DOI] [PubMed] [Google Scholar]
- 70.Adachi M, Ishizaka K. IgD-binding factors from mouse T lymphocytes. Proc Natl Acad Sci U S A. 1986;83:7003–7. doi: 10.1073/pnas.83.18.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Z, Almogren A, Furtado PB, Chowdhury B, Kerr MA, Perkins SJ. Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering. J Mol Biol. 2005;353:155–73. doi: 10.1016/j.jmb.2005.07.072. [DOI] [PubMed] [Google Scholar]
- 72.Osborne BA, Golde TE, Scwartz RL, Rudikoff S. Evolution of the IgA heavy chain in the genus Mus. Genetics. 1988;119:925–31. doi: 10.1093/genetics/119.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCumber LJ, Capra JD. The complete amino-acid sequence of a canine Mu chain. Mol Immunol. 1979;16:565–70. doi: 10.1016/0161-5890(79)90119-6. [DOI] [PubMed] [Google Scholar]