Abstract
Interleukin (IL)-5 induces CD38-activated splenic B cells to differentiate into immunoglobulin M-secreting cells and undergo µ to γ1 class switch recombination (CSR) at the DNA level, resulting in immunoglobulin G1 (IgG1) production. Interestingly, IL-4, a well-known IgG1-inducing factor does not induce immunoglobulin production or µ to γ1 CSR in CD38-activated B cells. In the present study, we implemented complementary DNA microarrays to investigate the contribution of IL-5-induced gene expression in CD38-stimulated B cells to immunoglobulin-secreting cell differentiation and µ to γ1 CSR. IL-5 and IL-4 stimulation of CD38-activated B cells induced the expression of 418 and 289 genes, respectively, that consisted of several clusters. Surprisingly, IL-5-inducible 78 genes were redundantly regulated by IL-4. IL-5 and IL-4 also suppressed the gene expression of 319 and 325 genes, respectively, 97 of which were overlapped. Genes critically regulated by IL-5 include immunoglobulin-related genes such as J chain and immunoglobulinκ, and genes involved in B-cell maturation such as BCL6, activation-induced cytidine deaminase (Aid) and B lymphocyte-induced maturation protein-1 (Blimp-1) and tend to be induced slowly after IL-5 stimulation. Intriguingly, among genes, the retroviral induction of Blimp-1 and Aid in CD38-activated B cells could induce IL-4-dependent maturation to Syndecan-1+ antibody-secreting cells and µ to γ1 CSR, respectively, in CD38-activated B cells. Taken together, preferential Aid and Blimp-1 expression plays a critical role in IL-5-induced immunoglobulin-secreting cell differentiation and µ to γ1 CSR in CD38-activated B cells.
Keywords: B cells, cytokine receptors/interleukin receptors/chemokine receptors, cytokines/interleukins, isotypes/isotype switching
Introduction
Interleukin (IL)-5 was originally identified by its activity as a B-cell growth and differentiation factor,1 an immunoglobulin A (IgA)-enhancing factor2,3 and a differentiation factor for eosinophil lineage.4 IL-5, mainly produced by activated T helper (Th) 2 cells and mast cells and, to a certain extent, by eosinophils and non-haematopoietic cells5–7, acts on various target cells by binding to IL-5 receptor (R) and induces the cell survival and differentiation of B cells, eosinophils and basophils, and activated T cells.8–12 Although neither IL-5Rα nor βc have any kinase activity, IL-5 activates a number of cytoplasmic kinases, including Src family kinase, Fyn13,14 and Lyn,15 Bruton's tyrosine kinase,16,17 and Jak family kinase, Jak1 and Jak2.18,19 The activation of intracellular signalling molecules modifies the expression of genes in the target cells, leading to their proliferation, survival, and differentiation.
The terminal differentiation of activated B cells to immunoglobulin-secreting cells is a crucial step in humoral immunity.20 Immunoglobulin class switch recombination (CSR) in B cells also provides functional diversity of the humoral immune response.21,22 CSR results in the replacement of the Cµ heavy chain constant region (CH) of the immunoglobulin gene with other CH sequences. In addition to the transcriptional activation of germline CH sequence, CSR between Sµ and another S region 5′ to a CH sequence is mediated by a DNA recombination event that moves the VDJ-segments to a new position upstream of the isotype being expressed.21,23 The deleted DNA forms circular structures termed ‘switch circles’ that may contain reciprocal recombination products consisting of the 3′ section of an S region joined to the 5′ section of the S region of the new isotype.24,25 While the molecular mechanisms of CSR are still obscure, the involvement of activation-induced cytidine deaminase (Aid), a potential RNA editing enzyme, has been shown in the regulation or catalysis of the DNA modification step of CSR.26 The switch of the immunoglobulin isotype from IgM to IgG, IgE or IgA, is highly regulated by cytokines such as transforming growth factor (TGF)-β, IL-4 and interferon-γ (IFN-γ), B cell activators such as CD40 ligand and lipopolysaccaride (LPS), or both.23 IL-4 is a prominent cytokine for CSR to IgG1 and IgE in B cells stimulated with CD40 or with LPS.23 Like IL-4, IL-5 is able to elicit the maturation of CD40-activated B cells to IgM- and IgG1-secreting cells and induce µ to γ1 CSR and IgG1 production in LPS-activated B cells (unpublished observation).27
CD38 is a type II transmembrane glycoprotein that is widely expressed in both haematopoietic and non-haematopoietic lineage cells.28 We previously reported that the binding of an anti-mouse CD38 monoclonal antibody (mAb, clone CS/2) to splenic B-2 cells induces a potent proliferative response associated with the expression of IL-5Rα and the prevention of B-cell apoptosis.29,30 Furthermore, CD38 stimulation induces sterile γ1 transcript through nuclear factor-κB (NF-κB) activation.31 IL-5 stimulation of CD38-activated splenic B-2 cells induces µ to γ1 CSR and IgG1 production in an IL-4-independent manner.32 We also reported that IL-5-dependent immunoglobulin secretion and µ to γ1 CSR were severely impaired in both Stat5a–/– and Stat5b–/– B cells.33 The impaired immunoglobulin production and µ to γ1 CSR in Stat5b–/–, but not in Stat5a–/– B cells, are rescued in part by IL-4.
As described, IL-5 exerts biological activities on various types of cell expressing IL-5R. Genes induced by IL-5 have been extensively studied in various types of cell line, including c-Myc, c-Fos, c-Jun,19Cis,34Cish1/Jab35 and pim-1.36,37 Although the results of microarray analysis of IL-5-induced genes in eosinophils have been previously reported36,38 there are no available data on IL-5 target genes in primary B cells despite their intrinsic roles in B-cell development and function. We applied DNA microarray technology to investigate the molecular basis for IL-5-dependent B-cell maturation.
In the present study, we conducted comprehensive analysis of the expression of IL-5-inducible genes in mouse B cells using a microarray system. A variety of genes were up-regulated by IL-4 and IL-5 in CD38-stimulated B cells, although a set of genes was critically regulated by IL-5. Genes critically regulated by IL-5 alone are prone to be induced slowly after IL-5 stimulation. In this report, we demonstrate that B lymphocyte-induced maturation protein-1 (Blimp-1) and Aid induction are critical for IgM production and µ to γ1 CSR in CD38-activated B cells.
Materials and methods
Mice, antibodies and reagents
C57BL/6 mice were purchased from Japan SLC (Hamamatsu, Japan) and were used for experiments at 8–12 weeks of age. The experiments were performed according to the guidelines for animal treatment at the Institute of Medical Science, the University of Tokyo.
Purified CS/2 (agonistic anti-mouse CD38 mAb) was prepared as previously described.30 Mouse IL-4 was purchased from R & D Systems (Minneapolis, MN). Mouse IL-5 was purified as previously described.13 Biotinylated anti-CD43, anti-Syndecan-1, and anti-IgG1 antibodies were purchased from BD PharMingen (San Diego, CA).
B-cell culture
Three different sets of experiments were carried out. In each experiment, splenic B cells were isolated from 8 to 12 weeks old C57BL/6 mice by negative selection with biotinylated-CD43 antibody and streptavidin magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). Purity was analysed by flow cytometry and was routinely >95%. Purified B cells were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS), 2 mm l-glutamine, 50 µm 2-mercaptoethanol, penicillin (50 U/ml), and streptomycin (50 µg/ml). Anti-CD38 mAb (CS/2, 1 µg/ml), IL-5 (100 U/ml), IL-4 (25 ng/ml), or selected combinations of these agents, were added when the cells were plated.
Generation of cRNA and microarray hybridization
Total RNAs were isolated using Trizol (Invitrogen, Gaithersburg, MD) or RNeasy (QIAGEN, Valencia, CA) according to the manufacturer's instructions. Poly A RNAs were purified from total RNA by PolyA Tract® mRNA Isolation Systems (Promega, Madison, WI). Double-stranded complementary DNA (cDNA) was generated from 1 µg of poly A RNA using a poly dT containing a T7 sequence and a SuperScript Choice System kit (Invitrogen). Biotinylated cRNA was generated by in vitro transcription using the Bio Array Highyield RNA transcript labelling kit (ENZO Diagnostics Inc, Farmingdale, NY). Purified cRNA was fragmented, and 10 µg of labelled cRNA was hybridized to Mouse Expression Set 430A and 430B chips (Affymetrix, Santa Clara, CA) for 16 hr at 45°. After washing according to Affymetrix's protocol, the DNA chips were scanned by a scanner from Affymetrix.
Data processing
To standardize the average signal of each experiment, the expression data from the Affymetrix GeneChips were processed as follows. First, data on CD38-stimulated B cells in three experiments were adjusted to a specified target signal value by all probe sets scaling (target signal = 500). Then, data on CD38+ IL-4- and CD38+ IL-5-stimulated B cells were normalized to each item of data on CD38-stimulated B cells as a baseline signal to trim the mean signal of each experiment. Our criterion for genes induced or repressed by IL-5 is that genes are more than twofold changed and expected to be present or marginal based on Affymetrix's algorithm in at least two of the three experiments.
Hierarchy clustering was conducted by Genes@work software.39 The Euclidean correlation coefficient was used as the distance metric. To determine the differences in gene expression, genes regulated by IL-5 were compared by clustering the selected genes according to the given criterion described above. All genes were annotated using available public databases. The GenBank accession numbers of representative sequences were described in genes with no available information.
Semi-quantitative reverse transcription–polymerase chain reaction (RT–PCR) analysis
Total RNAs were extracted from splenic B cells before or 3 days after the stimulation using Trizol (Invitrogen). cDNA synthesis was conducted in 20 µl aliquots of reaction mixture containing 1 or 5 µg total RNA, oligo-dT primer and SuperScript II RNase H– reverse transcriptase (Invitrogen). For semiquantitation, serial dilutions of the cDNA templates were subjected to PCR amplification using the primer described in Primers. PCR products were separated by electrophoresis on 2% agarose gel and visualized by ethidium bromide staining.
PCR analysis of γ1 to µ reciprocal DNA recombination products
PCR analysis of γ1 to µ reciprocal DNA recombination products was performed according to procedures previously described.32,33
Retroviral transfection
pMXs-IRES-green fluorescent protein (GFP) vector and the packaging cell line, Plat-E cells40,41 were kindly provided by Dr Toshio Kitamura (Tokyo University). The Plat-E cells were maintained in Dulbecco's modified Eagle's minimal essential medium containing 10% fetal calf serum (FCS), 1 µg/ml puromycin and 10 µg/ml blastcidin. The full-length cDNAs for Blimp-1 and Aid were amplified by Pfx DNA polymerase (Invitrogen) and confirmed by sequencing. Transfection of retrovirus vectors was performed by Fugene 6 Transfection Reagent (Roche Molecular Biochemicals, Indianapolis, IN) according to the manufacturer's protocol. At 24 hr after transfection, the culture medium was replaced with B-cell culture medium without antibiotics. The retroviral supernatants were harvested after an additional 24 hr of incubation and were then used for the infection of the target cells.
Isolated primary B cells were cultured with anti-CD38 mAb (1 µg/ml) for 24 hr. The activated cells were recovered and resuspended in retroviral supernatant containing 10 µg/ml DOTAP (Roche Molecular Biochemicals) at a concentration of 1–2 × 106 cells/ml.42 The cells were plated in 6- or 12-well tissue culture plates and centrifuged at 2000 r.p.m. (900 g) for 90 min at room temperature. Spin-inoculated cells were washed and incubated with B-cell culture medium containing anti-CD38 mAb. IL-4 (25 ng/ml) or IL-5 (100 U/ml) was added 12–24 hr after incubation. Flow cytometric and enzyme-linked immunosorbent assay (ELISA) analyses were performed 36–48 hr after cytokine addition.
Flow cytometric analysis
The cultured cells were recovered and suspended in staining buffer (phosphate-buffered saline, 2% FCS and 0·05% sodium azide) containing anti-FcyR mAb (2.4G2; 10 µg/ml). The cells were incubated for 15–20 min with biotinylated anti-Syndecan-1 or anti-IgG1 antibody (both from PharMingen). After being washed, the cells were stained with streptavidin phycoerythrin (Molecular Probes, Gaithersburg, MD) for 15–20 min. After washing twice with the staining buffer, the cells were suspended in staining buffer containing 2 µg/ml of 7-amino-actinomycin D (Sigma Fine Chemical Co., St. Louis, MO) to exclude dead cells from the analysis. Analyses were conducted using FACSCalibur instrument (BD Bioscience, Mountain View, CA).
Primers
The primers used in this paper are the following:
HPRT RTS1: AGTCCCAGCGTCGTGATTAG
HPRT RTA1: CGAGAGGTCCTTTTCACCAG
XBP1 RTS1: CTTCCCATGGACTCTGACACTG
XBP1 RTS2: CGCAGACTGCTCGAGATAGAAA
XBP1 RTA1: AGGTCCCCACTGACAGAGAAAG
XBP1 RTA2: GACAGAGAAAGGGAGGCTGGTA
BCL6 RTS1: CACTTAAACCTCCCCGTGAA
BCL6 RTA1: AAATGCAGGGCAATCTCATC
Blimp-1 RTS1: TGACGGGGGTACTTCTGTTC
Blimp-1RTA2: TGGGGACACTCTTTGGGTAG
AID S1: ATATGGACAGCCTTCTGATGAAGC
AID AS1: TCAAAATCCCAACATACGAAATGC
Blimp-1 S3: GGACGCGTCTGGGATCATGAAAATGGACATG
Blimp-1 S4: AAACGCGTACCATGGGTATCAACAACTTCAGCCTC
Blimp-1 AS2: TTCTCGAGATCTTAAGGATCCATCGGTTCAAC
Blimp-1 MS2: ACGCGTACCATGGGGGGTACTTCTGTTCAAGCC
Blimp-1 MS3: ACGCGTACCATGGATGGCTTTCGGAAAAATG.
Results
Expression analysis of genes regulated by IL-5
We reported that IL-5 stimulation on CD38-activated B cells induced IgM production and µ to γ1 CSR, leading to a large amount of IgG1 production.32 In contrast, IL-4 does not have any activity to induce immunoglobulin production or µ to γ1 CSR on CD38-stimulated B cells, while the further addition of IL-4 in CD38+IL-5 stimulation enhances IL-5-dependent µ to γ1 CSR. We also showed the essential roles of Stat5 on IL-5-dependent B-cell terminal maturation.33 The results suggest that IL-5 induces genes required for B-cell terminal differentiation through Stat5 activation.
To elucidate molecular mechanisms in IL-5-dependent B-cell terminal maturation, we used DNA microarray to analyse genes regulated by IL-5. We stimulated three groups of purified primary B cells with anti-CD38, anti-CD38 plus IL-4, or anti-CD38 plus IL-5 and then extracted RNA from each group of culture at 3 days after stimulation. Biotinylated cRNAs were generated from extracted RNAs and hybridized to Affymetrix GeneChip Mouse 430A and 430B. The chip sets allowed us to scrutinize the expression level for over 39 000 transcripts and variants. Our criterion for genes induced or repressed by IL-5 was that their expressions, compared to only CD38-stimulated B cells, are more than twofold changed and expected to be present or marginal based on Affymetrix's algorithm in at least two of three different experiments. As a result of 45 000 probe sets, we identified 289 probe sets as genes inducible by IL-5 (Fig. 1a, Table 1) and 325 probe sets as genes repressible by IL-5 (Fig. 1b). We found numerous IL-5-inducible genes, many of which have been previously shown to be regulated by IL-5, such as those encoding IL-2Rα chain, cyclin D2, Cis, Cish1, Cish2/Socs2,33Pim-1, Blimp-1 (Prdm1),29,33,43 and so on, indicating the validity of our analysis.
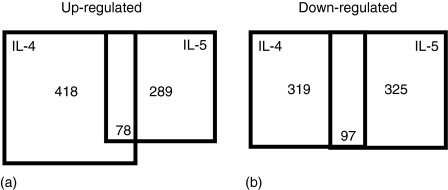
Figure 1.
Number of genes regulated by IL-4 and/or IL-5 on CD38-stimulated B cells. (a) IL-5 induced 289 probe sets (228 and 63 probe sets from 430A and 430B, respectively), 78 of which were shared with the genes up-regulated by IL-4 (430A:288, B:130). (b) IL-5 repressed 325 probe sets (430A:251, B:74), while IL-4 down-regulated 319 probe sets (430A:219, B:100).
Table 1.
The genes induced by IL-5. The values show the average of gene expression changes (log2) from the three independent experiments. The 178 prove sets (the mean fold induction is > two-fold) are shown.
| Gene symbol | Affymetrix ID | IL-4 | IL-5 |
|---|---|---|---|
| Apoptosis/Antiapoptosis | |||
| Bnip3** | 1422470_at | 1·7 | 2·3 |
| Bnip3l** | 1416923_a_at | 0·7 | 1·0 |
| Bnip3l* | 1448525_a_at | 0·9 | 1·1 |
| Cell cycle | |||
| 2600016F06Rik** | 1434745_at | 0·0 | 1·2 |
| Ccnd2* | 1416123_at | 0·4 | 1·7 |
| Ccnd2** | 1416124_at | −0·1 | 1·1 |
| Ccnd2** | 1430127_a_at | 0·3 | 1·4 |
| Ccnd2** | 1448229_s_at | 0·1 | 1·5 |
| Ccnd2** | 1455956_x_at | 0·1 | 1·3 |
| Ccne1** | 1416492_at | 0·7 | 1·0 |
| Gadd45g** | 1453851_a_at | 5·1 | 1·2 |
| Mkrn3** | 1448612_at | −0·1 | 1·5 |
| Slfnl* | 1418612_at | 2·2 | 3·3 |
| Class switch recombination | |||
| Aicda** | 1420577_at | 0·1 | 1·6 |
| Cytokine and receptor | |||
| Ccl22** | 1417925_at | −0·4 | 2·0 |
| Csf2rb1** | 1450200_s_at | −0·4 | 1·8 |
| Csf2rb1** | 1421326_at | −0·2 | 2·4 |
| Csf2rb2** | 1449360_at | −0·6 | 2·9 |
| Il1r2** | 1419532_at | −0·8 | 1·4 |
| Il2ra** | 1420691_at | −0·5 | 2·2 |
| Il2ra** | 1420692_at | −0·9 | 2·5 |
| Lta** | 1420353_at | −1·3 | 1·2 |
| Sdf2l1 | 1418206_at | 0·3 | 1·0 |
| Sema4a** | 1448110_at | 2·4 | 2·2 |
| Thy1** | 1423135_at | 2·5 | 1·7 |
| Immunoglobulin | |||
| Igh-4** | 1425247_a_at | 3·4 | 2·2 |
| Igh-4** | 1425324_x_at | 4·8 | 3·0 |
| Igh-4** | 1427756_x_at | 3·3 | 2·2 |
| Igh-4** | 1427870_x_at | 1·9 | 1·5 |
| Igj** | 1424305_at | −1·8 | 2·4 |
| Metabolism | |||
| Aldo1* | 1416921_x_at | 0·6 | 1·2 |
| Aldo1* | 1433604_x_at | 0·7 | 1·4 |
| Aldo1* | 1434799_x_at | 0·8 | 1·4 |
| Aldo1 | 1439375_x_at | 0·5 | 1·1 |
| Aldo3** | 1451461_a_at | 1·6 | 1·6 |
| Chst12* | 1448477_at | −0·1 | 1·3 |
| Glk* | 1417177_at | 0·6 | 1·1 |
| Gstt2** | 1417883_at | 0·7 | 1·0 |
| Pfkl | 1439148_a_at | 0·8 | 1·3 |
| Pfkp** | 1416069_at | 1·0 | 1·2 |
| Pfkp* | 1430634_a_at | 1·0 | 1·3 |
| Pk3** | 1417308_at | 0·6 | 1·3 |
| Psat** | 1451064_a_at | 0·3 | 1·2 |
| Psat** | 1454607_s_at | 0·3 | 1·3 |
| Pycr1** | 1424556_at | 1·3 | 2·4 |
| Scd2** | 1415823_at | 2·0 | 1·7 |
| Scd2** | 1415822_at | 1·7 | 1·5 |
| Scd2** | 1415824_at | 2·0 | 1·5 |
| Tpi* | 1415918_a_at | 0·9 | 1·6 |
| Tpi* | 1435659_a_at | 1·1 | 1·8 |
| Tpi** | 1452927_x_at | 0·9 | 1·6 |
| Ak4** | 1450387_s_at | 1·4 | 1·9 |
| Armet** | 1428112_at | 0·6 | 1·1 |
| Asns** | 1433966_x_at | 0·9 | 2·6 |
| Asns** | 1451095_at | 0·6 | 1·6 |
| Ass1** | 1416239_at | 2·4 | 1·9 |
| Miscellaneous | |||
| Anxa2** | 1419091_a_at | −0·1 | 1·1 |
| Anxa6 | 1429246_a_at | −0·2 | 1·1 |
| Crip** | 1416326_at | −0·5 | 1·5 |
| Crisp1** | 1416325_at | 0·0 | 2·9 |
| Eaf2** | 1422120_at | 0·3 | 1·2 |
| Egln1** | 1423785_at | 1·0 | 1·2 |
| Egln1 | 1451110_at | 1·2 | 1·4 |
| Egln3** | 1418649_at | 0·6 | 1·2 |
| Endog** | 1421097_at | 1·3 | 2·5 |
| Ero1l* | 1419030_at | 0·7 | 1·2 |
| Ero1l** | 1419029_at | 0·8 | 1·6 |
| Gbp2* | 1418240_at | 0·8 | 1·7 |
| Gbp2 | 1435906_x_at | 0·8 | 1·6 |
| Gbp4** | 1418392_a_at | 0·0 | 1·0 |
| Gcat** | 1417823_at | 3·6 | 4·1 |
| Glipr2** | 1428492_at | 2·2 | 1·3 |
| Gpam** | 1438040_a_at | 0·1 | 1·0 |
| Hbb-b2** | 1417184_s_at | −0·1 | 4·5 |
| Hig1 | 1416480_a_at | 0·8 | 1·1 |
| Hmmr** | 1450157_a_at | 0·6 | 1·0 |
| Ifi30** | 1422476_at | 2·0 | 1·6 |
| Klhdc2** | 1435086_s_at | 4·3 | 1·8 |
| Klhdc2** | 1460678_at | 4·5 | 2·1 |
| Lfng* | 1420643_at | 0·8 | 1·1 |
| Lmo7** | 1455056_at | 1·7 | 3·0 |
| mrg1** | 1452207_at | 1·6 | 1·6 |
| Mtdh* | 1434882_at | 1·0 | 1·3 |
| Mtdh* | 1455129_at | 1·1 | 1·1 |
| Myadm** | 1423321_at | 0·8 | 2·2 |
| Myadm** | 1439389_s_at | 1·0 | 2·0 |
| P4ha1** | 1452094_at | 0·7 | 1·2 |
| Pcp4** | 1460214_at | −0·8 | 3·5 |
| Pdcd2** | 1423534_at | 0·2 | 1·2 |
| Pdlim1** | 1416554_at | 1·1 | 1·5 |
| Prdx4 | 1416167_at | 0·1 | 1·0 |
| Rbpms** | 1425652_s_at | −0·7 | 1·0 |
| Rpn1* | 1448353_x_at | 0·5 | 1·1 |
| Scel** | 1422837_at | −0·4 | 2·4 |
| Spcs2 | 1423215_at | 0·0 | 1·0 |
| Spi2-1 | 1424923_at | 3·0 | 1·6 |
| St7** | 1418967_a_at | 4·5 | 2·0 |
| Txndc7** | 1423648_at | 0·6 | 1·0 |
| uchl3** | 1449855_s_at | 0·4 | 1·3 |
| Uck3** | 1448604_at | 0·3 | 1·1 |
| Upb1** | 1460244_at | 0·1 | 1·4 |
| Zdhhc2** | 1452654_at | 0·1 | 1·1 |
| Signalling | |||
| Cish** | 1448724_at | 3·2 | 2·0 |
| Cish2** | 1449109_at | 3·8 | 4·3 |
| Eps8* | 1422824_s_at | 0·4 | 1·1 |
| Ndr1** | 1450976_at | 1·5 | 1·9 |
| Ndr1** | 1456174_x_at | 1·0 | 1·4 |
| Ndrl** | 1420760_s_at | 1·0 | 1·2 |
| Nek2** | 1417299_at | 0·4 | 1·8 |
| Pim1** | 1423006_at | 0·9 | 1·3 |
| Pim1** | 1435872_at | 1·1 | 1·5 |
| Pkib** | 1421137_a_at | 3·1 | 1·7 |
| Pkib** | 1421138_a_at | 3·2 | 1·4 |
| Rap1ga1** | 1428443_a_at | 0·9 | 1·8 |
| Rras2** | 1417398_at | 0·0 | 1·3 |
| S100a4** | 1424542_at | 0·1 | 1·1 |
| S100a6** | 1421375_a_at | 2·5 | 5·3 |
| SSI-1** | 1450446_a_at | 4·0 | 1·6 |
| Stk39** | 1419550_a_at | 0·0 | 1·7 |
| Cytoskeltal protein | |||
| Ckap4** | 1426754_x_at | 0·8 | 1·8 |
| Ckap4 | 1426755_at | 1·2 | 2·2 |
| Ckap4** | 1452181_at | 0·6 | 1·5 |
| Ckap4* | 1455019_x_at | 0·9 | 1·7 |
| Itgb7** | 1418741_at | 0·2 | 1·4 |
| Vim** | 1438118_x_at | 0·6 | 1·2 |
| Vim** | 1450641_at | 0·6 | 1·3 |
| Vim** | 1456292_a_at | 0·8 | 1·4 |
| Fgl2* | 1421854_at | 0·6 | 1·0 |
| Fgl2* | 1421855_at | −0·5 | 1·0 |
| Transcription | |||
| Cited2** | 1421267_a_at | 1·5 | 1·6 |
| Ell2** | 1450744_at | 0·8 | 1·4 |
| Nfil3** | 1418932_at | 6·9 | 2·2 |
| Prdm1** | 1420425_at | −0·3 | 1·0 |
| Rog** | 1432459_a_at | −0·4 | 1·8 |
| Sap30* | 1417719_at | 0·1 | 1·1 |
| Xbp1** | 1420011_s_at | 1·6 | 1·1 |
| Xbp1** | 1420886_a_at | 1·5 | 1·0 |
| Xbp1** | 1437223_s_at | 1·3 | 1·0 |
| Transporter/channel | |||
| Atp1b1** | 1418453_a_at | −0·1 | 2·7 |
| Atp1b1** | 1423890_x_at | −0·2 | 3·1 |
| Atp1b1** | 1439036_a_at | −0·1 | 4·8 |
| Atp1b1** | 1451152_a_at | −0·1 | 3·0 |
| I2RF5 | 1416956_at | 0·2 | 1·1 |
| MCT4** | 1449005_at | 1·9 | 2·8 |
| Slc16a3** | 1452565_x_at | 1·7 | 2·7 |
| Slc2a1** | 1426600_at | 0·8 | 1·5 |
| Slc2a1** | 1426599_a_at | 1·3 | 2·0 |
| Slc2a1** | 1434773_a_at | 1·1 | 1·9 |
| Slc39a8** | 1416832_at | 1·1 | 1·7 |
| Slc7a5** | 1418326_at | −0·1 | 1·5 |
| Translation | |||
| Eif4ebp1** | 1417562_at | 0·7 | 1·4 |
| Eif4ebp1** | 1434976_x_at | 0·5 | 1·3 |
| Unknown | |||
| 1500032D16Rik** | 1424628_a_at | 0·6 | 1·1 |
| 1810014L12Rik** | 1416376_at | 0·6 | 1·3 |
| 2310056P07Rik** | 1451385_at | 0·5 | 1·5 |
| 2410129E14Rik** | 1452679_at | −0·6 | 1·8 |
| 2610020G18Rik** | 1451149_at | 0·6 | 1·0 |
| 2810433K01Rik* | 1450496_a_at | 0·5 | 1·0 |
| 5730592L21Rik** | 1452754_at | 0·3 | 1·1 |
| 5830411E10Rik** | 1426806_at | 1·8 | 1·7 |
| 5830411E10Rik** | 1452203_at | 1·9 | 1·7 |
| 5830411E10Rik** | 1460521_a_at | 2·0 | 1·6 |
| 9630044O09Rik* | 1424823_s_at | 1·0 | 1·5 |
| 9830147J24Rik** | 1425156_at | 0·7 | 1·9 |
| AI987662** | 1435939_s_at | −2·1 | 2·9 |
| BC003855.1** | 1425469_a_at | 1·0 | 1·5 |
| BC003855.1** | 1425470_at | 1·0 | 1·9 |
| BC003855.1** | 1425471_x_at | 0·8 | 1·6 |
| BC021921** | 1451518_at | −0·2 | 1·9 |
| BG070552** | 1455279_at | 0·3 | 2·3 |
| BM241271** | 1434380_at | 1·1 | 2·8 |
| D10Ertd214e | 1416849_at | 0·6 | 1·3 |
| D130043N08Rik** | 1418507_s_at | 4·0 | 4·5 |
| D530020C15Rik** | 1423747_a_at | 1·2 | 1·6 |
| D530020C15Rik** | 1423748_at | 1·1 | 1·3 |
| E330009J07Rik** | 1449056_at | 0·4 | 1·0 |
| E430036I04Rik** | 1423614_at | 0·9 | 1·3 |
| LOC381240** | 1438932_at | 0·4 | 1·3 |
Significant differences as determined by one-way anova are represented as
P < 0·05
P < 0·01
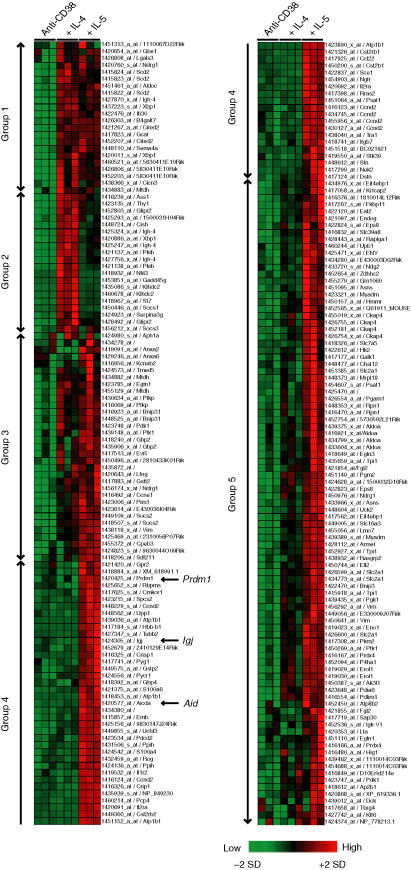
To group genes based on the degree of similarity of their expression patterns, average linkage hierarchical clustering was performed for the gene expression data on selected genes induced by IL-5. The upper half of the left-hand column in Fig. 2 indicates genes equally (Group 1 and 3) or preferentially (Group 2) up-regulated by IL-4. These genes included Xbp1, IgG1 (Igh-4), and the Cis/Socs family (Cish, Cish2 and Socs3). The preferential up-regulation of IgG1 by IL-4 can be explained by the induction of sterile γ1 transcript by IL-4. The lower part of the left-hand column and the upper part of the right-hand column (Group 4) show that these genes are exclusively induced by IL-5 and include Blimp-1, Aid (Aicda), and J chain. The results indicate that these genes in Group 4 are deeply involved in IL-5-dependent B-cell terminal maturation.
Figure 2.
Hierarchical clustering of the genes regulated by IL-5. Hierarchical clustering was performed using Genes@work software based on the average-linked algorithm. The colour scale indicates relative gene expression changes normalized by SD, with 0 representing the mean expression level.
IL-5, but not IL-4, induced the expression of Blimp-1 and Aid
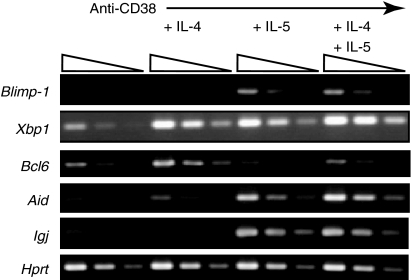
The expression of IL-5-inducible genes was confirmed by semiquantitative RT–PCR. Consistent with the microarray data, the expressions of Blimp-1, Aid, and J chain were elevated in B cells stimulated with anti-CD38 and IL-5- as well as anti-CD38, IL-5, and IL-4 (Fig. 3). Xbp1 expression was up-regulated in B cells stimulated with both anti-CD38 plus IL-5 and anti-CD38 plus IL-4, indicating that its expression is not sufficient to induce B-cell terminal maturation. Iwakoshi et al.44 showed the exclusive up-regulation of Xbp1 by IL-4 using purified naive B cells. Unlike IL-4R, IL-5R is expressed only in a subpopulation of splenic B cells, and the expression level of IL-5R in naive splenic B cells is quite low. The discrepancy can be mainly explained by the source of the experiments, naive or activated cells.
Figure 3.
Blimp-1 and Aid induced by IL-5, but not IL-4, in CD38-stimulated B cells. Purified splenic B cells were stimulated with anti-CD38 mAb and different cytokines as indicated for 72 hr. After incubation, RNA was extracted from activated B cells and was subjected to semiquantitative RT–PCR (fourfold dilutions) to confirm the induction of genes regulated by IL-5. The housekeeping gene, Hprt, was used to control sample variation in RNA isolation and integrity, RNA input, and reverse transcription.
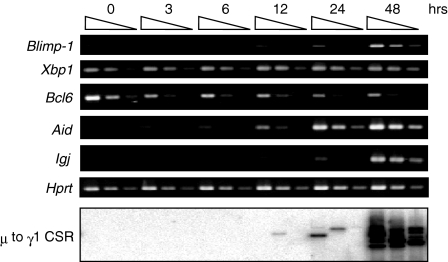
We then examined the kinetics of gene expression regulated by IL-5. As IL-5Rα expression is observed in a small population of primary B cells in the spleen and up-regulated by anti-CD38 stimulation29 we stimulated purified splenic B cells with anti-CD38 mAb for 2 days to induce IL-5Rα expression on a large proportion of the B cells. The B cells were then harvested at various periods of time after IL-5 stimulation and subjected to semiquantitative RT–PCR analysis (Fig. 4). Blimp-1 and Aid expressions were up-regulated from 24 h after IL-5 stimulation. Significant levels of J chain expression and γ1 to µ reciprocal circular DNA were detectable around 48 hr after IL-5 stimulation. These results suggest that the co-ordinated expression of Blimp-1 and Aid genes can be induced with B-cell maturation to immunoglobulin-secreting cells and µ to γ1 CSR, respectively.
Figure 4.
Kinetics of expression of genes induced or repressed by IL-5.Purified splenic B cells were stimulated with anti-CD38 Ab (1 µg/ml) for 48 hr and then stimulated with IL-5 (100 U/ml). After the addition of IL-5, RNA and DNA were prepared from activated B cells at the indicated times. Semiquantitative RT–PCR (fourfold dilutions) was performed with sets of primers specific for genes induced or repressed by IL-5. For µ to γ1 switch recombination (bottom lane), γ1 to µ switch circles were amplified by LA-Taq polymerase with 5′Sγ1 and 3′Sµ primers and hybridized with 5′Sγ1 probes.
Bcl6 has been reported to regulate Blimp-1 expression.45Bcl6 expression was down-regulated upon stimulation with IL-5, while was not affected upon IL-4 stimulation (Fig. 3). Moreover, Bcl6 expression started along with a reduction in B cells about 3 hr after IL-5 stimulation and then gradually diminished until 48 hr (Fig. 4). These results suggest that the down-regulation of Bcl6 by IL-5 stimulation is co-ordinated with Blimp-1 expression in anti-CD38-activated B cells.
Retroviral transduction of Blimp-1 forced activated B cells to differentiate to plasma cells
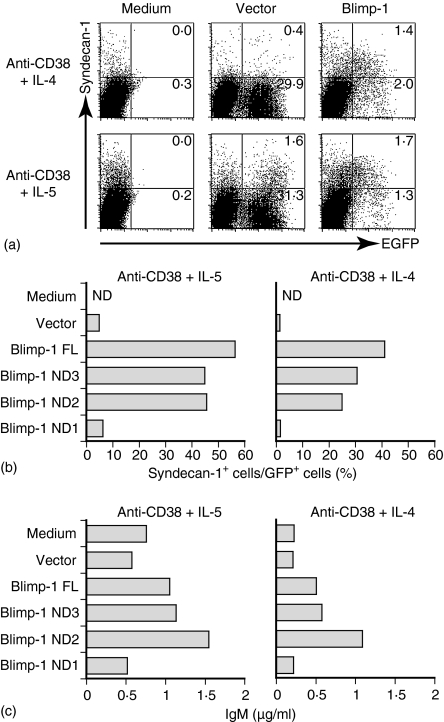
The preferential expression of Blimp-1 in CD38-activated B cells stimulated with IL-5 raises the possibility that Blimp-1 is a crucial transcriptional factor to activate B cells for IL-5-dependent terminal maturation to immunoglobulin-secreting cells. To address this issue, we transferred the Blimp-1 gene with retroviral expression system in CD38-activated primary B cells. The bicistronic retrovirus vector containing GFP enabled infected cells to be analysed using flow cytometry. Thus, purified B cells were stimulated with anti-CD38 mAb for 24 hr, and then activated B cells were harvested and spin-inoculated for 90 min with retroviral supernatant containing a cationic liposome, DOTAP. Then, IL-4 or IL-5 was added at 24 hr after the transfection and cultured for another 48 hr. After the culture, Syndecan-1+ cells in GFP+ cells were analysed using flow cytometry.
The population of Syndecan-1+ cells in GFP+ B cells was approximately 4·8% in anti-CD38 and IL-5-stimulated B cells transfected with a control vector (Fig. 5a). In contrast, enforced expression of Blimp-1 gene significantly enhanced the proportion of Syndecan-1+ cells in GFP+ cells up to 56%. Intriguingly, the proportion of Syndecan-1+ cells in GFP+ B cells dramatically increased up to 40% in anti-CD38 and IL-4 compared with cells transfected with the vector, indicating that the enforced expression of the Blimp-1 gene in anti-CD38-stimulated B cells induces B cell differentiation to immunoglobulin-secreting cells in response to IL-4. The ELISA data confirmed that the retrovirus-transfected B cells were producing IgM (Fig. 5c).
Figure 5.
Retroviral transduction of Blimp-1 forcing activated B cells to differentiate to Syndecan-1-positive cells. (a) Primary B cells were stimulated with anti-CD38 mAb (1 µg/ml) for 24 hr. Retrovirus expressing the control or Blimp-1 was transduced into activated cells. The infected cells were stimulated with IL-5 (upper) or IL-4 (lower) 12 hr after the retroviral infection. At 48 hr after the cytokine addition, activated cells were stained with biotinylated antibody against Syndecan-1, followed by streptavidin-phycoerythrin. (b) The proline-rich domain, but not the SET domain, was required for plasma cell induction by Blimp-1. The abilities of Blimp-1 and its mutant to induce Syndecan-1+ cells were examined. The bars indicate the percentage of Syndecan-1+ cells in GFP+ cells. (c) The concentrations of IgM in the supernatant of infected cells were measured by ELISA.
To evaluate the role of the functional domain of Blimp-1, Blimp-1 gene and its mutant genes were then transduced into anti-CD38-primed B cells, and IL-5- or IL-4-induced differentiation of B cells to Syndecan-1+ cells was examined (Fig. 5b, c). The results revealed that the expression of Blimp-1 lacking the N-terminal acidic region (ND3) or the SET domain (ND2) retained the ability in our assay system to induce B-cell terminal maturation to immunoglobulin-secreting cells, similar to full-length Blimp-1. However, deletion mutant of Blimp-1 (ND1) that lacks a proline-rich domain, N-terminal acidic region, and SET domain resulted in loss of the activity. These results indicate that Blimp-1 has sufficient activity to induce IL-4-dependent differentiation of CD38-activated B cells to immunoglobulin-secreting cells, and that the proline-rich domain of Blimp-1 plays an essential role in its functional activity.
Positive and negative role of Aid and Blimp-1 in µ to γ1 CSR
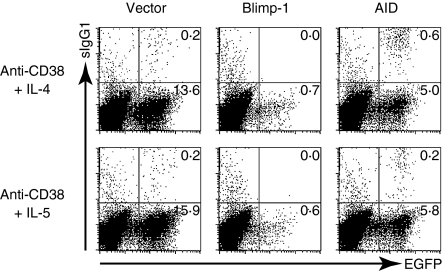
The results from microarray and RT–PCR showed that the expression of Aid is up-regulated by IL-5, but not IL-4 in CD38-stimulated B cells. The induction of Aid in B cells activated with anti-CD38 antibody upon IL-5, but not IL-4 stimulation, could account for IL-5-dependent µ to γ1 CSR. To examine this possibility, we examined the Aid expression in CD38-stimulated B cells by using retroviral gene transfer with a GFP marker and stimulated with IL-5 or IL-4 for 48 hr. The surface (s) IgG1+ cells in GFP+ cells were hardly detected in the control vector-transduced anti-CD38-stimulated B cells upon IL-5 or IL-4 stimulation (1·4% or 1·2%, respectively; Fig. 6). In contrast, the forced expression of Aid increased the population of sIgG1+ cells in GFP+ cells in the B cells stimulated with anti-CD38 mAb and IL-4 up to 11%. Only a slight increase of sIgG1+ cells in GFP+ cells was observed by the enforced expression of Aid in anti-CD38- and IL-5-stimulated B cells. This may be caused by the fact that Aid is adequately induced in CD38-activated B cells upon IL-5 stimulation.
Figure 6.
Retroviral induction of Aid helping CD38+IL-4-stimulated B cells to undergo µ to γ1 CSR. Retroviruses expressing Blimp-1 or Aid were transduced to CD38-stimulated B cells, followed by the addition of IL-4 or IL-5. At 3 days after retroviral infection, the transfected cells were stained with biotinylated antibodies against IgG1, followed by staining with streptavidin-phycoerythrin. The percentages of sIgG1-positive cells among GFP-positive cells are shown.
On the other hand, enforced Blimp-1 expression decreased the population of sIgG1+ cells in GFP+ cells. The population of sIgG1+ cells in GFP+ cells disappeared in B cells stimulated with anti-CD38 mAb and IL-5 by Blimp-1 overexpression, compared to the control virus-infected cells (the middle panel of Fig. 6). The negative role of Blimp-1 was also observed when Blimp-1 was overexpressed in B cells during stimulation with LPS and IL-4 in most of which µ to γ1 CSR were induced (data not shown). These results indicate that Blimp-1 plays negative role in CSR.
Discussion
The generation of immunoglobulin-secreting cells and CSR is one of hallmarks in adaptive immunity, although its molecular basis still remains unclear. The mechanisms of B-cell terminal differentiation are extensively studied by using in vitro culture systems due to the difficulty to visualize biochemical events in vivo. We previously found that IL-5 induced CD38-stimulated B cells to terminally differentiate to immunoglobulin-secreting cells and class-switched cells while IL-4, a well-known factor for CSR, failed to do so. In this paper, we performed expression analysis of IL-5 target genes using DNA microarray to elucidate the molecular mechanism of IL-5-dependent B-cell maturation. Our results provide a new aspect to the prevailing paradigm for B-cell terminal maturation.
The present molecular model for the development of immunoglobulin-secreting cells has been proposed based on studies on two knockout mice and the microarray result of Blimp-1. Xbp1-deficient B cells fail to produce all subtypes of immunoglobulin by the RAG-2 complementation system.46 The deficiency of Blimp-1 also showed impaired plasma cell differentiation.47 The expression analysis of Blimp-1 target genes linked these two transcriptional factors, proposing the attractive model that Blimp-1 induces Xbp-1 by relieving the repression of BSAP/Pax5.48 However, our finding that IL-4 induces Xbp1 but not Blimp-1 suggests that the expression of Xbp1 can be regulated independently of Blimp-1. In addition, B cells stimulated with anti-CD38 and IL-4 fail to differentiate to immunoglobulin-producing cells in spite of Xbp1 up-regulation. Furthermore, neither Xbp1 nor spliced Xbp1 with stronger transcriptional activity enhanced or induced the number of Syndecan-1+ positive cells in CD38+IL-5- or CD38+IL-4-stimulated B cells (data not shown). With these results taken together, we speculate that Xbp1 is a transcriptional factor required for plasma cell development, while its expression is not sufficient to drive activated B cells to differentiate to plasma cells.
Our microarray data and RT–PCR analysis showed that Blimp-1 was exclusively induced by IL-5 in CD38-stimulated B cells, while both IL-4 and IL-5 induced Xbp1. We also demonstrated that the retrovirus transfer of Blimp-1 enhanced or induced immunoglobulin-secreting cells differentiation in CD38+IL-5- or CD38+IL-4-stimulated cells, respectively. These results indicate that Blimp-1 is a transcriptional factor responsible for IL-5-dependent B cell terminal maturation to plasma cells. Xbp1 and Blimp-1 could act synergistically, rather than sequentially, for IL-5-dependent B-cell terminal maturation.
We also addressed the functional domain of Blimp-1 responsible for B-cell differentiation to immunoglobulin-secreting cells. As shown in Fig. 5(b, c), the proline-rich domain, but not the SET domain, of Blimp-1 is crucial for the induction of immunoglobulin-secreting cells. Because the proline-rich domain is thought to suppress the transcription of genes, Blimp-1 leads B-cell terminal maturation by suppressing the expression of its target genes, such as c-myc, CIITA, Pax5 and SpiB.48 Recent structural analyses revealed the catalytic activity of the SET domain to methyltransferase lysine residues on histone tails49,50 although no effects of the SET domain in B-cell maturation were observed in this study. The SET domain of Blimp-1 might be required to leave its repressive mark on its target genes.
Another difference in gene expression between IL-4 and IL-5 stimulation was the suppression of Bcl6 expression (Fig. 3). Bcl6 has been shown to repress Blimp-1 as well as many lymphocyte activation genes by microarray analyses.45 Furthermore, the forced expression of Bcl6 was reported to suppress the expression of Blimp-1 possibly by inhibiting the activity of Stat3, resulting in inhibiting the differentiation of transformed and primary B cells to plasma cells.51 In Fig. 4, we showed that Bcl6 expression is down-regulated from 3 hr after IL-5 stimulation. This implies that the down-regulation of Bcl6 is an initial step of B-cell terminal maturation by IL-5. Although Bcl6 expression is transcriptionally and post-transcriptionally regulated by B-cell receptor stimulation52,53 the mechanism of Bcl6 down-regulation by IL-5 is a subject of future study.
Similar to the induction of immunoglobulin-secreting cells, IL-5 but not IL-4 induces µ to γ1 CSR in CD38-stimulated B cells. It should be caused by the preferential expression of Aid26,54 by IL-5, because the retroviral gene transfer of Aid induced µ to γ1 CSR in CD38+IL-4-stimulated B cells in which µ to γ1 CSR could not be detected otherwise (Fig. 6). Further induction of Aid by retroviral transfer was not able to increase the number of sIgG1-positive cells in CD38+IL-5-stimulated B cells. This could be explained by IL-5 being unable to induce sterile γ1 transcript or the expression of the machineries for CSR being restricted in CD38+IL-5-stimulated B cells. In the retroviral experiment, the induction of Aid increased the percentage of sIgG-positive cells in CD38+IL-4-stimulated B cells much more than non-transfected B cells with CD38+IL-5 stimulation. This is likely because of enhanced accessibility to the γ1 locus by the increased sterile γ1 transcript resulting in CD38+IL-4-stimulated B cells. Recently, it was reported that the expression of the Aid gene is regulated by E-proteins and their inhibitory molecules, Id proteins.55 We examined the possible roles of these molecules by scrutinizing our microarray data. However, there was no significant expression change of Id (Id1, Id2 and Id3) or the E2A gene with IL-5 or IL-4 stimulation (data not shown). This suggests that IL-5 induces Aid gene through an unknown pathway.
The retroviral induction of Blimp-1 decreased the number of sIgG1-positive cells in CD38+IL-5-stimulated B cells. The suppressive role of Blimp-1 was also observed in LPS+IL-4-stimulated B cells (data not shown). Consistent with our data, Blimp-1 was reported to down-regulate genes involved in CSR, such as Aid, Ku86, and DNA-PKcs.48 The simultaneous expression of Blimp-1 and Aid was observed in CD38+IL-5-stimulated B cells, presumably because of heterogeneous populations containing B cells undergoing differentiation to plasma cells or µ to γ1 CSR. A recent division-linked analysis using 5-carboxyfluorescein diacetate succinimidyl ester clearly demonstrated that IL-5 increased the frequency of immunoglobulin-secreting cells and decreased the rate of CSR.56 The preferential Blimp-1 induction by IL-5 could account for this predisposition.
As we reported13,29,30,32,57, CD38 is expressed in follicular B cells but is down regulated in germinal centre B cells. Approximately 95% of B220+ B cells in the spleen express CD38. Among the B220+ B cells, about 4% of them are CD5+ B cells that respond poorly to anti-CD38 ligation for differentiation to immunoglobulin-secreting cells. It should be noted that gene expression data might be subject to differences in relative quantities of different B-cell subsets if those subsets reacted differently to CD38 stimulation. It should also be mentioned that we compared gene expression profiles at 3 days stimulation in this report. This is because we clearly found the biological effects of IL-5, such as immunoglobulin production, µ to γ1 CSR, and Syndecan-1 expression this time. However, the genes responsible for B cell maturation could be determined before or after 3 days. Analysing the kinetics of genes induced by IL-5 is a requirement of future work. We recently reported that Stat5 plays a crucial role in IL-5-dependent B-cell terminal maturation. Considering the function of Stat5 as a transcriptional factor, comparison of the gene expression profile between control and Stat5–/– B cells could be useful in identifying the genes required for B-cell terminal maturation.
In summary, we identified Blimp-1 and Aid as critical molecules for IL-5-dependent B-cell terminal maturation. The result from IL-5-target genes obtained in this paper should further contribute to providing a biochemical explanation of physiological roles of IL-5.
Acknowledgments
We thank Ms C. Usui for technical support, Dr S. Takaki for valuable suggestions, Dr T. Kitamura for providing pMXs-IRES-GFP vector and Plat-E cells, and Dr C.C. Goodnow for critical reading of the manuscript. We also acknowledge R. Matsumura and M. Kumagae for their secretarial assistance.
This works was supported by a Research Grant from the Human Frontier Science Program (K.T); by a Grant-in-Aid for Scientific Research on Priority Area (S) (K.T) from the Ministry of Education, Science, Sports and Culture, in Japan; and by a Grant-in-Aid for Encouragement of Young Scientists (K.H) from the Japan Society for the Promotion of Science.
References
- 1.Kinashi T, Harada N, Severinson E, et al. Cloning of complementary DNA encoding T-cell replacing factor and identity with B-cell growth factor II. Nature. 1986;324:70–3. doi: 10.1038/324070a0. [DOI] [PubMed] [Google Scholar]
- 2.Campbell HD, Sanderson CJ, Wang Y, et al. Isolation, structure and expression of cDNA and genomic clones for murine eosinophil differentiation factor. Comparison with other eosinophilopoietic lymphokines and identity with interleukin-5. Eur J Biochem. 1988;174:345–52. doi: 10.1111/j.1432-1033.1988.tb14104.x. [DOI] [PubMed] [Google Scholar]
- 3.Campbell HD, Tucker WQ, Hort Y, Martinson ME, Mayo G, Clutterbuck EJ, Sanderson CJ, Young IG. Molecular cloning, nucleotide sequence, and expression of the gene encoding human eosinophil differentiation factor (interleukin 5) Proc Natl Acad Sci USA. 1987;84:6629–33. doi: 10.1073/pnas.84.19.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokota T, Coffman RL, Hagiwara H, et al. Isolation and characterization of lymphokine cDNA clones encoding mouse and human IgA-enhancing factor and eosinophil colony-stimulating factor activities: relationship to interleukin 5. Proc Natl Acad Sci USA. 1987;84:7388–92. doi: 10.1073/pnas.84.21.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon BG, Takaki S, Miyake K, Takatsu K. The role of IL-5 for mature B-1 cells in homeostatic proliferation, cell survival, and Ig production. J Immunol. 2004;172:6020–9. doi: 10.4049/jimmunol.172.10.6020. [DOI] [PubMed] [Google Scholar]
- 6.Takatsu K. Interleukin-5. Curr Opin Immunol. 1992;4:299–306. doi: 10.1016/0952-7915(92)90080-x. [DOI] [PubMed] [Google Scholar]
- 7.Takatsu K, Tominaga A, Harada N, Mita S, Matsumoto M, Takahashi T, Kikuchi Y, Yamaguchi N. T cell-replacing factor (TRF)/interleukin 5 (IL-5): molecular and functional properties. Immunol Rev. 1988;102:107–35. doi: 10.1111/j.1600-065x.1988.tb00743.x. [DOI] [PubMed] [Google Scholar]
- 8.Sonoda E, Matsumoto R, Hitoshi Y, et al. Transforming growth factor β induces IgA production and acts additively with interleukin 5 for IgA production. J Exp Med. 1989;170:1415–20. doi: 10.1084/jem.170.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tominaga A, Takaki S, Koyama N, et al. Transgenic mice expressing a B cell growth and differentiation factor gene (interleukin 5) develop eosinophilia and autoantibody production. J Exp Med. 1991;173:429–37. doi: 10.1084/jem.173.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida T, Ikuta K, Sugaya H, et al. Defective B-1 cell development and impaired immunity against Angiostrongylus cantonensis in IL-5Rα-deficient mice. Immunity. 1996;4:483–94. doi: 10.1016/s1074-7613(00)80414-8. [DOI] [PubMed] [Google Scholar]
- 11.Hiroi T, Yanagita M, Iijima H, Iwatani K, Yoshida T, Takatsu K, Kiyono H. Deficiency of IL-5 receptor α-chain selectively influences the development of the common mucosal immune system independent IgA-producing B-1 cell in mucosa-associated tissues. J Immunol. 1999;162:821–8. [PubMed] [Google Scholar]
- 12.Takatsu K, Kikuchi Y, Takahashi T, Honjo T, Matsumoto M, Harada N, Yamaguchi N, Tominaga A. Interleukin 5, a T-cell-derived B-cell differentiation factor also induces cytotoxic T lymphocytes. Proc Natl Acad Sci USA. 1987;84:4234–8. doi: 10.1073/pnas.84.12.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasue T, Nishizumi H, Aizawa S, et al. A critical role of Lyn and Fyn for B cell responses to CD38 ligation and interleukin 5. Proc Natl Acad Sci USA. 1997;94:10307–12. doi: 10.1073/pnas.94.19.10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Appleby MW, Kerner JD, Chien S, Maliszewski CR, Bondada S, Perlmutter RM, Bondadaa S. Involvement of p59fynT in interleukin-5 receptor signaling. J Exp Med. 1995;182:811–20. doi: 10.1084/jem.182.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yousefi S, Hoessli DC, Blaser K, Mills GB, Simon HU. Requirement of Lyn and Syk tyrosine kinases for the prevention of apoptosis by cytokines in human eosinophils. J Exp Med. 1996;183:1407–14. doi: 10.1084/jem.183.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato S, Katagiri T, Takaki S, et al. IL-5 receptor-mediated tyrosine phosphorylation of SH2/SH3-containing proteins and activation of Bruton's tyrosine and Janus 2 kinases. J Exp Med. 1994;180:2101–11. doi: 10.1084/jem.180.6.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitoshi Y, Sonoda E, Kikuchi Y, Yonehara S, Nakauchi H, Takatsu K. IL-5 receptor positive B cells, but not eosinophils, are functionally and numerically influenced in mice carrying the X-linked immune defect. Int Immunol. 1993;5:1183–90. doi: 10.1093/intimm/5.9.1183. [DOI] [PubMed] [Google Scholar]
- 18.Ogata N, Kouro T, Yamada A, Koike M, Hanai N, Ishikawa T, Takatsu K. JAK2 and JAK1 constitutively associate with an interleukin-5 (IL-5) receptor α and βc subunit, respectively, and are activated upon IL-5 stimulation. Blood. 1998;91:2264–71. [PubMed] [Google Scholar]
- 19.Takaki S, Kanazawa H, Shiiba M, Takatsu K. A critical cytoplasmic domain of the interleukin-5 (IL-5) receptor α chain and its function in IL-5-mediated growth signal transduction. Mol Cell Biol. 1994;14:7404–13. doi: 10.1128/mcb.14.11.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–8. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 21.Stavnezer J. Antibody class switching. Adv Immunol. 1996;61:79–146. doi: 10.1016/s0065-2776(08)60866-4. [DOI] [PubMed] [Google Scholar]
- 22.Coffman RL, Lebman DA, Rothman P. Mechanism and regulation of immunoglobulin isotype switching. Adv Immunol. 1993;54:229–70. doi: 10.1016/s0065-2776(08)60536-2. [DOI] [PubMed] [Google Scholar]
- 23.Snapper CM, Marcu KB, Zelazowski P. The immunoglobulin class switch: beyond ‘accessibility’. Immunity. 1997;6:217–23. doi: 10.1016/s1074-7613(00)80324-6. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka M, Yoshida K, Maeda T, Usuda S, Sakano H. Switch circular DNA formed in cytokine-treated mouse splenocytes: evidence for intramolecular DNA deletion in immunoglobulin class switching. Cell. 1990;62:135–42. doi: 10.1016/0092-8674(90)90247-c. [DOI] [PubMed] [Google Scholar]
- 25.Iwasato T, Arakawa H, Shimizu A, Honjo T, Yamagishi H. Biased distribution of recombination sites within S regions upon immunoglobulin class switch recombination induced by transforming growth factor β and lipopolysaccharide. J Exp Med. 1992;175:1539–46. doi: 10.1084/jem.175.6.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 27.Strom L, Laurencikiene J, Miskiniene A, Severinson E. Characterization of CD40-dependent immunoglobulin class switching. Scand J Immunol. 1999;49:523–32. doi: 10.1046/j.1365-3083.1999.00539.x. [DOI] [PubMed] [Google Scholar]
- 28.Shubinsky G, Schlesinger M. The CD38 lymphocyte differentiation marker: new insight into its ectoenzymatic activity and its role as a signal transducer. Immunity. 1997;7:315–24. doi: 10.1016/s1074-7613(00)80353-2. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi Y, Yasue T, Miyake K, Kimoto M, Takatsu K. CD38 ligation induces tyrosine phosphorylation of Bruton tyrosine kinase and enhanced expression of interleukin 5-receptor α chain: synergistic effects with interleukin 5. Proc Natl Acad Sci USA. 1995;92:11814–8. doi: 10.1073/pnas.92.25.11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashita Y, Miyake K, Kikuchi Y, Takatsu K, Noda S, Kosugi A, Kimoto M. A monoclonal antibody against a murine CD38 homologue delivers a signal to B cells for prolongation of survival and protection against apoptosis in vitro: unresponsiveness of X-linked immunodeficient B cells. Immunology. 1995;85:248–55. [PMC free article] [PubMed] [Google Scholar]
- 31.Kaku H, Horikawa K, Obata Y, Kato I, Okamoto H, Sakaguchi N, Gerondakis S, Takatsu K. NF-κB is required for CD38-mediated induction of Cγ1 germline transcripts in murine B lymphocytes. Int Immunol. 2002;14:1055–64. doi: 10.1093/intimm/dxf072. [DOI] [PubMed] [Google Scholar]
- 32.Mizoguchi C, Uehara S, Akira S, Takatsu K. IL-5 induces IgG1 isotype switch recombination in mouse CD38-activated sIgD-positive B lymphocytes. J Immunol. 1999;162:2812–9. [PubMed] [Google Scholar]
- 33.Horikawa K, Kaku H, Nakajima H, et al. Essential role of Stat5 for IL-5-dependent IgH switch recombination in mouse B cells. J Immunol. 2001;167:5018–26. doi: 10.4049/jimmunol.167.9.5018. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya S, Stout BA, Bates ME, Bertics PJ, Malter JS. Granulocyte macrophage colony-stimulating factor and interleukin-5 activate STAT5 and induce CIS1 mRNA in human peripheral blood eosinophils. Am J Respir Cell Mol Biol. 2001;24:312–6. doi: 10.1165/ajrcmb.24.3.4238. [DOI] [PubMed] [Google Scholar]
- 35.Zahn S, Godillot P, Yoshimura A, Chaiken I. IL-5-Induced JAB–JAK2 interaction. Cytokine. 2000;12:1299–306. doi: 10.1006/cyto.2000.0718. [DOI] [PubMed] [Google Scholar]
- 36.Temple R, Allen E, Fordham J, et al. Microarray analysis of eosinophils reveals a number of candidate survival and apoptosis genes. Am J Respir Cell Mol Biol. 2001;25:425–33. doi: 10.1165/ajrcmb.25.4.4456. [DOI] [PubMed] [Google Scholar]
- 37.Sato N, Sakamaki K, Terada N, Arai K, Miyajima A. Signal transduction by the high-affinity GM-CSF receptor: two distinct cytoplasmic regions of the common β subunit responsible for different signaling. Embo J. 1993;12:4181–9. doi: 10.1002/j.1460-2075.1993.tb06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bystrom J, Wynn TA, Domachowske JB, Rosenberg HF. Gene microarray analysis reveals interleukin-5-dependent transcriptional targets in mouse bone marrow. Blood. 2004;103:868–77. doi: 10.1182/blood-2003-08-2778. [DOI] [PubMed] [Google Scholar]
- 39.Califano A, Stolovitzky G, Tu Y. Analysis of gene expression microarrays for phenotype classification. Proc Int Conf Intell Syst Mol Biol (ISMB) 2000;8:75–85. [PubMed] [Google Scholar]
- 40.Morita S, Kojima T, Kitamura T. Plat-E. an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–6. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 41.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–14. [PubMed] [Google Scholar]
- 42.Quong MW, Harris DP, Swain SL, Murre C. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–18. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 44.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–9. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 45.Shaffer ALYuX, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 46.Reimold AM, Iwakoshi NN, Manis J, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–7. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 47.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–20. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 48.Shaffer AL, Lin KI, Kuo TC, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 49.Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marmorstein R. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem Sci. 2003;28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- 51.Reljic R, Wagner SD, Peakman LJ, Fearon DT. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J Exp Med. 2000;192:1841–8. doi: 10.1084/jem.192.12.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 1998;12:1953–61. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allman D, Jain A, Dent A, Maile RR, Selvaggi T, Kehry MR, Staudt LM. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–68. [PubMed] [Google Scholar]
- 54.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–6. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 55.Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–93. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 56.Hasbold J, Corcoran LM, Tarlinton DM, Tangye SG, Hodgkin PD. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat Immunol. 2004;5:55–63. doi: 10.1038/ni1016. [DOI] [PubMed] [Google Scholar]
- 57.Yasue T, Baba M, Mori S, Mizoguchi C, Uehara S, Takatsu K. Expression and function of CD38 on mouse follicular mantle B cells than germinal center B cells. Int Immunol. 1999;11:915–23. doi: 10.1093/intimm/11.6.915. [DOI] [PubMed] [Google Scholar]