Abstract
Intestinal epithelial cells (IECs) and dendritic cells (DCs) play a pivotal role in antigen sampling and the maintenance of gut homeostasis. However, the interaction of commensal bacteria with the intestinal surface remains incompletely understood. Here we investigated immune cell responses to commensal and pathogenic bacteria. HT-29 human IECs were incubated with Bifidobacterium infantis 35624, Lactobacillus salivarius UCC118 or Salmonella typhimurium UK1 for varying times, or were pretreated with a probiotic for 2 hr prior to stimulation with S. typhimurium or flagellin. Gene arrays were used to examine inflammatory gene expression. Nuclear factor (NF)-κB activation, interleukin (IL)-8 secretion, pathogen adherence to IECs, and mucin-3 (MUC3) and E-cadherin gene expression were assayed by TransAM assay, enzyme-linked immunosorbent assay (ELISA), fluorescence, and real-time reverse transcriptase–polymerase chain reaction (RT-PCR), respectively. IL-10 and tumour necrosis factor (TNF)-α secretion by bacteria-treated peripheral blood-derived DCs were measured using ELISA. S. typhimurium increased expression of 36 of the 847 immune-related genes assayed, including NF-κB and IL-8. The commensal bacteria did not alter expression levels of any of the 847 genes. However, B. infantis and L. salivarius attenuated both IL-8 secretion at baseline and S. typhimurium-induced pro-inflammatory responses. B. infantis also limited flagellin-induced IL-8 protein secretion. The commensal bacteria did not increase MUC3 or E-cadherin expression, or interfere with pathogen binding to HT-29 cells, but they did stimulate IL-10 and TNF-α secretion by DCs. The data demonstrate that, although the intestinal epithelium is immunologically quiescent when it encounters B. infantis or L. salivarius, these commensal bacteria exert immunomodulatory effects on intestinal immune cells that mediate host responses to flagellin and enteric pathogens.
Keywords: commensal bacteria, flagellin, interleukin-8, intestinal epithelium, mucins
Introduction
The human gastrointestinal tract is divergently challenged in that it must be able to tolerate dietary antigens and endogenous microflora, and simultaneously recognize and signal the presence of pathogens. The epithelium plays a crucial role in the maintenance of intestinal homeostasis1 and actively samples resident bacteria, pathogens and other antigens.2,3 The epithelium is covered by mucus that protects the mucosal surface by limiting pathogen access.4 Mucus is mainly composed of complex glycoproteins called mucins which are encoded by various mucin (MUC) genes.4,5 Intestinal epithelial cells (IECs) secrete many mediators involved in immune responses to potentially pathogenic organisms, including antibacterial peptides such as defensins,6 mucins including MUC3,4 and chemokines and cytokines such as interleukin (IL)-8.7
IL-8, a C-X-C chemokine that is transcriptionally regulated by nuclear factor (NF)-κB,8 shows potent chemotactic activity for neutrophils.9 IL-8 is secreted by IECs in response to various pathogenic bacteria10 and pro-inflammatory cytokines such as tumour necrosis factor (TNF)-α. Furthermore, it has been reported that certain non-pathogenic commensal bacteria, such as Escherichia coli Nissle 1917,11,12 but not Lactobacillus reuteri, Lactobacillus rhamnosus GG or the probiotic cocktail VSL#3,11–13 induce the secretion of IL-8 by IECs.
Antigen-presenting dendritic cells (DCs) survey and sample commensal and pathogenic bacteria at mucosal interfaces.14,15 Moreover, intestinal DCs can directly sample the contents of the gut lumen by extending dendrites between IECs.16 Host pattern recognition receptors, including Toll-like receptors (TLRs), play a fundamental role in immune cell activation in response to specific microbial-associated molecular patterns that are associated with a variety of organisms including bacteria, fungi and viruses.17 IECs and DCs constitutively express several TLRs, including TLR5 which responds to the monomeric flagellin subunits of bacterial flagella.18–20 In the epithelium, TLR5 is a key mediator of pro-inflammatory responses to flagellin from pathogenic and commensal bacteria.21–23 Flagellin also stimulates the maturation of responsive DCs.18 Together with DCs and IECs, TLRs thus represent integral components of the mucosal innate immune system.
Probiotics, commensal organisms that can be harnessed for health benefits, have demonstrated therapeutic effects in murine models of colitis24,25 and in patients with inflammatory bowel diseases (IBDs),26–28 diarrhoea29,30 and, most recently, irritable bowel syndrome.31 Despite increased recognition of the importance of luminal flora in the development of colitis32,33 and the almost paradoxical benefits conferred by certain probiotics in these conditions, therapeutic approaches that modulate the bacterial load are hampered by a limited understanding of host flora interactions at the epithelial interface. In this study, we examined immune cell responses to Salmonella typhimurium, flagellin, and Bifidobacterium infantis and Lactobacillus salivarius, two commensal strains demonstrated to have probiotic properties.24,31,34 We demonstrate that, although the epithelium is immunologically unresponsive to B. infantis and L. salivarius, these commensal bacteria induce the secretion of regulatory cytokines by DCs. Furthermore, the data show that B. infantis and L. salivarius functionally modulate the epithelium by attenuating S. typhimurium- and flagellin-induced pro-inflammatory responses.
Materials and methods
Bacteria and growth conditions
Salmonella typhimurium UK1 (kindly provided by R. Curtiss III, Washington University in St Louis, MO), Bifidobacterium infantis 35624 and Lactobacillus salivarius subspecies salivarius UCC11834 35 were stored in 50% glycerol at −70°. Prior to use in experiments, S. typhimurium was cultured at 37° in tryptic soy broth (TSB) (Merck, Darmstadt, Germany) for 18 hr under aerobic conditions, B. infantis was cultured anaerobically at 37° in de Man Rogosa Sharpe (MRS) (Merck) broth supplemented with 0·05% cysteine (Sigma-Aldrich, St Louis, MO) for 48 hr, and L. salivarius was cultured anaerobically at 37° in MRS broth for 18 hr. The stationary-phase bacteria were centrifuged, resuspended in sterile phosphate-buffered saline (PBS), and Gram-stained to confirm purity. Bacterial number was estimated by measuring the absorbance at 600 nm, and relating the absorbance value to a standard curve of colony-forming units (CFU) on MRS agar or tryptic soy agar (TSA) (Merck).
Epithelial cell culture
In this study, the HT-29 human colonic epithelial cell line (American Type Culture Collection, Manassas, VA) was chosen as this IEC model has been extensively used by many groups investigating epithelial responses to bacteria.11,13,36,37 HT-29 cells were cultured in modified McCoy's 5A medium (Gibco-BRL, Grand Island, NY) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Sigma-Aldrich) in the presence or absence of 100 U/ml penicillin G and 100 µg/ml streptomycin (Gibco-BRL). The HeLa (human cervix epithelial-like) cell line (ATCC) was cultured in minimal essential medium (Gibco-BRL) supplemented with 0·1 mm non-essential amino acids (Gibco-BRL), 1·5 mm l-glutamine, 10% heat-inactivated FCS, 100 U/ml penicillin G and 100 µg/ml streptomycin. The cells were routinely propagated in 75-cm2 tissue culture flasks at 37° in a humidified, 5% CO2 incubator until they approached 80–90% confluency. Subsequently, the cells were trypsinized and used in experimental investigations as specified below.
HT-29 cell treatments
For all assays, cell viability was determined by trypan blue exclusion, and a known number of HT-29 cells were seeded into 25-cm2 culture flasks, 9·6-cm2 six-well plates, or 3·8-cm2 12-well plates. In some experiments, after 24 hr HT-29 cells were incubated for varying times with B. infantis, L. salivarius or S. typhimurium at a bacterial to epithelial cell ratio of 10 : 1, a dose used previously by others.13 In other investigations, HT-29 cells were grown to confluence, and confluent monolayers were treated with 1 × 105, 1 × 106 or 1 × 107 CFU/ml B. infantis or L. salivarius. Dose–response studies were performed to determine the optimal concentrations of flagellin and TNF-α to use for stimulation of IECs and, subsequently, HT-29 cells were treated with 0·5 µg/ml purified S. typhimurium flagellin (InvivoGen Corp., San Diego, CA) or 5 ng/ml TNF-α (R & D Systems, Minneapolis, MN). Bacterial survival following 1, 2, 6 or 11 hr of incubation with HT-29 cells was assessed by plating serial dilutions of cell culture supernatants on MRS agar or TSA. Following a 24-hr incubation (for S. typhimurium and L. salivarius) or a 48-hr incubation (for B. infantis), the CFU were quantified. In some experiments, HT-29 cells were pretreated for 2 hr with a known dose of commensal bacteria and subsequently were infected with an equivalent dose of S. typhimurium or treated with 0·5 µg/ml flagellin or 5 ng/ml TNF-α for varying times.
Cell viability assay
IEC viability was assessed using propidium iodide. Briefly, cell culture supernatants were removed from untreated and bacteria-treated HT-29 cells. The cells were scraped, washed in PBS, and resuspended in 50 µg/ml propidium iodide (Sigma-Aldrich) and 5 Kunitz units/ml ribonuclease A in PBS (Sigma-Aldrich). The stained cells were analysed using an Epics Elite flow cytometer (Beckman Coulter, Inc., Fullerton, CA), and propidium iodide fluorescence was detected at 675 nm. The pH of the corresponding cell culture supernatants was measured using a PHM61 Laboratory pH Meter (Radiometer A/S, Copenhagen, Denmark).
Gene arrays
Human Cytokine Expression Arrays (R & D Systems) were used to examine inflammatory gene expression in bacteria-treated HT-29 cells. Each array membrane comprised 847 cloned cDNAs representing immune-related genes including various cytokines, chemokines, and other immunoregulatory factors, printed as polymerase chain reaction (PCR) products on a positively charged nylon membrane. Briefly, HT-29 cells were seeded in six-well plates in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 10% heat-inactivated FCS and 50 µg/ml gentamycin (Gibco-BRL) and were grown to confluence. HT-29 monolayers were treated with approximately 1 × 107−108 cells/well stationary-phase B. infantis, L. salivarius or S. typhimurium for 0·5, 2 or 6 hr. Untreated cell monolayers served as controls. Following treatment, the cells were scraped and centrifuged at 400 g for 10 min. PolyA mRNA was extracted from the cell pellets using the Dynalbeads® mRNA Direct™ kit (Dynal, Oslo, Norway) according to the manufacturer's recommendations, and was treated with RNase-free DNase (Ambion, Cambridgeshire, UK). mRNA integrity was assessed by 2% agarose gel electrophoresis and ethidium bromide staining. Subsequently, human cytokine-specific primers (R & D Systems) were annealed to 600 ng of mRNA in 11 µl of diethyl pyrocarbonate (DEPC)-treated water. The mRNA was reverse-transcribed using a cDNA labelling and hybridization kit (R & D Systems) according to the manufacturer's instructions. The reaction comprised 20 µCi [α-33P]dCTP (Amersham Pharmacia, Buckinghamshire, UK), dNTP mix (333 µm dATP, dTTP and dGTP, and 1·67 µm dCTP) (R & D Systems), 30 Units of RNase inhibitor (Ambion), and 50 Units of avian myeloblastosis virus reverse transcriptase (RT). Unincorporated [α-33P]dCTP was removed from the cDNA using a Sephadex® G-25 Spin column (Sigma-Aldrich).
Hybridization of the [α-33P]dCTP-labelled cDNA probe to the Human Cytokine Expression Array membrane was performed according to the manufacturer's protocol. Following hybridization, array membranes were exposed to high-intensity phosphor screens at room temperature overnight. Screens were examined using Molecular Dynamics Storm Phosphorimager (Amersham Biosciences, Piscataway, NJ), and arrays were analysed using phoretix™ Array 2 (Nonlinear Dynamics, Newcastle-upon-Tyne, UK) and focus© (Steve Cole, Los Angeles, CA) software. Each array was performed in duplicate in each of two independent experiments.
Real-time RT-polymerase chain reaction (RT-PCR) analysis of IL-8 mRNA expression
The 2-hr mRNA samples from the gene array experiments were selected for RT-PCR analysis of IL-8 mRNA. mRNA (250 ng) was reverse-transcribed to yield cDNA using 40 U/µl RNase inhibitor (Ambion) and the Expand Reverse Transcriptase system (Roche Diagnostics Ltd, East Sussex, UK) according to the manufacturer's protocol. Real-time RT-PCR for human IL-8 was performed in a LightCycler 1·5 (Roche) using a previously described IL-8 primer set38 (MWG-Biotech Ltd, Ebersberg, Germany) and a Fast Start DNA Master SyBr Green I kit as specified by the manufacturer (Roche). Calculations were performed using GAPDH as a relative standard, and the GAPDH primers (forward, 5′-ACCACAGTCCATGCCATCAC-3′; reverse, 5′-TCCACCACCCTGTTGCTGTA-3′) were synthesized by Clontech (BD Biosciences, San Jose, CA).
Analysis of MUC3A, MUC3B and E-cadherin mRNA expression
In humans, MUC3 is among the predominant ileocolonic mucins4 and E-cadherin is a cell surface protein involved in cell adhesion.39 We used real-time RT-PCR to examine the expression of E-cadherin and two MUC3 genes, MUC3A and MUC3B,40 in commensal-treated HT-29 cells. In the absence of antibiotics, confluent HT-29 monolayers were treated with 1 × 107 cells/ml B. infantis or L. salivarius for 1 or 2 hr. The cells were harvested by trypsinization, and total RNA was isolated using the Absolutely RNA® Miniprep kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Samples were treated twice with RNase-free DNase I. Total RNA (1 µg) was reversed-transcribed from random primers (Roche) using ImProm-II RT (Promega, Madison, WI) and RNasin Plus RNase inhibitor (Promega). Real-time RT-PCR analysis was performed on 5 µl of the resulting cDNA using LightCycler TaqMan Master (Roche), and LightCycler Uracil-DNA glycosylase (Roche) as specified by the manufacturer, together with Universal ProbeLibrary probes (Exiqon, Vedbaek, Denmark) in a final reaction volume of 20 µl. Primers designed using the Universal ProbeLibrary Assay design centre (http://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp) were synthesized by MWG Biotech Ltd. PCR amplification was performed using a LightCycler 1·5 instrument (Roche). Thermal cycling conditions comprised 2 min at 40° and 10 min at 95°, followed by 45 amplification cycles at 95° for 10 seconds, 60° for 30 seconds, and 72° for 1 second, and a 40° cooling cycle for 30 seconds. Calculations were performed using GAPDH as the endogenous control reference gene. Fold difference in gene expression was calculated according to the standard formula 2(Ec–Rn) – (Et–Rt), where Ec is the crossing threshold (ct) of the experimental gene in untreated control samples, Rn is the ct of GAPDH in untreated samples, Et is the ct of the experimental gene in treated samples, and Rt is the ct of GAPDH in treated samples. Data are presented as the mean ± standard error (SE) of three independent experiments.
Immunohistochemical staining for intracellular NF-κB
HeLa cells were selected as an epithelial cell model to visualize intracellular NF-κB localization as they have an optimal nucleus-to-cytoplasm ratio and are epithelial cells.13 Intracellular NF-κB activation in the model was assessed using immunohistochemistry as described previously.13 Briefly, HeLa cells (1 × 107 cells/ml) were grown on coverslips in six-well plates for 3 days, and incubated with 1 × 107 B. infantis or 20 ng/ml TNF-α for 30 min. The coverslips were washed, fixed, blocked, and incubated with 6 µg of mouse anti-NF-κB antibody (BD Pharmingen, Mississauga, Ontario, Canada), followed by fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) (1 : 64) (Sigma-Aldrich) as described previously.13 The coverslips were mounted in glycerol-PBS, and slides were viewed using a LSM510 (Carl Zeiss, Jena, Germany) confocal microscope.
NF-κB p65 transcription factor assay
HT-29 cells (3 × 106) were seeded in 25-cm2 tissue culture flasks in 5 ml of antibiotic-free medium. After 24 hr of incubation, the medium was replaced with antibiotic- and serum-free medium, and the cells were treated with S. typhimurium, flagellin or TNF-α in the presence or absence of probiotic pretreatment for varying times. Nuclear proteins were extracted using the Active Motif Nuclear Extract kit (Active Motif Europe, Rixensart, Belgium) according to the manufacturer's instructions, and the total protein concentration of the lysates was determined by Bradford assay (Bio-Rad, Hercules, CA). Activation of the NF-κB p65 subunit in 5 µg of HT-29 nuclear extracts was determined using an NF-κB p65 enzyme-linked immunosorbent assay (ELISA)-based transcription factor assay kit (TransAM assay) (Active Motif Europe) according to the manufacturer's protocol. The NF-κB detecting antibody recognizes an epitope on p65 that is accessible only when NF-κB is activated. The positive control Jurkat nuclear extract provided with the kit was used to assess assay specificity.
IL-8 ELISA
Subconfluent or confluent HT-29 cells grown in antibiotic- and serum-supplemented media were treated with known doses of probiotic bacteria or pro-inflammatory stimuli for 6 or 24 hr. Following treatment, immunoreactive IL-8 protein levels in cell-culture supernatants were quantified using an ELISA DuoSet kit (R & D Systems) according to the manufacturer's protocol. In the gene array assays, the IL-8 DuoSet kit was used also to assess extracellular IL-8 protein levels in cell-culture supernatants and intracellular IL-8 in these cells following lysis with ice-cold water.
Bacterial interference assays
Probiotic interference with pathogen association with HT-29 cells was assessed using two independent techniques, the plate dilution method and biofluorescence. For the plate dilution method, HT-29 cells (1 × 106) seeded in six-well plates in antibiotic-free medium were incubated for 24 hr. Subsequently, the medium was replenished, and the cells were pretreated with or without 10 : 1 B. infantis or L. salivarius for 2 hr prior to infection with 10 : 1 S typhimurium for a further 2 hr. After infection, the medium was discarded and the cells washed five times with 2 ml of warm medium to remove non-adherent bacteria. Cells with associated bacteria were lysed using sterile distilled water supplemented with 0·1% bovine serum albumin for 30 min at 4° with agitation as described previously.41 IEC-associated S. typhimurium were defined as adherent plus intracellular bacteria and were quantified by CFU counts of diluted cell lysates on TSA.
Independently, S. typhimurium grown overnight in TSB were pelleted by centrifugation and washed with sterile 0·85% NaCl. S. typhimurium (2 × 108) were labelled using 1·5 µl of SYTO 9® (Molecular Probes, Inc., Eugene, OR), a green-fluorescent nucleic acid stain, as specified by the manufacturer. HT-29 cells were then infected with 10 : 1 SYTO 9-labelled S. typhimurium for 2 hr in the presence or absence of probiotic pretreatment. Subsequently, the cells were washed five times with warm 0·85% NaCl and cells with associated bacteria were lysed as outlined above. Aliquots of lysate (200 µl) were transferred to wells of flat-bottom 96-well plates, and fluorescence was quantified by in vivo imaging using the IVIS™ 100 Imaging System and Living Image Software (Xenogen, Alameda, CA). The numbers of IEC-associated bacteria were determined from a standard curve of serial dilutions of SYTO 9-labelled S. typhimurium relative to fluorescence intensity. For both techniques, probiotic interference was evaluated by comparing S. typhimurium association with HT-29 cells in the presence or absence of probiotic pretreatment.
Bacterial treatment of DCs isolated from human peripheral blood mononuclear cells (PBMCs)
In accordance with a protocol approved by the Ethics Committee of Cork University Hospital, Cork, Ireland, peripheral blood from healthy volunteers (n = 3) was collected by venepuncture into sterile ethylenediaminetetraacetic acid (EDTA) vacutainer tubes. PBMCs were purified from buffy coats in histopaque tubes (Greiner Bio-one, Inc., Longwood, FL) using Ficoll-Hypaque (Pharmacia, Dübendorf, Switzerland) gradient centrifugation (400 g for 30 min). PBMCs were taken from the interface and washed four times with Ca2+- and Mg2+-free PBS, and were finally resuspended in Ca2+- and Mg2+-free degassed column buffer (PBS, pH 7·2, supplemented with 0·5% BSA and 2 mm EDTA). Subsequently, monocytes were isolated from mononuclear cells by negative selection using the Monocyte Isolation Kit II (Miltenyi Biotec) with MACS Column and MACS Separator (Miltenyi Biotec) according to the manufacturer's instructions. To generate DCs, monocytes were incubated for 5 days in DMEM containing 10% FCS, 100 000 IU/ml granulocyte–monocyte colony-stimulating factor, and 40 000 IU/ml IL-4 (BD Biosciences). Cell purity was assessed by cell surface staining and flow cytometry using antibodies obtained from BD Biosciences, and viability was determined by trypan blue exclusion. Cell purity as determined by HLA-DR-positive and CD3/CD14/CD16/CD19/CD20/CD56-negative cells was consistently >85%, and cell viability was consistently >98%. Subsequently, PBMC-derived DCs (1 × 106 cells/ml) were seeded in 1·9-cm2 24-well plates in DMEM supplemented with 10% heat-inactivated FCS, 100 U/ml penicillin G, 100 µg/ml streptomycin, and 2·5 µg/ml fungizone (Gibco-BRL). Cells were treated with medium alone (negative untreated controls), or with 10 : 1 B. infantis or L. salivarius for 48 hr. Subsequently, IL-10 and TNF-α protein levels in cell-culture supernatants were quantified using commercially available ELISA kits (R & D Systems) according to the manufacturer's protocols. In order to determine the levels of endotoxin in the commensal bacterial preparations we used a Limulus amebocyte lysate (LAL) gel-clot assay with a sensitivity of 0·03 Endotoxin units/ml (Charles River Laboratories, Inc., Charleston, SC). The assay was used according to the manufacturer's instructions and Escherichia coli control standard endotoxin (Charles River Laboratories, Inc.) was used to confirm LAL-reagent sensitivity. Tap water was used as a positive control and LAL-reagent water (Charles River Laboratories, Inc.) as a negative control.
Statistics
All data are expressed as mean ± SE. Statistical analyses were performed using unpaired two-tailed Student's t-tests or analysis of variance (ANOVA). P-values <0·05 were considered to be statistically significant, and n represents the number of independent experiments performed.
Results
IEC viability following bacterial treatment
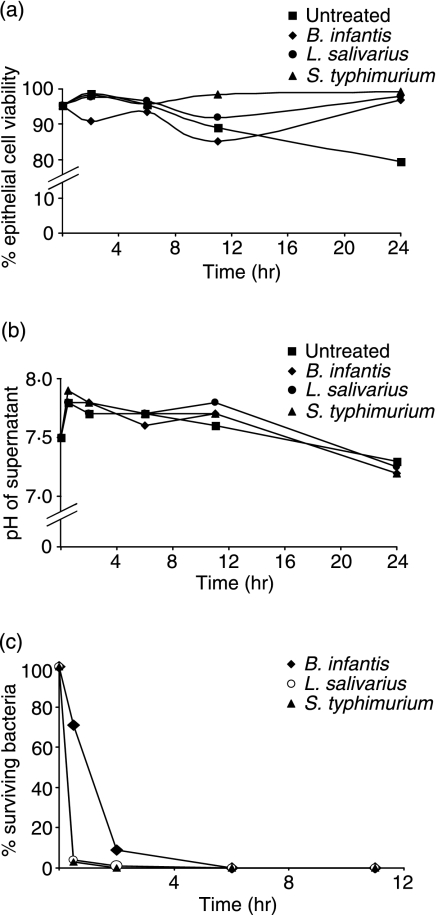
To determine whether the bacteria used in this study had any immediate direct effect on the cell line model, HT-29 cells were exposed to B. infantis, L. salivarius or S. typhimurium in the presence of antibiotics for varying times. Compared with untreated controls, neither the two probiotic strains nor S. typhimurium exerted a toxic effect on HT-29 cells over a 24-hr time-period (Fig. 1a). Parenthetically, compared with untreated cells, there was a marginal improvement in survival 24 hr after HT-29 cells were exposed to bacteria, although the biological significance of this is uncertain. Moreover, co-incubation of HT-29 cells with each individual bacterial strain did not adversely affect the pH of the growth media (Fig. 1b); no bacterial overgrowth was observed, and after 6 hr <0·02% of the bacteria remained viable (Fig. 1c). Therefore, under the assay conditions, IEC integrity was not compromised during co-incubation with B. infantis, L. salivarius or S. typhimurium.
Figure 1.
Effects of bacterial treatment on the HT-29 cell line model. HT-29 cells in antibiotic-supplemented media were untreated, or were exposed to Bifidobacterium infantis, Lactobacillus salivarius, or Salmonella typhimurium at a bacterial to epithelial cell ratio of 10 : 1 for up to 24 hr. (a) Epithelial cell viability was measured at the indicated time-points and results are expressed as per cent viable HT-29 cells detected at each time-point. (b) The pH of the corresponding cell culture supernatants recorded at each indicated time-point is shown. (c) Bacterial survival at various times post treatment was determined by quantifying colony-forming units and results are expressed as per cent surviving bacteria. The graphs are representative of n = 3 independent experiments.
Inflammatory gene expression following bacteria incubation
Human Cytokine Expression Arrays were used to examine inflammatory gene expression in bacteria-treated HT-29 cells. Treatment with B. infantis or L. salivarius for 0·5, 2 or 6 hr did not augment the expression of any of the 847 immune-related genes assayed, whereas infection with S. typhimurium increased expression of 36 genes associated with pro-inflammatory responses (Table 1). These included a repertoire of signal transducers, immunoreceptors, chemokines, and transcription factors, among them NF-κB and the chemokine IL-8.
Table 1.
Immunoregulatory genes stimulated by Salmonella typhimurium
| Maximum induction | ||||
|---|---|---|---|---|
| Gene (official nomenclature) | Function | n = 1* | n = 2* | Time (hr) |
| CXCL8, interleukin-8 (IL-8) | CXC chemokine | 17 | 20 | 2 |
| CXCL1, growth related oncogen protein-alpha (Gro-α) | CXC chemokine | 17 | 21 | 2 |
| CXCL2, growth related oncogene protein-beta (Gro-β) | CXC chemokine | 15 | 20 | 2 |
| CXCL3I, growth related oncogene protein-gamma (Gro-γ) | CXC chemokine | 15 | 21 | 2 |
| CCL20, macrophage inflammatory protein 3-alpha (MIP-3-α) | CC chemokine | 5 | 7 | 2 |
| CXCL5, epithelial neutrophil activating peptide-78 (ENA-78) | CXC chemokine | 7 | 4 | 6 |
| Tumour necrosis factor-alpha, (TNF-α) | Pro-inflammatory cytokine | 7 | 4 | 2 |
| Interleukin-1-beta (IL-1β) | Pro-inflammatory cytokine | 5 | 6 | 2 |
| CSF2, Colony stimulating factor 2 (GM-CSF) | Pro-inflammatory cytokine | 3 | 3 | 2 |
| Bone morphogenetic protein 4 (BMP-4) | Growth and differentiation factor | 1·5 | 1·7 | 0·5 |
| Amphiregulin (AREG) | Growth factor and mitogen | 2 | 3 | 2 |
| Cyclin-dependent kinase inhibitor 1A (p21) (CDKN1A (p21)) | Kinase inhibitor | 4 | 4 | 2 |
| v-myc myelocytomatosis viral oncogene homolog (avain) MYC (c-Myc) | Transcription factor activity | 3 | 2 | 6 |
| Signal recoginition particle 72 kDa (SRP72) | Signal transduction | 1·6 | 2 | 0·5 |
| Myeloid differentiation primary response gene (88) (MYD88) | Signal transduction | 2 | 1·7 | 0·5 |
| Tumour necrosis factor receptor-associated factor-2 (TRAF2) | Signal transduction | 2·75 | 3·5 | 2 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells (p105) (NF-κB-1 (p50)) | Transcription factor | 1·8 | 2·6 | 2 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) (NF-κB-2 (p100)) | Transcription factor | 2·6 | 2·8 | 2 |
| Interferon regulatory factor 1 (IRF-1) | Transcription activator | 2·8 | 2·6 | 2 |
| B-cell CLL/lymphoma 3 (BCL3) | Transcription coactivator | 2·7 | 4 | 2 |
| Interleukin 6 signal transducer (IL-6ST (gp130)) | Signal transduction | 2·1 | 2·2 | 0·5 |
| Toll-like receptor 3 (TLR3) | Receptor activity | 2 | 2·3 | 0·5 |
| Interferon gamma receptor 1 (IFNGR1) | Receptor activity | 3 | 2 | 6 |
| G protein-coupled receptor 19 (GPR19) | Receptor activity | 8 | 10 | 2 |
| Interleukin 18 receptor 1 (IL-18R1) | Receptor activity | 2 | 2 | 6 |
| Interleukin 10 receptor, beta (IL-10RB) | Receptor activity | 2·4 | 2·5 | 6 |
| Intercellular adhesion molecule type 1 (ICAM-1) | Receptor activity/adhesion | 6 | 5·75 | 2 |
| Integrin-alpha-4 (ITGA4) | Receptor activity | 4 | 8 | 2 |
| CD3G antigen, gamma polypeptide (TTT3 complex) (CD3G) | T-cell receptor binding | 2·4 | 1·5 | 0·5 |
| CD40 antigen (TNF receptor superfamily member 5) (CD40) | Receptor activity | 3 | 2 | 6 |
| Lymphocyte adhesion molecule 1 (SELL (l-selectin)) | Receptor activity | 3·5 | 2·7 | 6 |
| Expressed in non-metastatic cells 6, protein (nucleoside diphosphate kinase) NME6 (NM23-H6) | Kinase activity | 1·7 | 1·7 | 0·5 |
| Ephrin receptor A2 (EPHA2) | Enzymatic and receptor activity | 1·9 | 1·9 | 2 |
| Prostaglandin-endoperoxide synthase 2 (Cox-2) | Enzymatic activity | 1·7 | 2·8 | 2 |
| Matrix metallopeptidase 7 (MMP-7) | Enzymatic activity | 2·8 | 2·5 | 6 |
| Matrix metallopeptidase 8 (MMP-8) | Enzymatic activity | 2·3 | 2·2 | 6 |
Inflammatory gene expression was evaluated using Human Cytokine Expression Arrays.
Each array was performed in duplicate, and the mean result of each of two independent experiments expressed as maximum fold induction compared with baseline gene expression in untreated HT-29 cells is indicated. The time postinfection at which maximum gene induction occurred is also shown. See Supplementary Table S1 for a complete list of the genes examined in the cDNA array.
NF-κB activation following treatment of epithelial cells
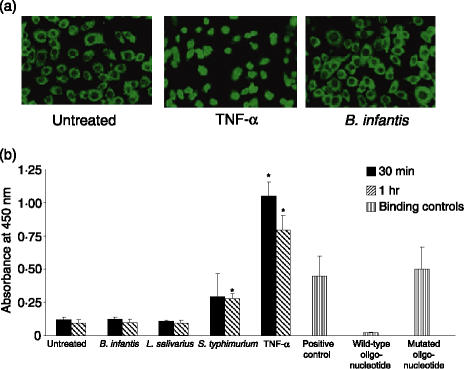
In order to confirm the gene array results, we used HeLa cervical epithelial cells as an independent epithelial model to assess NF-κB activation by immunohistochemistry. Untreated HeLa cells expressed NF-κB p65 constitutively in the cytoplasm (Fig. 2a), and treatment with 20 ng/ml TNF-α caused a rapid cytoplasmic to nuclear translocation of NF-κB, and increased levels of NF-κB p65 were detected in the nucleus within 30 min. In contrast, treatment with B. infantis for the same time did not induce nuclear translocation and NF-κB remained localized to the cytoplasm (Fig. 2a). Similarly, in HT-29 cells, treatment with B. infantis or L. salivarius for 30 min or 1 hr did not stimulate the DNA-binding activity of NF-κB p65 compared with untreated cells, whereas infection with S. typhimurium or treatment with TNF-α significantly increased NF-κB DNA-binding activity at 1 hr or 30 min, respectively (Fig. 2b). NF-κB DNA-binding activity was detected in the positive control Jurkat nuclear extract, and the specificity of NF-κB binding in the assay was confirmed by competition with free wild-type NF-κB consensus oligonucleotide or mutated NF-κB oligonucleotide (Fig. 2b). The data demonstrate that epithelial cells respond differently to various antigens, and, in contrast to S. typhimurium and TNF-α, the commensal bacteria used in this study did not induce NF-κB nuclear translocation or DNA-binding activity.
Figure 2.
Bifidobacterium infantis or Lactobacillus salivarius do not activate nuclear factor (NF)-κB in epithelial cells. (a) Immunohistochemical staining for NF-κB p65 demonstrated a constitutive expression of NF-κB p65 in the cytoplasm of untreated HeLa cells. Incubation with tumour necrosis factor (TNF)-α (20 ng/ml) for 30 min caused p65 nuclear translocation, whereas B. infantis did not induce translocation of p65. (b) HT-29 cells were untreated, or were treated with 10 : 1 B. infantis, L. salivarius, Salmonella typhimurium, or 5 ng/ml TNF-α for 30 min or 1 hr. The DNA-binding activity of NF-κB p65 in HT-29 nuclear extracts was determined using an enzyme-linked immunosorbent assay (ELISA)-based transcription factor assay. The positive control Jurkat nuclear extract provided with the kit was used to verify assay specificity in competition assays with wild-type or mutated NF-κB oligonucleotides. The data represent mean absorbance readings ± standard errors of five separate experiments. *P < 0·05 relative to untreated HT-29 cells.
S. typhimurium, but not B. infantis or L. salivarius, induces IL-8 expression
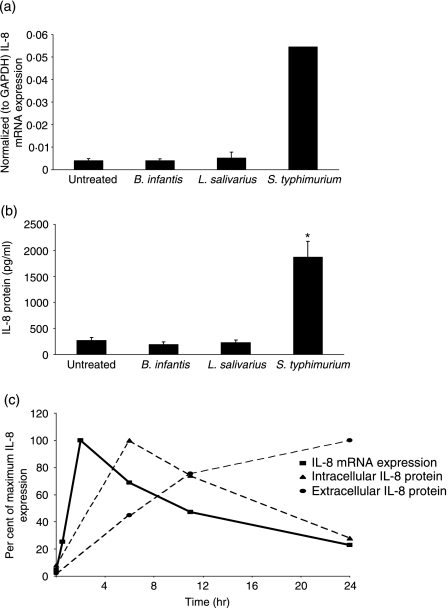
In subconfluent HT-29 cells treated with B. infantis or L. salivarius, the levels of IL-8 mRNA or protein detected after 2 or 24 hr, respectively, were similar to those found at baseline in untreated cells (Figs 3a and b). Conversely, infection with S. typhimurium stimulated a significant increase in IL-8 mRNA expression after 2 hr (13·3-fold) and IL-8 protein secretion after 24 hr (7-fold) compared with untreated cells. As shown in Fig. 3(c), S. typhimurium quickly induced IL-8 mRNA and maximum levels were detected within 2 hr of infection. Infection also resulted in a time-dependent accumulation of extracellular IL-8 that was associated with a corresponding decrease in the amount of detectable intracellular IL-8 protein (Fig. 3c).
Figure 3.
Salmonella typhimurium-induced interleukin (IL)-8 expression. Subconfluent HT-29 cells were treated with 10 : 1 Bifidobacterium infantis, Lactobacillus salivarius or S. typhimurium for varying times. Untreated cells served as controls. (a) IL-8 mRNA expression was determined after 2 hr by real-time reverse transcriptase–polymerase chain reaction (RT-PCR). The data are presented as normalized (to GAPDH) IL-8 mRNA expression levels and are mean ± standard error (SE) (n = 3). *P < 0·05 relative to untreated cells. (b) IL-8 protein levels (pg/ml) in cell culture supernatants were measured after 24 hr by enzyme-linked immunosorbent assay (ELISA). The data represent mean ± SE (n = 3). *P < 0·05 relative to untreated cells. (c) Time–course of S. typhimurium-induced IL-8 mRNA expression (normalized to GAPDH), intracellular IL-8 protein depletion, and extracellular IL-8 protein accumulation in HT-29 cells. The data are expressed as percentage of maximum IL-8 mRNA/protein expression and are representative of two independent experiments.
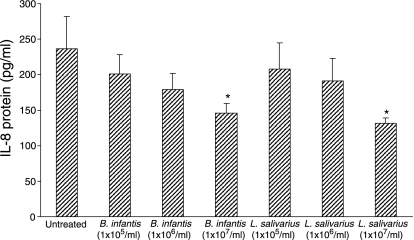
B. infantis and L. salivarius inhibit IL-8 secretion at baseline
We next examined whether treatment with increasing doses of B. infantis or L. salivarius (1 × 105, 1 × 106, or 1 × 107 CFU/ml) affected IL-8 secretion by confluent HT-29 cell monolayers. IL-8 protein levels were measured after 6 hr. A dose-dependent inhibition of baseline IL-8 secretion by confluent HT-29 monolayers was observed (Fig. 4). At a concentration of 1 × 107 CFU/ml, which under the experimental conditions was equivalent to approximately 10 bacteria per epithelial cell, both B. infantis and L. salivarius significantly inhibited basal IL-8 secretion by 38 and 44%, respectively. This suggests that these commensal bacteria exert anti-inflammatory effects on the epithelium by down-regulating the secretion of IL-8.
Figure 4.
Probiotic pretreatment down-regulates interleukin (IL)-8 secretion at baseline. Confluent HT-29 cells were treated with Bifidobacterium infantis or Lactobacillus salivarius at doses of 1 × 105, 1 × 106, or 1 × 107 colony-forming units (CFU)/ml, and IL-8 protein levels were measured after 6 hr. B. infantis and L. salivarius caused a dose-dependent inhibition of baseline IL-8 secretion by confluent HT-29 monolayers (*P < 0·05 compared with untreated monolayers). The data are expressed as pg/ml IL-8 and represent the mean ± standard error (n = 3 independent experiments).
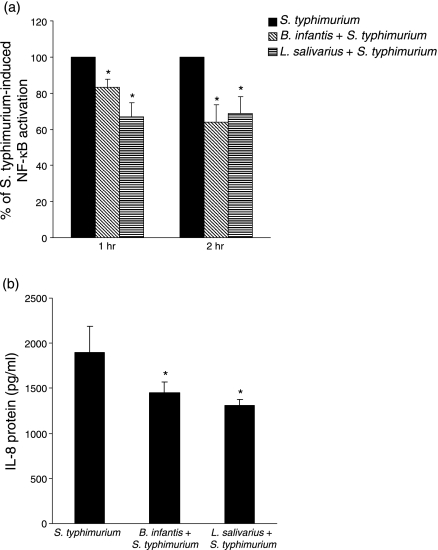
Probiotic bacteria attenuate S. typhimurium-induced pro-inflammatory responses
IL-8 mRNA expression is under the regulatory control of NF-κB,8 and our data indicated that B. infantis or L. salivarius do not activate NF-κB or elicit IL-8 production by HT-29 cells. Considering this and the finding that the probiotic bacteria affected IL-8 secretion at baseline, we explored whether B. infantis or L. salivarius might inhibit NF-κB activation or IL-8 secretion in response to S. typhimurium. Subconfluent HT-29 cells were pretreated with 10 : 1 B. infantis or L. salivarius for 2 hr prior to stimulation with S. typhimurium or TNF-α. The levels of activated NF-κB in nuclear extracts were determined after 0·5, 1 and 2 hr, and IL-8 protein was measured after 24 hr. Prior exposure to either B. infantis or L. salivarius significantly reduced S. typhimurium-induced NF-κB binding activity following 1 or 2 hr of infection (Fig. 5a). Within 30 min, TNF-α stimulated a 8·9-fold increase in activated NF-κB compared with untreated cells, and, although pretreatment with L. salivarius or B. infantis did not affect NF-κB activation by TNF-α at 30 min, a significant reduction (16% decrease) was observed in B. infantis-treated cells 1 hr post TNF-α-stimulation (data not shown).
Figure 5.
Probiotic pretreatment attenuates Salmonella typhimurium-induced pro-inflammatory responses. Subconfluent HT-29 cells were pretreated with 10 : 1 Bifidobacterium infantis or Lactobacillus salivarius for 2 hr prior to infection with 10 : 1 S. typhimurium. The levels of activated nuclear factor (NF)-κB in nuclear extracts were determined after 1 and 2 hr, and interleukin (IL)-8 protein levels were measured after 24 hr. (a) Pretreatment with B. infantis or L. salivarius reduced NF-κB activation by S. typhimurium after 1 and 2 hr (*P < 0·05 compared with S. typhimurium-infected HT-29 cells). The data are expressed as per cent of S. typhimurium-induced NF-κB activation, and represent the mean ± standard error (SE) (n = 5 independent experiments). (b) Pretreatment with B. infantis or L. salivarius inhibited S. typhimurium-induced IL-8 secretion (*P < 0·05 relative to S. typhimurium-infected HT-29 cells). The data are expressed as pg/ml IL-8 and represent the mean ± SE (n = 6 independent experiments).
As shown in Fig. 5(b), S. typhimurium induced 1897 (± 292) pg/ml IL-8, and pretreatment with B. infantis or L. salivarius significantly attenuated S. typhimurium-induced IL-8 secretion by 23·5 or 31%, respectively. Although TNF-α potently induced 50 550 (± 9933) pg/ml IL-8, probiotic pretreatment had no significant effect on TNF-α-induced IL-8 secretion (data not shown). Collectively, these data show that B. infantis and L. salivarius significantly attenuate S. typhimurium-induced pro-inflammatory responses.
S. typhimurium association with HT-29 cells is not blocked by the probiotic bacteria
We next determined whether the modulation of S. typhimurium-induced pro-inflammatory responses by the probiotic bacteria was attributable to interference with pathogen binding to IECs. Binding of S. typhimurium to HT-29 cells was evaluated in the presence or absence of probiotic pretreatment using biofluorescence and the plate dilution method. The two methods yielded complementary data and, as shown in Table 2, pretreatment with neither B. infantis nor L. salivarius affected the number of IEC-associated S. typhimurium detected 2 hr after infection. These data indicate that the immunoregulatory effects of the probiotic bacteria observed in this study are not attributable to competitive inhibition of S. typhimurium association with HT-29 cells.
Table 2.
Probiotic bacteria do not interfere with Salmonella typhimurium–HT-29 cell association
| Plate dilution method | Biofluorescence method | |
|---|---|---|
| Sample | Cell-associated S. typhimurium (log CFU/ml)* | % of cell-associated S. typhimurium† |
| S. typhimurium | 3·24E + 05 ± 1·25E + 05 | 100 |
| B. infantis + S. typhimurium | 3·25E + 05 ± 8·85E + 04 | 90·02 ± 13·75 |
| L. salivarius + S. typhimurium | 3·61E + 05 ± 7·23E + 04 | 112·04 ± 10·85 |
Probiotic interference with pathogen binding was evaluated by quantifying the numbers of S. typhimurium associated with HT-29 cells in the presence or absence of probiotic pretreatment. Data represent the mean ± standard error of three independent experiments.
Intestinal epithelial cell (IEC)-associated S. typhimurium were quantified by colony-forming units (CFU) counts of diluted cell lysates on tryptic soy agar (TSA) and results are expressed as log CFU/ml.
IEC-associated S. typhimurium were quantified by in vivo imaging fluorescence using a standard curve of serial dilutions of SYTO 9-labelled S. typhimurium relative to fluorescent intensity. Results are expressed as per cent HT-29 cell-associated S. typhimurium.
B. infantis, Bifidobacterium infantis; L. salivarius, Lactobacillus salivarius.
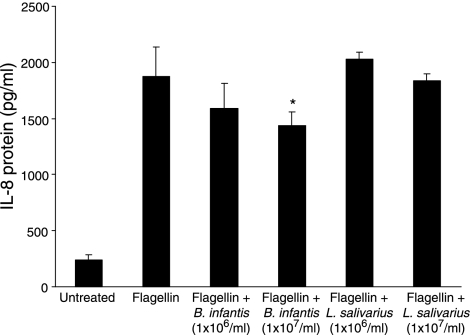
B. infantis inhibits flagellin-induced IL-8 secretion
Flagellin is a key activator of pro-inflammatory responses to Salmonella in IECs.22,42 Therefore, we explored whether the probiotic bacteria could inhibit IEC responses to Salmonella flagellin. Firstly, in dose–response studies, we incubated various doses of flagellin from S. typhimurium (0·1, 0·5 and 1·0 µg/ml) with confluent HT-29 monolayers, and found that, at a concentration of 0·5 µg/ml, flagellin significantly induced NF-κB activation and IL-8 secretion. Treatment with 0·5 µg/ml flagellin for 1 hr resulted in a 3·15 (± 0·3)-fold increase in NF-κB binding activity compared with untreated cells, and 1876 (± 262) pg/ml IL-8 was detected after 6 hr (P < 0·05) (data not shown). HT-29 monolayers were treated for 2 hr with either B. infantis or L. salivarius (1 × 106 or 1 × 107 CFU/ml), followed by 0·5 µg/ml flagellin for 6 hr. As shown in Fig. 6, pretreatment with L. salivarius did not affect flagellin-induced IL-8 at either dose tested. However, flagellin-induced IL-8 was significantly inhibited in cells that were pretreated with 1 × 107 CFU/ml B. infantis. The results suggest strain-specific antagonistic effects on inducible IL-8 expression in IECs.
Figure 6.
Bifidobacterium infantis attenuates flagellin-induced interleukin (IL)-8 secretion. Confluent HT-29 cells were pretreated for 2 hr with B. infantis or Lactobacillus salivarius at doses of 1 × 106 or 1 × 107 colony-forming units (CFU)/ml prior to stimulation with 0·5 µg/ml flagellin for 6 hr. Pretreatment with 1 × 107 CFU/ml B. infantis, but not L. salivarius, significantly inhibited flagellin-induced IL-8 secretion (*P < 0·05 relative to flagellin-treated HT-29 monolayers). The data are expressed as pg/ml IL-8 and represent the mean ± standard error of three independent experiments.
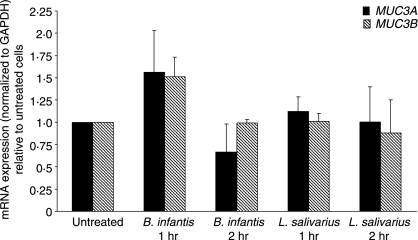
MUC3A, MUC3B and E-cadherin expression following probiotic treatment
It has been proposed that some strains of probiotic bacteria exert beneficial effects on the intestinal epithelium by up-regulating the expression of mucin genes.36,43 We used real-time RT-PCR to determine the expression of MUC3A and MUC3B mRNA in confluent HT-29 cells treated for 1 or 2 hr with 1 × 107 cells/ml probiotic bacteria. MUC3A and MUC3B mRNA was detected in untreated HT-29 cells, and, as shown in Fig. 7, treatment with B. infantis or L. salivarius had no significant effect on the expression of these genes. Similarly, no difference was observed in E-cadherin mRNA expression following treatment with the commensal bacteria (data not shown).
Figure 7.
MUC3 mRNA expression is unaltered by probiotic treatment. Confluent HT-29 monolayers were treated with 1 × 107/ml Bifidobacterium infantis or Lactobacillus salivarius for 1 or 2 hr. Real-time reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of MUC3A and MUC3B mRNA expression was performed and the fold difference in gene expression compared with that of untreated samples was calculated using GAPDH as the reference gene. Data are presented as mean ± standard error of three separate experiments.
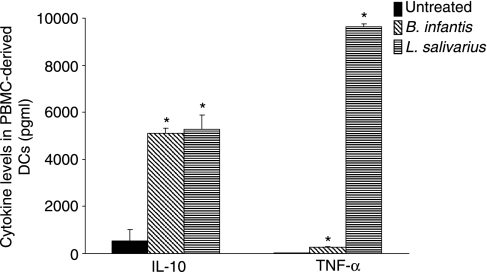
DC cytokine responses following probiotic incubation
We examined the effect of the commensal bacteria used in this study on cytokine secretion by myeloid DCs. DCs were treated with B. infantis or L. salivarius for 48 hr, and IL-10 and TNF-α protein levels were quantified. Both bacteria stimulated a significant increase in IL-10 and TNF-α secretion by PBMC-derived DCs compared with untreated DCs (Fig. 8). These effects were not a result of endotoxin contamination of the bacterial suspensions as <0·03 Endotoxin units/ml, the detection limit of the LAL assay, were detected in the medium controls and bacterial preparations.
Figure 8.
Commensal bacteria stimulate interleukin (IL)-10 and tumour necrosis factor (TNF)-α secretion by myeloid dendritic cells (DCs). DCs isolated from peripheral blood mononuclear cells (PBMCs) of healthy volunteers (n = 3) were exposed to Bifidobacterium infantis or Lactobacillus salivarius for 48 hr, and IL-10 and TNF-α levels were determined. B. infantis and L. salivarius stimulated IL-10 and TNF-α secretion (*P < 0·05 relative to untreated PBMC-derived DCs). The data are expressed as mean pg/ml ± standard error of three independent experiments.
Discussion
In the present study, we have shown that IECs respond differentially to various bacterial species. In contrast to S. typhimurium, B. infantis or L. salivarius did not induce pro-inflammatory responses by IECs. However, B. infantis and L. salivarius functionally modulated the epithelium by inhibiting the constitutive secretion of IL-8 and attenuating S. typhimurium-induced NF-κB activation and IL-8 secretion. Moreover, B. infantis inhibited flagellin-induced IL-8 protein, and both commensal strains stimulated the secretion of IL-10 and TNF-α by myeloid DCs. Taken together, these data demonstrate that B. infantis and L. salivarius exert immunomodulatory effects that mediate intestinal immune cell responses to enteric pathogens and their antigenic components.
Commensal bacteria, their ligands and commensal-derived symbiosis factors have been shown to be essential for normal development of the mucosal immune system and the maintenance of gut homeostasis,2,44,45 but their role in intestinal physiology is incompletely understood. Here we have demonstrated that the intestinal epithelium mounts a robust pro-inflammatory response to S. typhimurium, but not to B. infantis or L. salivarius. In particular, B. infantis and L. salivarius did not stimulate NF-κB activation or IL-8 secretion. Similarly, other commensal bacteria, including L. rhamnosus GG, L. reuteri, Lactobacillus plantarum, and the VSL#3 cocktail, do not activate NF-κB or IL-8.11–13,46 The current study considerably extends these observations, and the gene array analysis demonstrated that, in addition to NF-κB and IL-8, B. infantis or L. salivarius did not augment the expression of any of the other 845 immune-related genes assayed. However, immune cells were not universally unresponsive to these commensal bacteria and they induced the secretion of regulatory cytokines by myeloid DCs. This supports previous studies that showed that probiotic bacteria modulate DC function and promote tolerance-inducing DCs.47,48 Furthermore, certain non-pathogenic bacteria can activate gut inflammatory processes and induce IL-8 secretion by IECs.11–13,49 The non-pathogenic commensal strain Bacteroides vulgatus has been shown to activate NF-κB and pro-inflammatory gene expression in IECs.49 However, these effects were inhibited by the presence of immune cells37 and immune–IEC cross-talk differentially affects pro-inflammatory gene expression in response to commensal bacteria.50
Our data indicate that both B. infantis and L. salivarius limited the constitutive secretion of IL-8. This effect was only observed in confluent HT-29 monolayers, demonstrating the importance of an intact barrier for normal epithelial immune function. Moreover, pretreatment with B. infantis and L. salivarius inhibited S. typhimurium-, but not TNF-α-induced IL-8 secretion, although B. infantis delayed TNF-α-induced NF-κB activation. This suggests the involvement of different intracellular signalling pathways between commensal-mediated affects on S. typhimurium- and TNF-α-mediated pro-inflammatory responses. Considering the crucial role of IL-8 in inflammatory processes and the implication of enteric bacteria in the pathogenesis of IBD,51 our finding that B. infantis and L. salivarius attenuated IL-8 secretion at baseline and in response to S. typhimurium may be of particular physiologic relevance in IBD and other inflammatory conditions. Other commensal strains, including L. reuteri, Bacteroides thetaiotaomicron, Salmonella pullorum and VSL#3, have exhibited similar effects on pathogen-induced IL-8 secretion in IECs,11,13,52,53 and L. reuteri was shown to inhibit the constitutive synthesis, but not secretion, of IL-8.13
It has been established that flagellin is a prerequisite for Salmonella-induced TLR5-mediated pro-inflammatory responses in IECs.22,42 In our study B. infantis, but not L. salivarius, attenuated flagellin-induced IL-8, suggesting that different modes of action mediate strain-specific antagonistic effects on inducible IL-8 expression. To our knowledge, only one other commensal strain, B. thetaiotaomicron, has been reported to restrict flagellin-mediated signalling.52 Although the responsible mechanism(s) remains unknown, it is possible that commensal surface structures act through host cell receptors to modulate inflammatory responses,54,55 but our data do not implicate the TNF receptor in this process. It has been proposed that receptor systems antagonistic to TLR action, analogous to SIGIRR (single Ig IL-1R-related molecule),56 are involved in mediating protective effects of the commensal flora.57 Moreover, the composition of cell surface structures such as lipotechoic acid of Lactobacillus plantarum has been shown to differentially impact on the host immune system.58 The ability of the gut to tolerate large numbers of potentially pro-inflammatory commensal bacteria could, in part, be attributed to the anti-inflammatory effects of bacteria such as B. thetaiotaomicron and B. infantis that limit the signalling induced by flagellin and Salmonella. The possibility that B. infantis exerts similar effects on other flagellated pathogens is being investigated.
The data suggest that the inhibitory effects of B. infantis and L. salivarius were mediated, at least in part, through the NF-κB pathway. B. infantis and L. salivarius inhibited S. typhimurium-induced NF-κB activation, and B. infantis did not stimulate nuclear translocation of p65 in HeLa cells. Several distinct modes of action by which commensal bacteria impinge on NF-κB signalling to limit inflammation have been elucidated. These include inhibition of IκB-α ubiquitination or epithelial proteasome function53,59 or the nuclear export of transcriptionally active p65.52 Mitogen-activated protein kinase and protein kinase B pathways have also been implicated in the protective effects mediated by various commensal bacteria.11,60 In addition, several host homeostatic mechanisms that limit inflammatory responses in the gut have been described.1,57,61
We queried whether the inhibitory effects of B. infantis and L. salivarius on S. typhimurium-induced pro-inflammatory responses were attributable to competitive exclusion at the epithelial surface. Such a defensive layer could comprise the commensal bacteria themselves, or the induction of mucin genes. The data indicate that these commensal bacteria did not hinder the association of S. typhimurium with HT-29 cells. Consistent with this observation, B. infantis and L. salivarius did not induce the expression of MUC3A or MUC3B. VSL#3 and other Lactobacillus strains have been reported to induce mucin expression,11,36,43,62 thereby preventing adhesion of pathogens to the epithelial surface.36,43 However, commensal-induced mucin up-regulation is strain-specific, and Lactobacillus acidophilus does not stimulate MUC3 expression.36 Of note, mucin expression is affected by the growth medium, and in other studies HT-29 cells were progressively transferred to glucose-free medium to preferentially increase MUC3 mRNA expression.36,43 Although commensal bacteria have been shown to modify pathogen-induced alterations in cytoskeletal and tight junction proteins such as zonula occludens, occludin and actinin,11,41 we have shown that B. infantis or L. salivarius do not induce alterations in E-cadherin mRNA expression in HT-29 cells.
In conclusion, we have demonstrated that IECs display immunological quiescence when exposed to B. infantis or L. salivarius, but these bacteria can modulate epithelial responses to limit inflammatory signals. Functional capabilities appear to be strain-specific, and the precise mechanisms by which B. infantis and L. salivarius limit inflammatory responses are being explored. Collectively, the data demonstrate that B. infantis and L. salivarius exert immunomodulatory effects on IECs that mediate host responses to flagellin and enteric pathogens.
Acknowledgments
The authors' work is supported, in part, by Science Foundation Ireland in the form of a centre grant (Alimentary Pharmabiotic Centre), the Irish Health Research Board, the Higher Education Authority of Ireland, and the European Union.
Abbreviations
- CFU
colony-forming units
- ct
crossing threshold
- DCs
dendritic cells
- DMEM
Dulbecco's modified Eagle's medium
- FCS
fetal calf serum
- IBD
inflammatory bowel disease
- IECs
intestinal epithelial cells
- IL
interleukin
- MRS
de Man Rogosa Sharpe
- NF
nuclear factor
- PBMCs
peripheral blood mononuclear cells
- RT
reverse transcriptase
- TLR
Toll-like receptor
- TNF
tumour necrosis factor
- TSA
tryptic soy agar
- TSB
tryptic soy broth
Supplementary material
The following supplementary material is available for this article online:
Table S1. The 847 cloned cDNAs representing immunerelated genes on the Human Cytokine Expression Arrays used to examine inflammatory gene expression in bacteria-treated HT29 cells.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- 1.Otte JM, Cario E, Podolsky DK. Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. 2004;126:1054–70. doi: 10.1053/j.gastro.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Kagnoff MF, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corfield AP, Myerscough N, Longman R, Sylvester P, Arul S, Pignatelli M. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut. 2000;47:589–94. doi: 10.1136/gut.47.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang SK, Dohrman AF, Basbaum CB, Ho SB, Tsuda T, Toribara NW, Gum JR, Kim YS. Localization of mucin (MUC2 and MUC3) messenger RNA and peptide expression in human normal intestine and colon cancer. Gastroenterology. 1994;107:28–36. doi: 10.1016/0016-5085(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi A, Wada A, Ogushi K, et al. Production of beta-defensin-2 by human colonic epithelial cells induced by Salmonella enteritidis flagella filament structural protein. FEBS Lett. 2001;508:484–8. doi: 10.1016/s0014-5793(01)03088-5. [DOI] [PubMed] [Google Scholar]
- 7.Stadnyk AW. Intestinal epithelial cells as a source of inflammatory cytokines and chemokines. Can J Gastroenterol. 2002;16:241–6. doi: 10.1155/2002/941087. [DOI] [PubMed] [Google Scholar]
- 8.Jobin C, Sartor RB. The I kappa B/NF-kappa B system: a key determinant of mucosalinflammation and protection. Am J Physiol Cell Physiol. 2000;278:C451–62. doi: 10.1152/ajpcell.2000.278.3.C451. [DOI] [PubMed] [Google Scholar]
- 9.Harada A, Sekido N, Akahoshi T, Wada T, Mukaida N, Matsushima K. Essential involvement of interleukin-8 (IL-8) in acute inflammation. J Leukoc Biol. 1994;56:559–64. [PubMed] [Google Scholar]
- 10.Jung HC, Eckmann L, Yang SK, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff MF. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G613–26. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 12.Lammers KM, Helwig U, Swennen E, et al. Effect of probiotic strains on interleukin 8 production by HT29/19A cells. Am J Gastroenterol. 2002;97:1182–6. doi: 10.1111/j.1572-0241.2002.05693.x. [DOI] [PubMed] [Google Scholar]
- 13.Ma D, Forsythe P, Bienenstock J. Live Lactobacillus reuteri is essential for the inhibitory effect on tumor necrosis factor alpha-induced interleukin-8 expression. Infect Immun. 2004;72:5308–14. doi: 10.1128/IAI.72.9.5308-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niess JH, Brand S, Gu X, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–8. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 15.Kelsall BL, Biron CA, Sharma O, Kaye PM. Dendritic cells at the host–pathogen interface. Nat Immunol. 2002;3:699–702. doi: 10.1038/ni0802-699. [DOI] [PubMed] [Google Scholar]
- 16.Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–7. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 17.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 18.Means TK, Hayashi F, Smith KD, Aderem A, Luster AD. The Toll-like receptor 5 stimulus bacterial flagellin induces maturation and chemokine production in human dendritic cells. J Immunol. 2003;170:5165–75. doi: 10.4049/jimmunol.170.10.5165. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 20.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–5. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 21.Zeng H, Carlson AQ, Guo YYuY, Collier-Hyams LS, Madara JL, Gewirtz AT, Neish AS. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J Immunol. 2003;171:3668–74. doi: 10.4049/jimmunol.171.7.3668. [DOI] [PubMed] [Google Scholar]
- 22.Tallant T, Deb A, Kar N, Lupica J, de Veer MJ, DiDonato JA. Flagellin acting via TLR5 is the major activator of key signaling pathways leading to NF-kappa B and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol. 2004;4:33. doi: 10.1186/1471-2180-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bambou JC, Giraud A, Menard S, et al. In vitro and ex vivo activation of the TLR5 signaling pathway in intestinal epithelial cells by a commensal Escherichia coli strain. J Biol Chem. 2004;279:42984–92. doi: 10.1074/jbc.M405410200. [DOI] [PubMed] [Google Scholar]
- 24.McCarthy J, O'Mahony L, O'Callaghan L, et al. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut. 2003;52:975–80. doi: 10.1136/gut.52.7.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, Tonkonogy SL, Sartor RB. Lactobacillus plantarum 299V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis. 2002;8:71–80. doi: 10.1097/00054725-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 26.Gionchetti P, Rizzello F, Venturi A, et al. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology. 2000;119:305–9. doi: 10.1053/gast.2000.9370. [DOI] [PubMed] [Google Scholar]
- 27.Lammers KM, Vergopoulos A, Babel N, et al. Probiotic therapy in the prevention of pouchitis onset: decreased interleukin-1beta, interleukin-8, and interferon-gamma gene expression. Inflamm Bowel Dis. 2005;11:447–54. doi: 10.1097/01.mpa.0000160302.40931.7b. [DOI] [PubMed] [Google Scholar]
- 28.Bibiloni R, Fedorak RN, Tannock GW, Madsen KL, Gionchetti P, Campieri M, De Simone C, Sartor RB. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol. 2005;100:1539–46. doi: 10.1111/j.1572-0241.2005.41794.x. [DOI] [PubMed] [Google Scholar]
- 29.Correa NB, Peret Filho LA, Penna FJ, Lima FM, Nicoli JR. A randomized formula controlled trial of Bifidobacterium lactis and Streptococcus thermophilus for prevention of antibiotic-associated diarrhea in infants. J Clin Gastroenterol. 2005;39:385–9. doi: 10.1097/01.mcg.0000159217.47419.5b. [DOI] [PubMed] [Google Scholar]
- 30.Wullt M, Hagslatt ML, Odenholt I. Lactobacillus plantarum 299v for the treatment of recurrent Clostridium difficile-associated diarrhoea: a double-blind, placebo-controlled trial. Scand J Infect Dis. 2003;35:365–7. doi: 10.1080/00365540310010985. [DOI] [PubMed] [Google Scholar]
- 31.O'Mahony L, McCarthy J, Kelly P, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–51. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 32.Macpherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365–75. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sartor RB. The influence of normal microbial flora on the development of chronic mucosal inflammation. Res Immunol. 1997;148:567–76. doi: 10.1016/s0923-2494(98)80151-x. [DOI] [PubMed] [Google Scholar]
- 34.Dunne C, O'Mahony L, Murphy L, et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73(Suppl. 2):386S–92S. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 35.Dunne C, Murphy L, Flynn S, et al. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Van Leeuwenhoek. 1999;76:279–92. [PubMed] [Google Scholar]
- 36.Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 2003;52:827–33. doi: 10.1136/gut.52.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haller D, Holt L, Parlesak A, Zanga J, Bauerlein A, Sartor RB, Jobin C. Differential effect of immune cells on non-pathogenic Gram-negative bacteria-induced nuclear factor-kappaB activation and pro-inflammatory gene expression in intestinal epithelial cells. Immunology. 2004;112:310–20. doi: 10.1111/j.1365-2567.2004.01874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckmann L, Smith JR, Housley MP, Dwinell MB, Kagnoff MF. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J Biol Chem. 2000;275:14084–94. doi: 10.1074/jbc.275.19.14084. [DOI] [PubMed] [Google Scholar]
- 39.Zbar AP, Simopoulos C, Karayiannakis AJ. Cadherins: an integral role in inflammatory bowel disease and mucosal restitution. J Gastroenterol. 2004;39:413–21. doi: 10.1007/s00535-004-1335-8. [DOI] [PubMed] [Google Scholar]
- 40.Pratt WS, Crawley S, Hicks J, Ho J, Nash M, Kim YS, Gum JR, Swallow DM. Multiple transcripts of MUC3: evidence for two genes, MUC3A and MUC3B. Biochem Biophys Res Commun. 2000;275:916–23. doi: 10.1006/bbrc.2000.3406. [DOI] [PubMed] [Google Scholar]
- 41.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52:988–97. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed KA, Hobert ME, Kolenda CE, et al. The Salmonella typhimurium flagellar basal body protein FliE is required for flagellin production and to induce a proinflammatory response in epithelial cells. J Biol Chem. 2002;277:13346–53. doi: 10.1074/jbc.M200149200. [DOI] [PubMed] [Google Scholar]
- 43.Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol. 1999;276:G941–50. doi: 10.1152/ajpgi.1999.276.4.G941. [DOI] [PubMed] [Google Scholar]
- 44.Garg S, Bal V, Rath S, George A. Effect of multiple antigenic exposures in the gut on oral tolerance and induction of antibacterial systemic immunity. Infect Immun. 1999;67:5917–24. doi: 10.1128/iai.67.11.5917-5924.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 46.McCracken VJ, Chun T, Baldeon ME, Ahrne S, Molin G, Mackie RI, Gaskins HR. TNF-alpha sensitizes HT-29 colonic epithelial cells to intestinal lactobacilli. Exp Biol Med (Maywood) 2002;227:665–70. doi: 10.1177/153537020222700817. [DOI] [PubMed] [Google Scholar]
- 47.Hart AL, Lammers K, Brigidi P, et al. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut. 2004;53:1602–9. doi: 10.1136/gut.2003.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drakes M, Blanchard T, Czinn S. Bacterial probiotic modulation of dendritic cells. Infect Immun. 2004;72:3299–309. doi: 10.1128/IAI.72.6.3299-3309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haller D, Russo MP, Sartor RB, Jobin C. IKK beta and phosphatidylinositol 3-kinase/Akt participate in non-pathogenic Gram-negative enteric bacteria-induced RelA phosphorylation and NF-kappa B activation in both primary and intestinal epithelial cell lines. J Biol Chem. 2002;277:38168–78. doi: 10.1074/jbc.M205737200. [DOI] [PubMed] [Google Scholar]
- 50.Haller D, Bode C, Hammes WP, Pfeifer AM, Schiffrin EJ, Blum S. Non-pathogenic bacteria elicit a differential cytokine response by intestinal epithelial cell/leucocyte co-cultures. Gut. 2000;47:79–87. doi: 10.1136/gut.47.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 52.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 53.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000;289:1560–3. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 54.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–81. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 55.Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthesy-Theulaz IE. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immun. 2004;72:2160–9. doi: 10.1128/IAI.72.4.2160-2169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wald D, Qin J, Zhao Z, et al. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol. 2003;4:920–7. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- 57.Kelly D, Conway S. Bacterial modulation of mucosal innate immunity. Mol Immunol. 2005;42:895–901. doi: 10.1016/j.molimm.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Grangette C, Nutten S, Palumbo E, et al. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–6. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petrof EO, Kojima K, Ropeleski MJ, Musch MW, Tao Y, De Simone C, Chang EB. Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127:1474–87. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002;277:50959–65. doi: 10.1074/jbc.M207050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cario E, Brown D, McKee M, Lynch-Devaney K, Gerken G, Podolsky DK. Commensal-associated molecular patterns induce selective toll-like receptor-trafficking from apical membrane to cytoplasmic compartments in polarized intestinal epithelium. Am J Pathol. 2002;160:165–73. doi: 10.1016/S0002-9440(10)64360-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattar AF, Teitelbaum DH, Drongowski RA, Yongyi F, Harmon CM, Coran AG. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr Surg Int. 2002;18:586–90. doi: 10.1007/s00383-002-0855-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The 847 cloned cDNAs representing immunerelated genes on the Human Cytokine Expression Arrays used to examine inflammatory gene expression in bacteria-treated HT29 cells.