Abstract
Introducing double-stranded DNA (dsDNA) into the cytoplasm of macrophages and dendritic cells triggers the activation of these professional antigen-presenting cells (APCs). This process is characterized by the up-regulation of costimulatory molecules and the production of various cytokines, chemokines, and antibacterial/viral factors. Current findings indicate that interferon-β (IFN-β) plays a key role in the stimulatory cascade triggered by dsDNA. Both immune and non-immune cells respond to intracytoplasmic dsDNA by up-regulating IFN-β) expression, a process that reduces host susceptibility to infection. The immune activation induced by dsDNA is independent of MyD88, TRIF and DNA-PKcs, indicating that a Toll-like receptor-independent mechanism underlies the cellular activation mediated by intracytoplasmic dsDNA.
Keywords: type 1 IFNs, Toll-like receptor, DNA, host protection
Introduction
Dendritic cells (DCs) and macrophages transfected with dsDNA up-regulate expression of major histocompatibility complex (MHC) class II, CD40, CD54, and produce factors (such as nitric oxide) that reduce host susceptibility to infection.1–4 These effects suggest that self DNA released by injured host cells may stimulate antigen-presenting cells (APCs), improving the clearance of necrotic debris and reducing the risk of opportunistic infection.5–8 Previous studies showed that the recognition of dsDNA was sequence independent and required that the DNA be present in the cytoplasm of target cells.1,2 Thus, immune stimulation was promoted by agents that improved dsDNA uptake (such as liposomal transfection reagents) and inhibited by agents that blocked DNA internalization.9,10 Little is known of the signalling cascade triggered by dsDNA recognition.
DCs and macrophages respond to pathogen associated molecular patterns (PAMPs) – highly conserved molecules expressed by a diverse array of infectious microorganisms.11 This recognition is typically mediated by members of the Toll-like family of receptors (TLR).12 For example, unmethylated CpG motifs in bacterial DNA stimulate an innate immune response by binding to TLR9, while viral nucleic acids are recognized via TLR7 and TLR8.13–15 Upon binding to their cognate PAMP, TLRs interact with adaptor molecules (MyD88 and/or TIR-domain-containing adapter-inducing IFN-β; TRIF), initiating a signalling cascade that culminates in cellular activation.16,17 Of note, APCs are also stimulated by certain self molecules (particularly components of dying host cells), although the receptors involved in that process are unknown.18,19
Type 1 interferons (IFNs; IFN-α/β) play an essential role in antiviral immunity by directly inhibiting viral replication in infected cells. Type 1 IFNs are induced by PAMPs, including dsRNA, ssRNA and CpG DNA.15,20 Thus, viral infection of professional APCs could trigger the secretion of type 1 IFNs through a PAMP/TLR mediated pathway. However, many non-immune cells that lack TLRs also produce type 1 IFNs in response to viral infection, suggesting that an alternative viral recognition pathway may exist.
This work examines the effect of dsDNA on the activation of immune and non-immune cells. Results suggest that type 1 IFNs, most notably IFN-β, are key to the immunomodulatory cascade stimulated by dsDNA. This cascade is TLR, MyD88 and TRIF independent, and is present in both immune and non-immune cells, and confers resistance to herpes simplex virus-2 (HSV-2) infection.
Materials and methods
Animals and cell lines
Female BALB/c C57BL/6 and SCID mice were obtained from the National Cancer Institute (Frederick, MD) and studied at 6–10 weeks of age. TLR9-, MyD88- and TRIF- knockout (KO) mice were kindly provided by Dr S. Akira (Osaka University, Osaka, Japan). All studies were approved by the Center for Biologics Evaluation and Research Animal Care and Use Committee. The RAW 264.7 mouse macrophage cell line, C2C12 mouse muscle cell line, TRAMP mouse prostate carcinoma and VK2/E6E7 human vaginal mucosa were purchased from American Type Culture Collection (Manassas, VA). MCA-106 mouse fibrosarcoma was kindly donated by Dr K. Kawakami. KYSE70 human oesophageal squamous cell carcinoma, HLE human hepatoma and Panc-1 human pancreatic adenocarcinoma were kindly donated by Dr T Shimamura.
Cell preparation
Bone marrow-derived DCs (BMDCs) were prepared from BALB/c mice as previously described.21 Briefly, 2 × 106 bone marrow cells obtained from the femur were seeded into a 100 mm Petri dish in 10 ml of RPMI supplemented with 10% fetal calf serum (FCS) and 20 ng/ml granulocyte–macrophage colony-stimulating factor (BD PharMingen, San Diego, CA). Medium was replaced on day 3, and cells harvested on day 7 by treatment with 3 mm ethylenediaminetetra-acetic acid (EDTA) for 5 min.
Peritoneal cells were isolated from mice injected i.p. with thioglycolate. These cells were cultured in RPMI plus 5% FCS for 4 hr, non-adherent cells removed, and peritoneal macrophages in the adherent monolayer used for further study.22
Murine embryonic fibroblasts (MEFs) from BALB/c mice were described previously.17 MEFs were cultured in RPMI supplemented with 10% FCS.
DNA preparation and antibodies
Calf thymus (CT) and Escherichia coli (EC) DNA were purchased from Sigma Chemical Co. (St. Louis, MO). HSV DNA was prepared from virions isolated from Vero cells infected with HSV-2 strain 333. The cells were lysed by freeze—thawing three times and then resuspended in lysis buffer (0·5% NP-40, 30 mm Tris [pH 7·4], 120 mm KCl, 5 mm Mg acetate, 3·6 mm CaCl2, 0·5 mm EDTA) containing 6 mmβ-mercaptoethanol and 0·5% deoxycholic acid (DOC). Contaminating cellular nucleic acids were removed by digestion for 30 min at 37° with DNase I and RNase A. Virions in the solution were pelleted on a 5–40% glycerol gradient by centrifugation for 1 h at 108 000 g. The virus pellet was resuspended in STEP buffer (1% sodium dodecyl sulphate, 0·5 m Tris [pH 7·4], 10 mm EDTA and proteinase K). HSV-2 virion DNA was extracted twice with an equal volume of phenol—chloroform (1 : 1) equilibrated with Tris-HCl (pH 7·5) and twice with chloroform—isoamyl alcohol (24 : 1).
Synthetic polynucleotides (PN) were purchased from Amersham Pharmacia (Piscataway, NJ). All DNA preparations were purified by repeat phenol-chloroform extractions, and treated with Triton-X-114 to remove endotoxin as previously described.1 Less than 0·01 U/µg endotoxin or protein was present in any of the DNA preparations as determined by a Limulus amoebocyte lysate assay (BioWhittaker, Walkersville, NJ). ssDNA was prepared by boiling dsDNA for 5 min and then plunging into an ice bath. Poly(I:C) was purchased from Invivogen (San Diego, CA). Murine anti-interleukin-6 (IL-6; clone MP5-20F3) and anti-tumour necrosis factor-α (TNF-α; clone TN3-19,12) Abs were purchased from BD Pharmingen (San Diego, CA). Anti-IFN-β antibody was purchased from Yamasa corporation (Tokyo, Japan).
Oligodeoxynucleotides (ODN)
Endotoxin-free phosphorothioate ODN were synthesized at the CBER core facility. The following ODNs were used: murine CpG ODN1555 (GCTAGACGTTAGCGT); Human CpG ODNK3 (ATCGACTCTCGAGCGTTCTC).
Transfection
DNA cytofectin complexes were prepared according to the manufacturers' instructions. Briefly, DNA was mixed 1 : 1 with the Fugene6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN) in 1 ml of serum-free OptiMEM (Life Technologies, Gaithersberg, MD) for 15 min at room temperature and then added to cells.
Fluorescence-activated cell sorting (FACS)
Cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 5 min at 37°, and stained with phycoerythrin (PE)-labelled anti-CD40 antibody (BD PharMingen) for 30 min at room temperature. Stained cells were washed, re-suspended in PBS/0·1% bovine serum albumin (BSA) plus azide, and analysed by FACSort (BD Biosciences, San Jose, CA).
Nitrite assay
NO levels in culture supernatants were assessed using the Griess reagent (Sigma-Aldrich). Nitrite concentration was calculated by comparison to a standard curve generated by sequentially diluting sodium nitrite.
Reverse transcription—polymerase chain reaction (RT—PCR)
Total RNA was extracted from target cells using TRIzol reagent (Life Technologies Inc., Carlsbad, CA) as recommended by the manufacturer. 1 µg of total RNA was reverse-transcribed in first strand buffer (50 mm Tris-HCl, pH 7·5, 75 mm KCl, and 2·5 mm MgCl2), containing 25 µg/ml oligo-(dT)12–18, 200 U Moloney leukaemia virus reverse-transcriptase, 2 mm dinucleotide triphosphate, and 10 mm dithiothreitol. The reaction was conducted at 42° for 1 hr. A standard PCR was performed on 1 µl of the cDNA synthesis using the following primer pairs in Table 1. Aliquots of the PCR reactions were separated on a 1·5% agarose gel and visualized with UV light after ethidium bromide staining. Images were processed using NIH image for densitometric analysis.
Table 1. PCR primers used in this study.
| Gene | Primers (sense/antisense) |
|---|---|
| Mouse | |
| mIL-6 | S: GACAAAGCCAGAGTCCTTCAGAGAG |
| AS: CTAGGTTTGCCGAGTAGATCTC | |
| mIP-10 | S: ACCATGAACCCAAGTGCTGCCGTC |
| AS: GCTTCACTCCAGTTAAGGAGCCCT | |
| mTNF-α | S: ATGAGCACAGAAAGCATGATC |
| AS: TACAGGCTTGTCACTCGAATT | |
| mIFN-β | S: CCACAGCCCTCTCCATCAACTATAAGC |
| AS: AGCTCTTCAACTGGAGAGCAGTTGAGG | |
| mβ-actin | S: GACATGGAGAAGATCTGGCAACCACA |
| AS: ATCTCCTGCTCGAAGTCTAGAGCAA | |
| mMCP-1 | S: ATGCAGGTCCCTGTCATGCTTCTGG |
| AS: CTAGTTCACTGTCACACTGGTCACTCC | |
| mMCP-6 | S: GCACATCAAAAGCCCACAGC |
| AS: TAGACAGGGGAGACAGAGGAC | |
| mlMP2 | S: GTTCCGGACGGAAGAAGTCC |
| AS: GCAGCTCATCTCCCAGGATG | |
| mlMP7 | S: GCTCCGGAGCTCGCACTTC |
| AS: TAGTTGTCTCTGTGGGTAG | |
| mTAP1 | S: GTCCAGATGCCTTCGCTATCAG |
| AS: GTTGGCTGTGTCCTCAGTCAC | |
| mPKR | S: TCCTCTGCCGTGGTTTTCCTTT |
| AS: ACAGGAGCCTGCCTTTCTCTTT | |
| m2′5′ OAS | S: TGGCTGAAGAGGCTGATGTGTG |
| AS: TGAGGAAGGCTGGCTGTGATTG | |
| Human | |
| hIFN-β | S: GAACTTTGACATCCCTGAGGAGATTAAGCAGC |
| AS: GTTCCTTAGGATTTCCACTCTGACTATGGTCC | |
| hGAPDH | S: ACCACCATGGAGAAGGCTGG |
| AS: CTCAGTGTAGCCCAGGATGC | |
TaqMan RT—PCR
IFN-β and reduced glyceraldehyde phosphate dehydrogenase (GAPDH) mRNA levels were quantified by TaqMan PCR. Primers and probes were used at a final concentration of 300 nm and 200 nm, respectively, according to the manufacturer's recommendation. All PCR reactions were performed in duplicate using the TaqMan Universal PCR Master Mix (Applied Biosystems). Amplifications, detection, and analysis were performed in an ABI PRISM 7700 system (Applied Biosystems). Relative mRNA levels were determined using the comparative cycle threshold (CT) method. This calculated the amount of target normalized to an endogenous reference (GAPDH) as described in the Perkin-Elmer Applied Biosystems user bulletin. mRNA levels in DNA transfected cells were compared to those in non-stimulated controls. Real-time PCR was performed with the following primers: ATGAGTGGTGGTTGCAGGC (mIFN-β-f), TGACCTTTCAAATGCAGTAGATTCA (mIFN-β-r). TaqMan probes consisted of an oligonucleotide labelled at its 5′ end with the reporter dye 6-carboxyfluorescein (FAM) and at the 3′ end with the quencher dye 6-carboxytetramethylrhodamine (TAMRA). Probe sequences were as follows: FAM-AAGCATCAGAGGCGGACTCTGGGA-TAMRA (mIFN-β). Primer-probe sets for the endogenous controls rodent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were obtained from Applied Biosystems and used according to the manufacturer's instructions.
Standard viral plaque assay
MEFs were placed into six-well plates and incubated at 37° for 1 day. Cells were pretreated by transfection with CT DNA and/or incubation with neutralizing antibody or CpG DNA. After 16 hr, HSV-2 strain 333 was prepared in MEM without FCS and diluted to generate 50–60 plaques/well. The medium was aspirated, and 0·2 ml of virus was added to each well in triplicate for 1 hr, aspirated, and replaced with 2 ml of minimal essential medium containing 10% FCS. Two days later the cells were fixed in formalin and stained with 0·8% crystal violet (Sigma). Plaque counts were performed in triplicate.
Statistical analysis
To facilitate analysis of data from multiple repetitions of individual experiments, results were standardized by calculating the fold change in test versus control groups in each experiment. Student's t-test was then used to evaluate whether changes between groups were statistically significant.
Results
Mammalian and synthetic dsDNA up-regulate cytokine mRNA expression by peritoneal macrophages
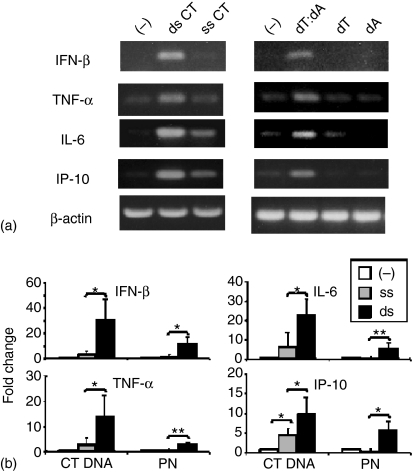
Previous studies examining the immunomodulatory properties of ds mammalian DNA showed that RAW 264.7 cells transfected with calf thymus (CT) DNA in Fugene6 were stimulated to secrete TNF-α.9 To confirm and extend those observations, murine peritoneal macrophages were transfected with ss or ds DNA. Whereas cells cultured with Fugene6 transfection reagent alone were unaffected, cells transfected with Fugene6 plus dsCT DNA significantly up-regulated IFN-β, TNF-α, IL-6, IL-10 and inducible protein-10 (IP-10) mRNA expression (Fig. 1a, b). DsDNA had no effect on IFN-β, IFN-α or IL-12 mRNA levels (data not shown). Transfection with ssCT DNA had a detectable but significantly smaller effect than dsDNA. This ssDNA was produced by heat denaturing dsDNA, raising the possibility that a small amount of dsDNA might remain. Thus, ds and ss polynucleotides were synthesized. Whereas the synthetic ds polynucleotides induced the same expected pattern of gene activation observed with dsDNA, the ss polynucleotides had no effect on cytokine mRNA levels (Fig. 1a, b). If dsDNA is digested by DNAse, these effects were not observed (data not shown).
Figure 1.
Mammalian and synthetic dsDNA up-regulate cytokine gene expression by murine macrophages. (a) Peritoneal macrophages were transfected with Fugene6 alone (–) or combined with 3 µg/ml of CT DNA or synthetic polynucleotides. mRNA expression was monitored by RT—PCR 3 hr later. PCR conditions for each cytokine were established such that a faint band was generated using control mRNA. (b) Quantitative analysis of RT—PCR data from three independent experiments performed as described in (a). mRNA levels were quantitated by analysing PCR image intensity using ‘NIH image’ software, and the data normalized to the endogenous β-actin reference. Values represent the mean ± SD increase in mRNA levels when compared to cells treated with Fugene6 alone. *P < 0·05; **P < 0·001 (compared with Fugene6 alone).
Contribution of IFN-β to the immune activation elicited by dsDNA
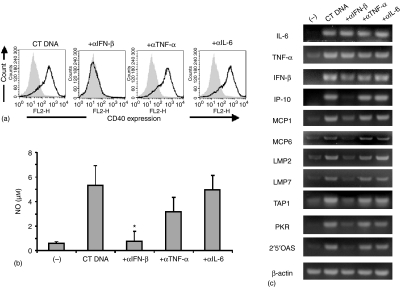
Time course studies showed that cells stimulated with dsDNA initially up-regulated their production of cytokines, and subsequently increased their expression of costimulatory molecules (such as CD40) and secreted antimicrobial factors (such as NO).1,13 To determine whether the early cytokine production contributed to subsequent cellular activation, cells incubated with dsDNA in Fugene6 were cocultured with neutralizing anticytokine antobodies. Anti-IFN-β antibody completely blocked the up-regulation of CD40 expression by RAW 264.7 cells, reduced by > 95% NO production by peritoneal macrophages (Fig. 2a, b) and blocked changes in gene expression involving chemokines IP-10, monocyte chemoattractant protein (MCP)1/6, antigen processing/presentation transporters associated with antigen processing 1 (TAP1), low molecular weight proteins 2/7 (LMP2/7) and antiviral pathways (2′,5′-oligoadenylate synthetases (OAS), RNA-activated protein kinase (PKR)) (Fig. 2c). Anti-IFN-β antobody did not alter the expression of IL-6, TNF-α or IFN-β mRNAs, however (Fig. 2c). In contrast, antibodies against TNF-α, IL-6, IL-10, IFN-α and IFN-γ had no significant impact on the activation cascade triggered by transfected dsDNA (Fig. 2).
Figure 2.
Role of IFN-β in dsDNA-induced cell activation. (a) RAW 264.7 cells were untreated (grey shadow) or transfected with 3 µg/ml dsCT DNA in Fugene6 (dark line). Cells were then incubated with 2·5 µg/ml of cytokine-neutralizing antibody for 16 hr, and CD40 expression analysed by FACS. Experiments were repeated three times with similar results. (b) Peritoneal macrophages were transfected with 3 µg/ml dsCT DNA in Fugene6. Cells were then incubated with 2·5 µg/ml of cytokine-neutralizing antibody for 24 hr, and NO levels in culture supernatants determined. (c) Peritoneal macrophages were transfected with 3 µg/ml dsCT DNA in Fugene6. Cells were then incubated with 2·5 µg/ml of cytokine-neutralizing antibody for 3 hr and mRNA expression monitored by RT—PCR. Data represent the mean ± SD response of three independent populations of macrophages, and were confirmed in two additional experiments. *P < 0·05 (compared with CT DNA transfected cells alone).
Kinetics and dose—response of IFN-β gene expression following dsDNA stimulation
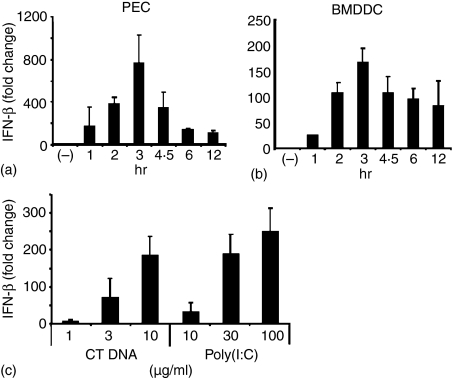
Given the apparent importance of IFN-β in mediating the down-stream effects of dsDNA activation, the kinetics and dose—response of this cytokine response were further examined. As seen in Fig. 3, dsDNA stimulated both macrophages and DC to up-regulate IFN-β mRNA expression by >100-fold. In both cell types, IFN-β mRNA levels peaked 3 hr after transfection, and persisted at elevated levels for at least 12 hr. In contrast, Fugene6 alone had no significant effect on IFN-β expression at any time. The magnitude of IFN-β induction by dsDNA with Fugene6 was compared to that induced by poly(I : C). As seen in Fig. 3, the level of cytokine production induced by 10 µg of dsDNA rivaled that elicited by 100 µg of poly(I : C).
Figure 3.
Effect of dsDNA on IFN-β mRNA expression by macrophages and dendritic cells. Peritoneal macrophages (a) and BMDCs (b) were transfected with 10 µg/ml dsCT DNA in Fugene6 for 1–12 hr. BMDCs (c) were stimulated with the indicated dose of dsCT DNA plus Fugene6 or poly(I:C) for 3 hr. IFN-β mRNA levels were determined by TaqMan PCR. Results represent the mean fold increase ± SD of mRNA levels from three independent experiments when compared to unstimulated cells cultured for the same period.
Non-mammalian dsDNA also up-regulates IFN-β expression
Previous studies examined the capacity of ds mammalian DNA to stimulate APCs.1 To determine whether DNA from other sources, including bacteria and viruses, might have the same effect, single and double stranded DNA from E. coli and HSV were evaluated. Of concern, the genomes of HSV and E coli contain a high frequency of immunostimulatory CpG motifs, that interact with TLR9 to stimulate an innate immune response characterized by the production of multiple cytokines, including IFN-β.20,23–26
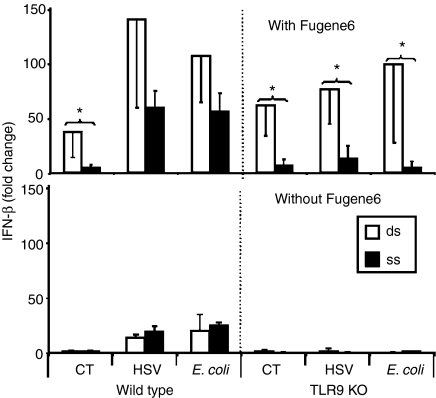
Two experiments were performed to examine the relative contribution of CpG- versus dsDNA-induced immune activation by DNA from E. coli and HSV. First, DNA from each source was rendered single stranded by heat denaturation. As expected, cells treated with heat-denatured CT DNA did not up-regulate IFN-β mRNA production, since ss mammalian DNA is non-stimulatory (Fig. 4). By comparison, heat denaturation of bacterial and viral DNA reduced IFN-β mRNA levels by approximately half (Fig. 4). Because CpG motifs are fully stimulatory in ss form24,25 this partial loss of activity suggests that E. coli and HSV DNA may stimulate through both CpG and dsDNA dependent mechanisms.
Figure 4.
Effect of DNA transfection on IFN-β mRNA expression. Peritoneal macrophages from from C57Bl/6 or TLR9 KO mice were transfected with 3 µg/ml of ds or ss DNA in the presence or absence of Fugene6. IFN-β mRNA levels were measured 3 hr later by quantitative RT—PCR. Results represent the mean fold increase ± SD in IFN-β mRNA levels compared to cells transfected with Fugene6 alone from three independent experiments. *P < 0·05 (compared with ssDNA group).
To examine this hypothesis, a second experiment was performed to evaluate the ability of ss and ds DNA to stimulate cells from TLR9 KO mice (which do not respond to CpG motifs). When the contribution of CpG motifs was eliminated, dsDNA from CT, E. coli and HSV induced similar levels of IFN-β mRNA production (Fig. 4). Stimulation of cells from TLR9 KO mice required that the DNA gain entry to the cytoplasm (no stimulation was seen in the absence of Fugene6), and that it be in double-stranded form (no stimulation was seen after heat denaturation).
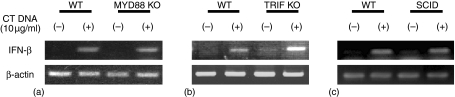
Neither MyD88, TRIF nor DNA-protein kinase C (PKC) are involved in the recognition of dsDNA
The above studies suggest that: (i) the immunostimulatory capacity of dsDNA from different organisms is equivalent; and (ii) dsDNA can trigger APCs in a TLR9-independent manner. To determine whether other TLR family members contribute to the recognition of dsDNA, APCs from MyD88 and TRIF KO mice were studied (because MyD88 and TRIF are critical elements of the signalling cascade elicited by all TLRs1). As seen in Fig. 5(a, b), cells from wild type and KO strains responded similarly to stimulation by dsCT DNA.
Figure 5.
Induction of IFN-β by dsDNA does not involve MyD88, TRIF or DNA—PKCs. BMDDC from WT, MyD88 KO, TRIF KO and SCID mice were transfected with 10 µg/ml dsCT DNA in Fugene6. IFN-β mRNA levels were determined 3 hr later by RT—PCR. Experiments were repeated three times with similar results.
Previous studies suggested that DNA-PKC might be involved in DNA mediated immune activation.27 Yet cells from severe combined immunodeficient (SCID) mice (defective in DNA-PKC function) responded normally to stimulation by dsCT DNA (Fig. 5c). The production of other cytokines elicited by dsDNA (including TNF-α, IL-6 and IL-10) was similarly up-regulated in cells from normal and KO donors (data not shown). These findings indicate that dsDNA recognition is not mediated by any member of the TLR family or by DNA-PKC.
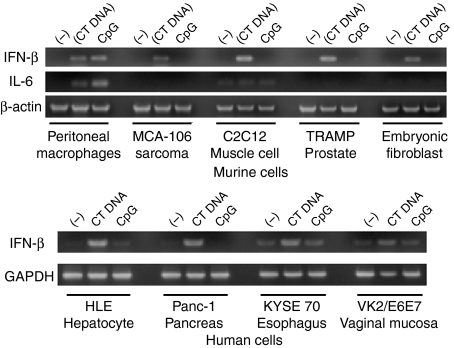
Non-immune cells can respond to dsDNA by producing IFN-β
If TLRs do not contribute to the recognition of dsDNA, cell types lacking those receptors might be responsive to intracytoplasmic dsDNA. To investigate this possibility, several murine cell lines that do not express TLR9 (including C2C12 muscle cells, embryonic fibroblasts, TRAMP prostate cells and MCA-106 sarcoma cells) were transfected with dsDNA in Fugene6. As seen in Fig. 6, dsCT DNA induced these non-immune lines to express IFN-β and various IFN-inducible genes, but not IL-6 or TNF-α (Fig. 6). These results were duplicated in studies of human cancer cell lines (VK2/E6E7 vaginal mucosa, KYSE70 oesophageal squamous cell carcinoma, HLE hepatoma and Panc-1 pancreatic adenocarcinoma cells).
Figure 6.
Stimulatory activity of dsDNA in murine and human cell lines. Non-immune cells of murine and human origin were transfected with 3 µg/ml of dsCT DNA in Fugene6 or 3 µm CpG DNA. IFN-β and IL-6 mRNA levels were determined 3 hr later by RT—PCR. Experiments were repeated twice with similar results.
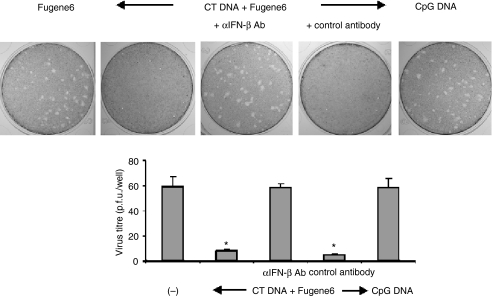
Murine embryonic fibroblasts transfected with dsDNA resist HSV-2 infection through an IFN-β-mediated process
To examine the ability of dsDNA to improve the resistance of non-immune cells to infection, MEFs were transfected with CT DNA for 16 hr prior to HSV-2 challenge. Whereas neither CpG DNA nor Fugene6 protected against infection, viral titres were reduced >85% in cells treated with CT DNA (P < 0·001). This antiviral effect was mediated by IFN-β, because antibodies against that cytokine abrogated dsDNA induced protection.
Discussion
Introducing dsDNA into the cytoplasm of immune and non-immune cells stimulates the production of cytokines, chemokines, and/or factors that reduce host susceptibility to infection.1–4 This work demonstrates that IFN-β plays a unique and critical role in this immunoprotective cascade (Figs 1 and 2). Virtually all aspects of dsDNA-dependent cellular activation, including the up-regulation of chemokines, of antigen processing/presentation molecules, and antiviral molecules, is abrogated by treatment with neutralizing anti-IFN-β antibodies (Fig. 2). Moreover, the increased resistance to viral infection induced by dsDNA is blocked by treatment with anti-IFN-β antibodies (Fig. 7).
Figure 7.
DsDNA reduces susceptibility to HSV-2 infection. Monolayers of MEFs were transfected with 1 µg/ml of dsCT DNA or 3 µm of CpG DNA. Cells were then incubated with 10 µg/ml of IFN-β-neutralizing or control antibody for 16 hr and challenged with HSV-2. Plaques were visualized 2 days after infection by staining with crystal violet. All studies were performed in triplicate, with the histogram showing the mean ± SD virus titre/treatment group. Experiments were repeated twice with similar results. *P < 0·001 (compared with Fugene6 treatment group).
DsDNA triggers a large (>100-fold) and rapid (<1 hr) increase in IFN-β mRNA expression (Fig. 3). This is consistent with findings from Yoshida et al. showing that the accumulation of dsDNA in the cytoplasm of DNAse II deficient macrophages (because of a defect in DNA degradation) culminates in the hypersecretion of IFN-β.28 Previous studies established the importance of IFN-β (and other type 1 IFNs) to APC function. They facilitate antigen processing/presentation by inducing the expression of TAP, LMP and MHC class I/II molecules.29 They also promote the differentiation of T helper 1 cells and contribute to immune protection by directly inhibiting viral replication in infected cells.30,31 Thus, the immunomodulatory activity of dsDNA is consistent with the physiological properties of IFN-β.
Early research in this field showed that APCs were activated when mammalian dsDNA was introduced into their cytoplasm.1 In the current study, this was accomplished experimentally with the aid of Fugene6, but could occur physiologically following the uptake of apoptotic/necrotic debris or chromatin–immunoglobulin complexes.18,19,32,33 Moreover, other agents that induce DNA internalization, including lipofectamine, effectene and LyoVec, were equally effective at promoting the immune response induced by dsDNA. We postulated that other sources of dsDNA might also be immunostimulatory, and found that dsDNA of bacterial and viral origin mimicked the immunostimulatory activity of self DNA (Fig. 4). These findings suggest that intracytoplasmic dsDNA of any form provides a ‘danger signal’ to the host. Consistent with that conclusion, the magnitude of the IFN-β response triggered by dsDNA rivalled that elicited by the immunoprotective CpG motifs expressed by bacterial DNA (Fig. 4).
The immunostimulatory activity of dsDNA and CpG DNA were additive, indicating that these two forms of activation utilize discrete signalling pathways. Supporting such a conclusion, APC from TLR9, MyD88 and TRIF KO mice responded to stimulation by dsDNA, indicating that TLR were not involved in the recognition of dsDNA. This finding is not entirely consistent with that of Yasuda et al. who reported that dsDNA mediated activation of APCs included both a TLR9-dependent and -independent component.34 Studies of cells from KO mice indicate that the immune activation induced by dsDNA is entirely TLR independent (Fig. 5 and data not shown).
Current results further establish that TLR-negative non-immune cells of both murine and human origin recognize intracytoplasmic dsDNA and respond by up-regulating expression of IFN-β and IFN-inducible genes. Consistent with this conclusion, Li et al. found that an epithelial tumour cell line transfected with dsDNA bacterial DNA (in the form of a plasmid DNA vaccine) up-regulated IFN-β and IFN-inducible gene expression.35 Thus, intracytoplasmic dsDNA may trigger the production of factors that reduce the susceptibility of even non-immune cells to intracellular infection. Indeed, murine embryonic fibroblasts treated with dsDNA were more resistant to infection by HSV (Fig. 7). Additional study is needed to identify the receptor(s) involved in the recognition of dsDNA and to clarify the nature of the signalling cascade responsible for such protection.
Acknowledgments
The authors thank Drs Akira Shizuo, Hiroaki Hemmi and Masahiro Yamamoto for kindly providing TLR9, MyD88 and TRIF KO mice and Dr Philip Krause and Amita Patel for kindly providing HSV DNA for these studies. The assertions herein are the private ones of the authors and are not to be construed as official or as reflecting the views of USAMRIID or the Food and Drug Administration at large.
Abbreviations
- ds
double-stranded
- ODN
oligodeoxynucleotide
- TRIF
TIR-domain-containing adapter-inducing IFN-β
- DC
dendritic cell
- bone-marrow-derived DCs
BMDCs
- CT
calf thymus
- PN
polynucleotides
- MEFs
murine embryonic fibroblasts
References
- 1.Ishii KJ, Suzuki K, Coban C, Takeshita F, Itoh Y, Matoba H, Kohn LD, Klinman DM. Genomic DNA released by dying cells induces the maturation of APCs. J Immunol. 2001;167:2602–7. doi: 10.4049/jimmunol.167.5.2602. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, Mori A, Ishii KJ, Saito J, Singer DS, Klinman DM, Krause PR, Kohn LD. Activation of target-tissue immune-recognition molecules by double-stranded polynucleotides. Proc Natl Acad Sci USA. 1999;96:2285–90. doi: 10.1073/pnas.96.5.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu FG, Reich CF, Pisetsky DS. Effect of cytofectins on the immune response of murine macrophages to mammalian DNA. Immunology. 2003;109:255–62. doi: 10.1046/j.1365-2567.2003.01653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang W, Reich CF, III, Pisetsky DS. Mechanisms of activation of the RAW264.7 macrophage cell line by transfected mammalian DNA. Cell Immunol. 2004;229:31–40. doi: 10.1016/j.cellimm.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Ibrahim MAA, Chain BM, Katz DR. The injured cell: the role of the dendritic cell system as a sentinel receptor pathway. Immunol Today. 1995;16:181–6. doi: 10.1016/0167-5699(95)80118-9. [DOI] [PubMed] [Google Scholar]
- 6.Galluci S, Matzinger P. Danger signals: SOS to the immune system. Curr Opin Immunol. 2001;13:114–9. doi: 10.1016/s0952-7915(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Zheng W, Rock KL. Cell injury releases endogenous adjuvant that stimulate cytotoxic T cell responses. Proc Natl Acad Sci USA. 2001;97:14590–5. doi: 10.1073/pnas.260497597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu S, Binder RJ, Suto R, Anderson KM, Srivastava PK. Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-κB pathway. Int Immunol. 2001;12:1539–46. doi: 10.1093/intimm/12.11.1539. [DOI] [PubMed] [Google Scholar]
- 9.Yasuda K, Ogawa Y, Kishimoto M, Takagi T, Hashida M, Takakura Y. Plasmid DNA activates murine macrophages to induce inflammatory cytokines in a CpG motif-independent manner by complex formation with cationic liposomes. Biochem Biophys Res Commun. 2002;293:344–8. doi: 10.1016/S0006-291X(02)00210-3. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda K, Ogawa Y, Yamane I, Nishikawa M, Takakura Y. Macrophage activation by a DNA/cationic liposome complex requires endosomal acidification and TLR9-dependent and -independent pathways. J Leukoc Biol. 2005;77:71–9. doi: 10.1189/jlb.0204089. [DOI] [PubMed] [Google Scholar]
- 11.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 12.Kaisho T, Akira S. Regulation of dendritic cell function through toll-like receptors. Curr Mol Med. 2003;3:759–71. doi: 10.2174/1566524033479366. [DOI] [PubMed] [Google Scholar]
- 13.Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 14.Heil F, Hemmi H, Hochrein H, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–9. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 15.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–31. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 16.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–22. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto M, Sato S, Hemmi H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signalling pathway. Science. 2003;301:640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 18.Gallucci S, Lolkema M, Matzinger P. Natural adjuvants: endogenous activators of dendritic cells. Nat Med. 1999;11:1249–55. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- 19.Sauter B, Albert ML, Francisco L, Larsson M, Somersan S, Bhardwaj N. Consequences of cell death. Exposure to necrotic tumor cells, but not primary tissue cells or apoptotic cells, induces the maturation of immunostimulatory dendritic cells. J Exp Med. 2000;191:423–34. doi: 10.1084/jem.191.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoshino K, Kaisho T, Iwabe T, Takeuchi O, Akira S. Differential involvement of IFN-beta in Toll-like receptor-stimulated dendritic cell activation. Int Immunol. 2002;14:1225–31. doi: 10.1093/intimm/dxf089. [DOI] [PubMed] [Google Scholar]
- 21.Shirota H, Sano K, Hirasawa N, Terui T, Ohuchi K, Hattori T, Shirato K, Tamura G. Novel roles of CpG oligodeoxynucleotides as a leader for the sampling and presentation of CpG-tagged antigen by dendritic cells. J Immunol. 2001;167:66–74. doi: 10.4049/jimmunol.167.1.66. [DOI] [PubMed] [Google Scholar]
- 22.Shirota H, Gursel I, Gursel M, Klinman DM. Suppressive oligodeoxynucleotides protect mice from lethal endotoxic shock. J Immunol. 2005;174:4579–83. doi: 10.4049/jimmunol.174.8.4579. [DOI] [PubMed] [Google Scholar]
- 23.Klinman DM, Yi A, Beaucage SL, Conover J, Krieg AM. CpG motifs expressed by bacterial DNA rapidly induce lymphocytes to secrete IL-6, IL-12 and IFNγ. Proc Natl Acad Sci USA. 1996;93:2879–83. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg AM, Yi A, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–8. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 25.Lundberg P, Welander P, Han X, Cantin E. Herpes simplex virus type 1 DNA is immunostimulatory in vitro and in vivo. J Virol. 2003;77:11158–69. doi: 10.1128/JVI.77.20.11158-11169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemmi H, Kaisho T, Takeda K, Akira S. The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J Immunol. 2003;170:3059–64. doi: 10.4049/jimmunol.170.6.3059. [DOI] [PubMed] [Google Scholar]
- 27.Dragoi AM, Fu X, Ivanov S, Zhang P, Sheng L, Wu D, Li GC, Chu WM. DNA-PKC, but not TLR9, is required for activation of Akt by CpG-DNA. EMBO J. 2005;24:779–89. doi: 10.1038/sj.emboj.7600539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- 29.Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–87. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 30.Brinkmann V, Geiger T, Alkan S, Heusser CH. Interferon alpha increases the frequency of interferon gamma producing human CD4+ cells. J Exp Med. 1993;1778:1655–63. doi: 10.1084/jem.178.5.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parronchi P, DeCarli M, Manetti R, et al. IL-4 and IFN (alpha and gamma) exert oppostie regulatory effects on the development of cytolytic potential by TH1 or TH2 human T cell clones. J Immunol. 1992;149:2977–83. [PubMed] [Google Scholar]
- 32.Decker P, Singh-Jasuja H, Haager S, Kotter I, Rammensee HG. Nucleosome, the main autoantigen in systemic lupus erythematosus, induces direct dendritic cell activation via a MyD88-independent pathway: consequences on inflammation. J Immunol. 2005;174:3326–34. doi: 10.4049/jimmunol.174.6.3326. [DOI] [PubMed] [Google Scholar]
- 33.Boule MW, Broughton C, Mackay F, Akira S, Marshak-Rothstein A, Rifkin IR. Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J Exp Med. 2004;199:1631–40. doi: 10.1084/jem.20031942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasuda K, Yu P, Kirschning CJ, et al. Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J Immunol. 2005;174:6129–36. doi: 10.4049/jimmunol.174.10.6129. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Wilkinson M, Xia X, David M, Xu L, Purkel-Sutton A, Bhardwaj A. Induction of IFN-regulated factors and antitumoral surveillance by transfected placebo plasmid DNA. Mol Ther. 2005;11:112–9. doi: 10.1016/j.ymthe.2004.09.008. [DOI] [PubMed] [Google Scholar]