Abstract
Macrophages play an essential role in the immune response to Mycobacterium tuberculosis (Mtb). Previous transcriptome surveys, by means of micro- and macroarrays, investigated the cellular gene expression profile during the early phases of infection (within 48 hr). However, Mtb remains within the host macrophages for a longer period, continuing to influence the macrophage gene expression and, consequently, the environment in which it persists. Therefore, we studied the transcription patterns of human macrophages for up to 7 days after infection with Mtb. We used a macroarray approach to study 858 human genes involved in immunoregulation, and we confirmed by quantitative real-time reverse transcriptase polymerase chain reaction (q-rt RT-PCR) and by enzyme-linked immunosorbent assay the most relevant modulations. We constantly observed the up-regulation in infected macrophages versus uninfected, of the following genes: interleukin-1β and interleukin-8, macrophage inflammatory protein-1α, growth-related oncogene-β, epithelial cell-derived neutrophil-activating peptide-78, macrophage-derived chemokine, and matrix metalloproteinase-7; whereas macrophage colony-stimulating factor-receptor and CD4 were down-regulated in infected macrophages. Mtb is able to withstand this intense cytokine microenvironment and to survive inside the human macrophage. Therefore we simultaneously investigated by q-rt RT-PCR the modulation of five mycobacterial genes: the alternative sigma factors sigA, sigE and sigG, the α-crystallin (acr) and the superoxide dismutase C (sodC) involved in survival mechanisms. The identified host and mycobacterial genes that were expressed until 7 days after infection, could have a role in the interplay between the host immune defences and the bacterial escape mechanisms.
Keywords: chemokines, cytokines, host–pathogen interplay, human phagocytes, virulent mycobacteria
Introduction
One-third of the world's population is infected with Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis (TB), and ratings of the TB epidemic estimate 2–3 million deaths per year.1 Despite the availability of an effective antitubercular therapy for over 40 years, the incidence of TB has increased in the last decade, especially in association with the human immunodeficiency virus pandemic. Although in most cases a vigorous immune response is mounted, Mtb can evade host defences and replicate inside macrophages, establishing a long-term-residence. The immune response against Mtb is complex, and macrophages play a peculiar role in the host response because they represent both the primary effector cells for bacterial killing and the primary habitat in which the persisting bacilli reside.2 Thus, during the infection a dynamic cross-talk occurs between the host and the pathogen, in which they reciprocally influence their gene expression profiles. The analysis of Mtb gene expression can reveal the strategies adopted by the bacteria to cope with the signals received from the host. In this context, the Mtb transcriptome has been investigated in different environmental conditions.3 Moreover, the study of the gene expression profile of human macrophages during Mtb infection may be crucial for the understanding of the regulation of antimycobacterial immunity.4–6 Analysis of the infected macrophage transcriptome revealed the induction of several genes, including those for chemokines, cytokines and intracellular signalling proteins. In particular, interleukin-10 (IL-10), IL-8, growth-related oncogene-β (GRO-β), macrophage inflammatory protein 1α (MIP-1α), RANTES and pro-inflammatory cytokines such as tumour necrosis factor-α (TNF-α), IL-1β and IL-6 were consistently up-modulated in human macrophages;5,7,8 as well as in THP-1 cells4 during virulent Mtb infection. Despite the amount of available data upon macrophage transcription, those studies elucidated regulatory networks at very early phases of mycobacterial infection (up to 48 hr post exposure). Conversely, Mtb is able to survive inside the host macrophages and create a long-lasting relationship; it continues to influence the cellular expression profile. Therefore, we investigated the immune response at the transcriptional level of human monocyte-derived macrophages (MDMs) up to 7 days of infection. We performed an array analysis on the host's genes involved in the immunoregulation and we quantified by real-time polymerase chain reaction (PCR) the modulation of the selected genes. We also investigated the concurrent expression of some Mtb genes involved in the bacterial response to the changes of the host environment. In particular, we studied three regulatory alternative sigma factors, the α-crystallin (acr) (Rv2051) and the superoxide dismutase C (sodC) (Rv0430) genes. The alternative sigma factors are fundamental regulators that re-direct the transcription machinery to a specific group of genes9 and they may be necessary for the survival of bacteria under hostile environmental conditions. We also selected the acr gene, encoding for a 16 000 molecular weight chaperonin which may protect Mtb from environmental stress, and sodC, encoding for the inducible superoxide dismutase, a protein that could participate in the detoxification of the reactive oxygen intermediates generated inside the macrophages.
Materials and methods
Human macrophage culture
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy donors by density gradient centrifugation using Lympholyte-H (Cederlane, Hornby, Ontario, Canada). Monocytes were positively separated by anti-CD14 magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). The percentage of monocytes was checked using FACscalibur with monoclonal antibodies specific for CD14 and CD3 (Becton Dickinson Bioscience, Mountain View, CA). These showed a degree of purity ≥99% without CD3 T-cell contamination (data not shown). The cells were then resuspended in RPMI-1640 (Gibco BRL Life Technologies, Paisley, UK) supplemented with 20% fetal calf serum (BioWhittaker, Verviers, Belgium), l-glutamine 2 mm, gentamicin 5 μg/ml (Gibco BRL) and cultured for 7 days at 37° in 5% CO2, to generate MDMs.
Mtb infections of human macrophages
Mtb H37Rv was transferred every 2 months, in Sauton's medium, and allowed to grow as a surface pellicle;10,11 as previously described.12 To infect macrophages, we harvested layers of bacilli, sonicated the mycobacteria to ensure a homogeneous suspension and then dicied them into aliquots before storing them at −80° until use. Representative samples were thawed and colony-forming units (CFUs) per ml were enumerated by plating on Middlebrook 7H10 (Becton Dickinson, Cockeysville, MD) supplemented with Middlebrook oleic acid-albumin-dextrose-catalase (OADC). Inocula were prepared by suspending mycobacteria in RPMI-1640 medium, which was sonicated for 2 min to allow any clumps to settle and was used to infect human MDMs at a multiplicity of infection (MOI) of 5. After 3 hr, the cells were washed extensively with phosphate-buffered saline (PBS), to remove all extracellular bacilli, and placed in culture for another 1, 3 or 7 days.
Determination of CFUs
Bacilli in the culture supernatants and intracellular bacteria were plated immediately after 3 hr of incubation with cells, to determine the percentage of phagocytosis, and at 1, 3, 7 and 11 days after infection. Intracellular bacteria were obtained by lysing the cells with sterile PBS containing 0·1% saponin (Sigma, St Louis, MO) and released bacilli were serially diluted in PBS containing 0·01% Tween-80 (Merck, Darmstadt, Germany). Finally, bacteria were plated on Middlebrook 7H10 with OADC in triplicate. CFUs were counted after 14 days of incubation at 37° and plates were maintained for 30 days to ensure that no additional CFUs appeared.
Detection of cytotoxicity
Mycobacterium-induced cell death was monitored by measuring the presence of lactate dehydrogenase (LDH) in cell culture supernatant using the CytoTox96 kit (Promega Corp., Madison, WI), in accordance with the manufacturer's instructions. The percentage cytotoxicity was calculated as follows: 100 × (experimental release − spontaneous release)/(total release − spontaneous release), where spontaneous release is the amount of LDH activity in MDM lysates.
Acridine orange staining
To determine the percentage of infected cells, the nucleic acids of the cells and of the mycobacteria were stained with acridine orange. This dye binds to both eukaryotic and prokaryotic nucleic acids and it also allows the differentiation of single (orange-red staining) and double (green-yellow staining) nucleic acid strands, thus indicating the replicative form of DNA. The medium overlying the infected cells attached on chambered slides (Becton Dickinson) was gently aspirated. Duplicate monolayers were prepared for each experimental condition. The monolayer was fixed in 4% paraformaldehyde for 10 min, permeabilized with Triton X-100 0·2% (Sigma) for 4 min and stained with acridine orange (Merck, Germany) solution for 3 min. Then, cells were analysed using a confocal fluorescence microscope.
RNA extraction
Cells were drained of medium and the adherent cells were resuspended in ice-cold 4-m guanidium isothiocyanate (GTC) lysis solution. Total RNA was extracted as described by Mangan et al.13 This protocol allowed the enrichment of mycobacterial RNA over total RNA and was used to perform real-time PCR on Mtb genes. The supernatant obtained after the first wash in GTC was used to re-extract RNA using the 4-m GTC single-step method.14 The RNA obtained by this second extraction is represented mainly by host RNA (data not shown) and was used to perform array and real time on the cytokine experiments. The RNA was examined in a 1·5% denaturing agarose gel for degradation and quantified by UV spectroscopy at 260/280 nm. Then digested with 1 unit DNase (Invitrogen, Carlsbad, CA) for 15 min at room temperature to avoid any genomic contamination. DNAse I-digested RNA was used for macroarray and for quantitative real-time PCR.
Preparation of 33P-radiolabelled probes
The 33P-radiolabelled cDNA probes for array hybridization were prepared by reverse transcription (Sigma-Genosys, Woodlands, TX, USA) according to manufacturer's instructions, using 1 μg DNaseI-treated total RNA.
Human cDNA expression arrays
Panorama human cytokine gene arrays (Sigma-Genosys, USA) consist of a matched set of charged nylon membranes containing PCR products spotted. Each array contains 858 different human cytokine-related genes and experiments are performed as previously described.4 Briefly the arrays were prehybridized for 1 hr at 65° with hybridization solution (Sigma-Genosys, USA). The filters were then incubated with the denatured, labelled cDNAs for 12–18 hr at 65° in a hybridization oven. The filters were washed and exposed to a phosphor-imager screen (Amersham Biosciences, Little Chalfont, UK) for 48 hr and the resulting hybridization signals were quantified using Phosphor-imager Typhoon (Molecular Dynamics, Amersham Biosciences) and Array Vision 7·0 software (Imaging Research Inc., St. Catharines, Canada). The intensity of each spot was corrected for background levels and normalized for differences in probe labelling using the average values for all genes. Genes showing a change of three-fold or more in intensity were considered to be up- or down-regulated following infection.
Reverse transcription (RT) and quantitative real-time PCR
Macrophages and Mtb-selected genes were examined for their expression levels by quantitative real-time PCR on the cDNA generated from the same RNA samples that were used for the macroarray experiments. We selected the following macrophage genes because their modulation was confirmed in all three array experiments: IL-1β, IL-8, epithelial cell-derived neutrophil-activating peptide 78 (ENA-78), macrophage-derived chemokine (MDC), GRO-β, MIP-1α, matrix metalloproteinase-7 (ΜMP-7), macrophage colony stimulating factor-receptor (M-CSF-R) and CD4. In addition we chose also TNF-α, interferon-γ (IFN-γ) and IL-10 for their importance in antimycobacterial immunity. L34 was used as an internal control because it was shown to be stable with differentiation induction. The chosen mycobacterium genes were sigA (Rv2703), sigE (Rv1221), sigG (Rv0182c), acr (Rv2031c), sodC (Rv0432) and 16S rRNA as an internal control. One microgram of DNAseI-treated total RNA was retro-transcribed using random examers and Molony murine leukaemia virus reverse transcriptase (Invitrogen, Paisley, UK), according to the manufacturer's instruction. Quantification of PCR products used the ABI PRISM 7000 SDS (Applied Biosystems, Foster City, CA, USA). The SYBR Green I PCR Core kit was used to produce fluorescently labelled PCR products, and we monitored increasing fluorescence during repetitive cycling of the amplification reaction. The only alteration of the manufacturer's protocol was to add 1·25% formamide15 in the reactions for Mtb genes.16 For all primers, the following temperature cycling profile was used: 2 min at 50° and 10 min at 95° followed by 10 seconds at 95° and 1 min at Tm for 40 cycles. The Tm temperature for each primer set and their sequence are reported in Table 1. All primers designed for eukaryotic genes were RNA-specific and non-reactive with DNA. PCR for Mtb genes were also performed on RT-negative control samples, to exclude any genomic DNA contamination (data not shown). Each condition was performed in duplicate. At the end of the amplification, a dissociation curve of the PCR product was performed to confirm the specificity of the product and each PCR product was also run on a 2% agarose gel. The results of the real time PCR are expressed as a threshold cycle (Ct). The Ct represents the number of reaction cycles at which the reporter fluorescence raises above a set baseline threshold, and indicates that the DNA amplicon is replicating exponentially. The expression level of each gene was normalized using the L34 housekeeping gene and the relative level of each transcript, was obtained by the 2–ΔCt method.17
Table 1.
Primers used for real-time polymerase chain reaction
| Gene | Sense | Antisense | Annealing temp. (°) |
|---|---|---|---|
| IL-1β | 5′-TGCCTTAGGGTAGTGCT-3′ | 5′-GCGGTTGCTCATCAGA-3′ | 58 |
| IL-8 | 5′-TGCTAAAGAACTTAGATGTCAGTGCAT-3′ | 5′-TGGTCCACTCTCTCAATCACTCTCA-3′ | 68 |
| ENA-78 | 5′-AGAGCTGCGTTGCGTTTGTT-3′ | 5′-TACCACTTCCACCTTGGAGCAC-3′ | 60 |
| GRO- β | 5′-GCTTATTGGTGGCTGTTCCTGA-3′ | 5′-GCTCAAACACATTAGGCGCAA-3′ | 60 |
| MMP-7 | 5′-AGCCAAACTCAAGGAGATGC-3′ | 5′-ACTCCACATCTGGGCTTCTG-3′ | 56 |
| MDC | 5′-TGCGCGTGGTGAAACACTT-3′ | 5′-TAGGCTCTTCATTGGCTCAGCT-3′ | 68 |
| MIP-1α | 5′-GCAACCAGTTCTCTGC-3′ | 5′-CTGGACCCACTCCTCA-3′ | 58 |
| IFN-γ | 5′-GGCTGTTACTGCCAGGACCCATATGT-3′ | 5′-GATGCTCTTGCACCTCGAAACAGCAT-3′ | 60 |
| TNF-α | 5′-AGGCGGTGCTTGTTCCTC-3′ | 5′-GTTCGAGAAGATGATCTGACTGCC-3′ | 60 |
| M-CSF-R | 5′-AGGCTTTCAATAGCACCTTGCC-3′ | 5′-CCCACACCTTCTTCGACTGTTG-3′ | 66 |
| CD4 | 5′-CTCCCCACTGCTCATTTGGAT-3′ | 5′-AACAGTCCCATGCTCCATGCT-3′ | 60 |
| L34 | 5′-GGCCCTGCTGACATGTTTCTT-3′ | 5′-GTCCCGAACCCCTGGTAATAGA-3′ | 60 |
| sig A | 5′-CGATGAGCCGGTAAAACGC-3′ | 5′-GAGCCACTAGCGGACTTCGC-3′ | 62 |
| sig E | 5′-CCAGCATGTCTCATCCCCAA-3′ | 5′-GGCCTTGTCCCCGGTG-3′ | 62 |
| sig G | 5′-CGTTCGAGGCTTATGACATCGACC-3′ | 5′-CGATCAACGAAATCAGGCGCATAT-3′ | 62 |
| acr | 5′-CGACACCCGGTTGATGC-3′ | 5′-CGCTCGGCCTTGATGGTC-3′ | 60 |
| sod C | 5′-GCAGGTACGCGGTGACGGTT-3′ | 5′-CGTAGCGTTCTGGCGGAATG-3′ | 64 |
| 16S rRNA | 5′-GCACCGGCCAACTACGTG-3′ | 5′-GAACAACGCGACAAACCACC-3′ | 64 |
Cytokine determination by enzyme-linked immunosorbent assay (ELISA)
TNF-α, IFN-γ, MDC and ENA-78 proteins released in uninfected and mycobacteria-infected MDM supernatants were tested by ELISA, using antibody pairs, as indicated by the supplier (Endogen and R & D Systems, Brebieres, France).
Statistical analysis
Results are expressed as mean ±SD. Statistical significance was determined using Student's t-test for normally distributed data with equal variances.
Results
Determination of the best experimental conditions for transcriptome analysis
The infectivity of Mtb (H37Rv strain) in human macrophage was determined to identify the time point at which Mtb still replicates inside the cells, without causing a massive cellular death. We generated MDMs from purified monocytes through a 7-day incubation in complete medium, and infected differentiated cells with Mtb at an MOI of 5. Three hours after Mtb exposure, about 10% of bacteria were phagocytosed, whereas the majority of bacteria remained in the extracellular compartment (Fig. 1a). We found that bacilli are able to replicate inside the macrophages and their release drastically increased after 11 days of infection (Fig. 1b). The cytotoxicity induced by bacteria within the cells, although displaying a significant increase at 7 days, was never above 11 ± 2% (Fig. 1c). In the same experiments, we measured by confocal microscopy the percentage of infected cells after 3 hr of infection (time 0) and 1, 3 and 7 days after infection, finding a peak of 40 ± 7% at 7 days, as reported in Fig. 1(d). The morphology of macrophages showed that, in the course of infection, cells were tightly adherent to the plastic surface. The number of both infected macrophages and internalized bacteria in active replication, increased over time, as shown by acridine-orange staining (Fig. 1d,e). From these observations, we chose to use 7 days of culture after infection in our experiments because at this time-point we observed the highest percentage of infected cells but still low levels of cytotoxicity.
Figure 1.
Survival of M. tuberculosis (Mtb) inside human macrophages. Human MDMs infected with Mtb H37Rv (MOI 5). The percentages of intracellular and not phagocytosed bacilli immediately after infection were determined by CFU assay and compared to inoculum (a). Lysed infected macrophages and released bacteria in the culture supernatants at 1, 3, 7 and 11 days after infection were plated on Middlebrook 7H10 with OADC and the CFU were counted after 14 days (b). Cytotoxicity induced by Mtb in human MDMs was measured by LDH activity (c). Values are the means ± SD from four independent experiments. The percentage of infected cells was measured, after 3 hr of infection (time 0) and 1, 3, 7 days after infection, by acridine-orange staining (d). and the representative pictures of infected MDMs, are shown (e). A significant difference was detected with respect to 1 and 3 days post-infection (*P < 0·05).
Transcription pattern of Mtb-infected human macrophages
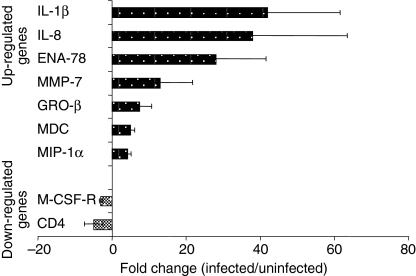
We performed a macroarray analysis to study the gene expression profile in human MDMs after 7 days of infection with Mtb. We used membranes that included 858 human genes belonging to different immunomodulatory factors and their receptors. Genes showing a change of at least threefold in normalized density values were considered as up- or down-regulated following Mtb infection. An overall analysis of results showed that less than 5% of the genes was differentially expressed by uninfected and infected macrophages in at least one experiment out of three (Table 2). Only a restricted number of genes showed the same modulation in all three independent array experiments, and this list is reported in Fig. 2. The genes consistently up-regulated included those for cytokines, chemokines and inflammatory factors. In particular IL-1β, IL-8 and ENA-78 genes showed about 30-fold induction ratio. Other chemokines such as MIP-1α, GRO-β and MDC were induced at lower levels (four- to sevenfold induction). Interestingly, MMP-7 was also induced 17-fold during the infection. Finally, only two genes among those investigated were always down-modulated: M-CSF-R and the CD4 antigen.
Table 2.
Overview of modulation of macrophages' genes at 7 days of Mtb infection obtained by array
| Fold change uninf./infected | Fold change uninf./infected | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Exp. | Exp. | ||||||||||
| Coexpressed genes | Down-regulated genes | 1 | 2 | 3 | Mean ± SD | Up-regulated genes | 1 | 2 | 3 | Mean ± SD | |
| β-actin | Telomerase-related | Cytokines and rec. | Chemokines | ||||||||
| GAPDH | TERT | M-CSF-R | + | + | + | 3·0 ± 0·5 | ENA-78 | + | + | + | 28·0 ± 13·6 |
| α-tubulin | TGF-β superfamily | SPARC | + | + | 6·9 ± 4·0 | IL-8 | + | + | + | 37·9 ± 25·6 | |
| β2microglobulin | BMP-2 | Cell surface proteins | + | MIP-1α | + | + | + | 3·9 ± 0·7 | |||
| CiclophilinA | BMP-7 | CD4 | + | + | 5·0 ± 2·5 | MDC | + | + | + | 4·6 ± 0·9 | |
| HLA-A0201 | BMP-9 | CD9 | + | + | 4·5 ± 1·9 | GRO-β | + | + | + | 7·3 ± 3·2 | |
| L19 | Eph family | TOSO | + | 3·4 ± 0·2 | GRO-γ | + | + | 19·4 ± 15·3 | |||
| Transferrin R | EphB3 | TNF superfamily | GRO-α | + | |||||||
| Cytokines and rec. | EphB4 | DR6 | + | Rantes | + | ||||||
| GAS1 | Ephrin-A2 | TNFSF14/LIGHT | + | I-309 | + | ||||||
| PDGF-B chain | Cell surface proteins | TAJ | + | + | + | MIP-1β | + | ||||
| SCGF | Endoglycan | Integrin | Interleukin | ||||||||
| Osteopontin | C3 | Integrin-α3 | + | IL-1β | + | + | + | 52·0 ± 19·1 | |||
| INSRR | CD14 | Integrin-αM | + | Protease or rel. factors | |||||||
| PD-ECGF | Weight regulation | TGF-β superfamily | MMP-7 | + | + | + | 17·0 ± 9·3 | ||||
| IFN-γ R2 | HCRTR1 | BMP-10 | TIMP-1 | + | |||||||
| GAS6 | BMCP1/SLC25A14 | BMP RIIA | + | Urokinase | + | + | 4·7 ± 1·7 | ||||
| WNT-16 | Cell cycle regulators | G-protein coupled rec. | Signals transduction | ||||||||
| CNTF Rα | CDKN1A | GPR-2 | + | + | TRAF1 | + | + | 6·7 ± 4·0 | |||
| Binding proteins | Cyclin D3 | GPR-3 | CARDIAK | + | |||||||
| LTBP4 | Cyclin D2 | Binding proteins | NFKB2 | + | |||||||
| Endoglin | Interleukin rec. | LTBP3 | STAT1 | + | |||||||
| IGF Binding Protein | IL-17R | Chemokines | + | Cytokines and rec | |||||||
| LTBP2 | IL-2Rγ | PARC | + | PBEF | + | + | 6·8 ± 3·2 | ||||
| LAMR1 | IL-22R | Cell cycle regulators | Interl. and chemok. rec. | ||||||||
| Galectin-3 | IL-15Rα | CDK9 | CCR-7 | + | |||||||
| Signals transduction | IL-1R antagonist | Cytokines and rec. | Adhesion molecule | ||||||||
| TRADD | Neurotrophic group | IL-4Rα | + | ICAM-1 | + | + | 4·3 ± 0·7 | ||||
| SKI | Neurturin | Weight regulation | Cell cycle regulators | ||||||||
| SOCS1 | GFRA4 | UCP2 | + | + | 4·5 ± 2·2 | Cyclin B1 | + | ||||
| IRS2 | APP | TGF-β superfamily | |||||||||
| JUN B | Ret | Activin A (bA subunit) | + | + | 137 ± 187 | ||||||
| PTPN 18 | TNF superfamily | TNF superfamily | |||||||||
| INPP5D | TNFSF3/LT-β | TNFSF2/TNFα | + | ||||||||
| FAST1 | TACI | TNFSF5/CD40 | + | ||||||||
| EP300 | Adhesion molecules | Apoptosis-related | |||||||||
| AATK | ICAM-3 | Granzyme B | + | ||||||||
| MAP3K3 | L-Selectin | Cox-2 | + | + | 33·4 ± 21·1 | ||||||
| PTPN6 | Integrin | ||||||||||
| YWHAZ | Integrin-β5 | ||||||||||
| CEBPB | Integrin-α5 | ||||||||||
| PKC-α | Integrin-β1 | ||||||||||
| JUN D | Integrin-β2 | ||||||||||
| GRB2 | Chemokines | ||||||||||
| FADD | Fractalkine | ||||||||||
| Proteases or rel. actors | Angiogenic factors | ||||||||||
| MMP-14 | VEGF-B | ||||||||||
| UBB | Interleukin | ||||||||||
| MMP-9 | IL-16 | ||||||||||
| TIMP-2 | Apoptosis-related | ||||||||||
| PLAU | TP73 | ||||||||||
| ADAM8 | Other factors | ||||||||||
| Developmental factors | GL50/B7RP-1 | ||||||||||
| NTN2L | MMCP-7-LIKE-2 | ||||||||||
| Notch-3 | AVP | ||||||||||
| Notch-1 | |||||||||||
| LFNG | |||||||||||
+ indicates fold-change of at least 3-fold.
Figure 2.
Transcription pattern of M. tuberculosis-infected human macrophages. Human MDMs were infected with Mtb H37Rv (MOI 5) for 3 hr, washed and total RNA was processed for cDNA expression array at 7 days after infection. Genes showing a change (infected/uninfected) of three-fold or more in intensity confirmed in three independent experiments were represented as up- or down-regulated following infection. The average fold-change values ± SD, observed in three independent experiments, are displayed.
Time–course analysis of selected macrophage genes
To confirm the results obtained by arrays and to investigate the expression in a time–course, we tested the same RNA samples at 1, 3 and 7 days post infection by quantitative real-time PCR, focusing only on the genes modulated in all three independent macroarray experiments. According to array analysis, the real-time quantitative assay confirmed that Mtb infection of MDMs had induced the up-regulation of IL-1β, IL-8, ENA-78, MMP-7, GRO-β, MDC and MIP-1α genes already after 1 and 3 days, and this lasted until the 7th day of culture. On the other hand, macrophages reduced the expression of M-CSF-R and CD4 genes in the same time period (Fig. 3a). The gene expression patterns obtained using both the experimental methods displayed the same trend (Fig. 3b). The time–course results indicate that the gene transcripts induced or repressed at the 7th day are similarly regulated during the earlier phases of infection.
Figure 3.
Confirmation of macroarray results by quantitative real-time PCR. Macrophage samples from three donors were prepared for quantitative real-time PCR as for the cDNA array hybridization experiments and expression levels of the indicated genes were measured, by SYBR Green incorporation during PCR. Results are shown as average fold-change values (uninfected/infected or infected/uninfected) ± SD, of two independent experiments, at 1, 3 and 7 days of infection (a) and results, of each gene, at 7 days of infection were compared with fold induction obtained by array (b).
To substantiate our findings, we validated at protein level the modulation of two chemokines associated with TB in this work for the first time: ENA-78 and MDC. We found that also at the protein level these two chemokines were up-regulated by Mtb-infected macrophages compared to non-infected cells (Fig. 4).
Figure 4.
Protein secretion of ENA-78, MDC, TNF-α and IFN-γ. Levels of TNF-α, IFN-γ, ENA-78 and MDC in the supernatants of MDMs at 3 hr (on day 0) and at 1, 3 and 7 days after infection were tested by ELISA and compared with cytokine secretion in the supernatants of uninfected MDMs. Data shown are the mean of triplicates ± SD, of one representative experiment out of five performed. Statistical significance within each experiment was determined by Student's t-test; *P < 0·05 infected versus uninfected, except for MDC, where *P < 0·05 T7 infected versus T1 infected.
Even though macroarrays represent a valuable screening method,18 some genes were detectable only in one experiment (TNF-α) (Table 2), or were undetectable (IFN-γ, IL-10). Since these molecules have an important and controversial role in the human immune response during TB infection,19,20 we investigated their expression by real-time PCR. The induction of TNF-α and IFN-γ mRNAs was detected at all time-points by real-time assay in Mtb-infected macrophages (Fig. 5). The lack of detection of the mentioned mRNA-relative signals in the array was probably because of the lower sensitivity of this technique in revealing low-abundance messages. We found that there was a threshold value of intensity, below which the transcript were undetectable by array, although they were assessable in real-time assay at a high number of Ct. As shown in Fig. 6, for example, IFN-γ expression in infected macrophages was detected at higher Ct compared to the other up-regulated genes from the same cDNA sample. We also confirmed the up-regulation of TNF-α and IFN-γ release in Mtb-infected macrophages by quantifying the amount of proteins secreted in the supernatants, by ELISA. The amount of TNF-α protein peaked early after infection and remained constant until day 7. In contrast, the release of IFN-γ protein dramatically increased only 7 days after infection (Fig. 4), although the mRNA fold induction was also evident at early time-points. Finally, the analysis of IL-10 expression revealed that Mtb-infected MDMs enhanced the relative mRNA transcription 1 day after infection, which then rapidly decreased, thus confirming the array experiment at 7 days, where IL-10 expression was always undetectable (Fig. 5).
Figure 5.
Time–course analysis of IL-10, TNF-α and IFN-γ transcription. Macrophage samples from three donors were prepared for quantitative real-time PCR as for the cDNA array hybridization experiments and expression levels of genes were measured by SYBR Green incorporation during PCR. Results are shown as average fold-change values (infected/uninfected) ± SD, of two independent experiments, at 1, 3 and 7 days of infection.
Figure 6.
Comparison of mRNA detection level measured by array and real time. The values of mRNA detection, obtained by array and real-time PCR, were compared on the same sample; the results are reported as normalized density and cycle threshold Ct, respectively, of a representative of three independent experiments.
Changes of Mtb transcripts during intracellular survival
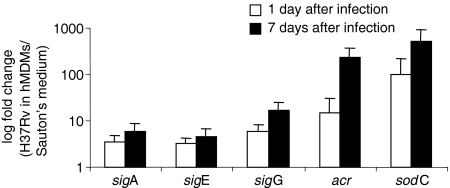
To understand the bacterial response to the hostile cellular environment, we investigated the Mtb gene expression in the same sample used for the macrophage profiling. We focused our analysis on a small group of mycobacterial genes, involved in different mechanisms of Mtb survival. We studied the expression of three RNA polymerase sigma factors (sigA, sigE, and sigG) and the acr and sodC genes by real-time RT-PCR. As shown in Fig. 7, the three sigma factors were up-regulated during mycobacterial growth into the cells at 1 and 7 days, compared to Mtb growth in synthetic medium. Similarly, the transcription of acr and sodC genes was increased during Mtb infection of human macrophages. For all the tested genes, we did not find any significant differences in the up-regulation between day 1 and day 7 post-infection.
Figure 7.
Intracellular change in M. tuberculosis (Mtb) RNA. The sigA, sigE, sigG, acr and sodC mRNA expression at the 1st and 7th days of intracellular infection were compared with the expression of the same genes in bacilli from in vitro culture. Individual values of each gene were corrected to the 16S rRNA level in the same sample. The average fold-change values ± SD, observed in two independent experiments, are shown.
Discussion
The different manifestations and outcomes of Mtb infection reflect the balance between the bacillus and the host defence mechanisms. Indeed, soon after the contact between Mtb and macrophages, the transcription programmes of both bacterial and host cells start to regulate their interaction. Studies in cultured macrophages can approximate many of the host–pathogen interactions occurring in vivo, and genes expressed selectively, in the early phase of infection (24–48 hr), have been reviewed.36 For this reason we investigated the gene expression profile of MDMs, infected with a virulent Mtb strain, during 7 days of infection. At this time-point, the MDM population studied includes cells at different stages of infection (infected early, infected later, or re-infected) and also uninfected cells; this situation could be similar to the heterogeneous microenvironment occurring at the site of infection, in the in vivo TB.
Macroarray analysis of macrophage genes revealed that 142 out of 858 genes were expressed and 64% of them were constitutively expressed. In contrast, 16% and 20% of the expressed genes, were, respectively, up- or down-regulated during Mtb infection in at least one experiment out of three. The large group of constitutively expressed genes includes several cytokines, signal transduction proteins and other immune-related genes, which suggests a preactivation of uninfected macrophages during the in vitro culture (Table 2). This study describes how human macrophages up-regulate, mainly, genes encoding for molecules with a chemotactic role, indicating that they maintain, after 7 days of Mtb–MDMs interaction, the capacity to recruit other cells at the site of infection. In particular MIP-1α, IL-8, GRO-β, ENA-78, IL-1β, and MDC were up-regulated from the first to the 7th day of infection. The bronchoalveolar lavage (BAL) fluid of patients with active pulmonary TB contains increased levels of several chemokines21 but not MIP-1α. This chemokine has never been associated with TB patients while it has been previously found in the BAL fluid of patients with other lung diseases.22,23 IL-8, instead, was induced by Mtb infection of alveolar macrophages, other than in lavage fluid of patients with TB.24 GRO-β and ENA-78, with IL-8, are potent neutrophil chemotactic factors and are defined as major mediators of inflammation.25 All these factors, produced by macrophages, could therefore play an important role during TB infection, together with IL-1β which is known to be involved in the granulomatous inflammation.26 Interestingly we observed the up-regulation of another chemoattractant, MDC, that has a crucial role for dendritic cell and natural killer cell functions, as well as for T helper type 2 cell activation,27,28 but has never been associated with TB. Furthermore, the role of this molecule could be related to the bactericidal activity of macrophages, by induction of both a respiratory burst and the release of lysosomal enzyme.29 The results of the ELISA for MDC, showed its up-regulation in the course of Mtb infection.
Therefore, the lasting production of such chemotactic factors after one week of infection suggests that one of the principal mechanisms of defence, exerted by the infected MDMs, is the recruitment of inflammatory cells at the site of infection. Other genes, such as IL-10, IL-6 and RANTES, were not constantly detected, in all the experiments, although their activation at early time-points has been reported elswehere.5 In our study, and in previous works,30 analysis of the IL-10 gene in Mtb-infected macrophages revealed a peak of induction at 1 day, but a decrease at the later stage of infection. IL-10 is known to be a potent suppressor of the functions of activated macrophages and it has been recently associated with the ability of Mtb to rapidly grow within macrophages.31,32 IL-10 could interfere with host defence mechanisms against Mtb down-regulating the production of IFN-γ and TNF-α.33 In this work, we did not point out an association between IL-10 induction and IFN-γ or TNF-α repression, at least at the transcriptional level. In our experiments, in fact, we observed a sustained induction of TNF-α transcripts and protein secretion, during the whole period of Mtb infection and the induction of IFN-γ in Mtb-infected macrophages, mainly at later time-points of infection. It has been shown that macrophages are an important source of IFN-γ, under physiological or pathological conditions,34 particularly in response to Mtb infection.35,36 Since it has been demonstrated that Mtb may counteract the host's defence mechanisms, inhibiting the macrophage response to the exogenous IFN-γ,37 it could be interesting to investigate the effect of the endogenous IFN-γ on the same macrophage's antimicrobial activities. Interestingly, among the up-regulated genes, we reported a significant increase of MMP-7 transcripts. Mycobacterial infection is known to activate also other matrix metalloproteinases, such as MMP-9, -11 and −14.7,38 Since MMP-7 was reported as an up-regulated gene in hypoxic macrophages in vitro39 we hypothesize that the hypoxic environment, generated during mycobacterial infection,40 could be related to persistent induction of the MMP-7 gene. The hypoxic intracellular condition, generated during infection, may, most probably, influence the transcription profile of both bacteria and host cells. The acr mycobacterial gene, for example, encodes a chaperonin that is induced by Mtb in hypoxic conditions;41,42 and we analysed the expression of this gene on the same infected sample. We reported that, actually, acr was strongly induced by mycobacteria grown within human macrophages, compared to those grown in synthetic medium culture. We might therefore argue that, in response to a hypoxic environment, macrophages induce a strong up-regulation of MMP-7, as a possible antimicrobial peptide,43 while Mtb induces an overexpression of the acr gene to withstand the action of toxic oxygen metabolites.41 To extend the analysis of Mtb gene induction during intracellular survival, we studied other genes correlated with Mtb growth in particular conditions. The sodC gene, which encodes a mycobacterial superoxide dismutase, was selected because the superoxide dismutase activity decreases during the non-replicating persistence (NRP) state of Mtb, in the Wayne model.40 The up-regulation of sodC that we observed in macrophages after 1 and 7 days of infection could suggest that Mtb persists and replicates inside human macrophages, without entering the NRP state, and the pathogen may induce sodC in response to reactive oxygen intermediate production by activated macrophages to destroy Mtb. We selected also three sigma factors, sigA, sigG and sigE, because they are involved in mycobacterial gene expression regulation under different growth conditions.9 In particular sigA seems to modulate the expression of genes that contribute to virulence, enhancing growth in human macrophages and during the early phases of pulmonary infection in vivo.44 Our results revealed that these three sigma factors were up-regulated by mycobacteria grown within human macrophages, compared to those grown in synthetic medium culture, suggesting their possible role to redirect the Mtb transcription, orchestrating its survival therein.
The array analysis on macrophages revealed also the down-regulation of some immunoregulator genes, after infection with a virulent Mtb. The M-CSF-R and CD4 membrane receptors were down-regulated in all the array experiments and their modulation was confirmed by real-time quantitative PCR. In particular M-CSF-R mediates the biological effects of macrophage colony-stimulating factor (M-CSF), which is involved in macrophage differentiation and survival45 besides being essential for the macrophage response against bacterial infections.46 Thus, the Mtb-induced down-regulation of M-CSF-R could be linked to mycobacterial survival mechanisms within human cells. The down-regulation of the CD4 gene is not easy to explain because the precise role of CD4 molecule on monocytes and macrophages is not clear.47 It has been previously described as a receptor for the chemotactic factor IL-1648 and it was shown that the recombinant IL-16 is able to down-regulate the CD4 expression on the membrane.49 In our experiments, the similar expression of IL-16 in both infected and uninfected macrophages led us to assert that the down-modulation of CD4 is not regulated by its ligand but is most likely dependent on Mtb infection.
The present study contributed to confirm the limited sensitivity of the macroarray assay. The genes subjected to more rigorous analysis by quantitative RT-PCR revealed that the array may estimate the relevant changes of the most expressed genes, while the real-time assay is able to detect the fine differences of genes that are only faintly expressed. These differences may depend on the cDNA amplification step of the PCR method, and on the fixed dynamic range of most array scanners, which renders it difficult to correctly measure the minimal value of intensity changes.50
Altogether our data identify a group of genes that are modulated during mycobacterial infection, and that could be relevant for the establishment of a cut-and-thrust interaction between macrophages and mycobacteria, the outcome of which is delicately balanced. After 7 days of infection, in fact, Mtb has not yet fully succeeded in damaging the macrophage, and, at the same time, the macrophage has not fully succeeded in controlling Mtb.
Acknowledgments
We are very grateful to our colleague Dr Giovanni Auricchio for critical comments on the paper. This work was supported by the Target Oriented Project, Ministry of Health, Vaccine for HCV and TB (1999–2003); by FIRB-MIUR 2001, RBNE01PPTTF Project; by Target Oriented Project, Ministry of Health, Human Anti-infectious Vaccines for the Third Millennium, 2002; and by Target Oriented Project, Ministry of Health, Epidemiology and Drug-resistance of TB in Disadvantaged Social Groups, 2003.
Abbreviations
- acr
α-crystallin
- BAL
bronchoalveolar lavage
- CFUs
colony-forming units
- Ct
threshold cycle
- ELISA
enzyme-linked immunosorbent assay
- ENA
epithelial cell-derived neutrophil-activating peptide
- GRO
growth-related oncogenes
- GTC
guanidium isothiocyanate
- IFN
interferon
- IL
interleukin
- LDH
lactate dehydrogenase
- M-CSF
macrophage colony-stimulating factor
- M-CSF-R
macrophage colony-stimulating factor-receptor
- MDC
macrophage-derived chemokine
- MDMs
monocyte-derived macrophages
- MIP
macrophage inflammatory protein
- MMP
matrix metalloproteinase
- MOI
multiplicity of infection
- Mtb
Mycobacterium tuberculosis
- NRP
non-replicating persistence
- OADC
oleic acid-albumin-dextrose-catalase
- PBMCs
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RT
reverse transcriptase
- sod
superoxide dismutase
- TB
tuberculosis
- TNF
tumour necrosis factor
References
- 1.Raviglione MC. The TB epidemic from 1992 to 2002. Tuberculosis (Edinb) 2003;83:4–14. doi: 10.1016/s1472-9792(02)00071-9. [DOI] [PubMed] [Google Scholar]
- 2.Co DO, Hogan LH, Kim SI, Sandor M. Mycobacterial granulomas: keys to a long-lasting host-pathogen relationship. Clin Immunol. 2004;113:130–6. doi: 10.1016/j.clim.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Schnappinger D, Ehrt S, Voskuil MI, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ragno S, Romano M, Howell S, Pappin DJ, Jenner PJ, Colston MJ. Changes in gene expression in macrophages infected with M. tuberculosis: a combined trasncriptome and proteomic approach. Immunology. 2001;104:99–108. doi: 10.1046/j.0019-2805.2001.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giacomini E, Iona E, Ferroni L, Miettinen M, Fattorini L, Orefici G, Julkunen I, Coccia EM. Infection of human macrophages and dendritic cells with Mycobacterium tuberculosis induces a differential cytokine gene expression that modulates T cell response. J Immunol. 2001;166:7033–41. doi: 10.4049/jimmunol.166.12.7033. [DOI] [PubMed] [Google Scholar]
- 6.Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA. 2002;99:1503–8. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JP, Rought SE, Corbeil J, Guiney DG. Gene expression profiling detects patterns of human macrophage responses following M. tuberculosis infection. FEMS Immunol Med Microbiol. 2003;39:163–72. doi: 10.1016/S0928-8244(03)00223-2. [DOI] [PubMed] [Google Scholar]
- 8.McGarvey JA, Wagner D, Bermudez LE. Differential gene expression in mononuclear phagocytes infected with pathogenic and non-pathogenic mycobacteria. Clin Exp Immunol. 2004;136:490–500. doi: 10.1111/j.1365-2249.2004.02490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, Smith I. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol. 2004;186:895–902. doi: 10.1128/JB.186.4.895-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen BW. Mycobacteria. General culture methodology and safety considerations. Meth Mol Biol. 1998;101:15–30. doi: 10.1385/0-89603-471-2:15. [DOI] [PubMed] [Google Scholar]
- 11.Petricevich VL, Ueda C, Alves RC, de Silva MA, Moreno C, Melo AR, Dias da Silva W. A single strain of Mycobacterium bovis bacillus Calmette–Guérin (BCG) grown in two different media evokes distinct humoral immune responses in mice. Braz J Med Biol Res. 2001;34:81–92. doi: 10.1590/s0100-879x2001000100010. [DOI] [PubMed] [Google Scholar]
- 12.Mariani F, Cappelli G, Riccardi G, Colizzi V. Mycobacterium tuberculosis H37Rv comparative gene-expression analysis in synthetic medium and human macrophages. Gene. 2000;253:281–91. doi: 10.1016/s0378-1119(00)00249-3. [DOI] [PubMed] [Google Scholar]
- 13.Mangan JA, Monahan IM, Butcher PD. Gene expression during host–pathogen interactions: approaches to bacterial mRNA extraction and labelling for micro array analysis. In: Wren BW, Dorrel N, editors. Functional Microbial Genomics. London: Academic Press; 2002. pp. 137–51. [Google Scholar]
- 14.Chomczynski P, Sacchi N. Single-step method of RNA isolation by guanidium thiocyanate phenol–chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar G, Kapelner S, Sommer SS. Formamide can dramatically improve the specificity of PCR. Nucl Acids Res. 1990;18:7465. doi: 10.1093/nar/18.24.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grassi M, Volpe E, Colizzi V, Mariani F. An improved real-time PCR assay for the detection of GC-rich and low abundance templates of Mycobacterium tuberculosis. J Microbiological Methods. 2006;64:406–10. doi: 10.1016/j.mimet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Hess S, Peters J, Bartling G, Rheinheimer C, Hegde P, Magid-Slav M, Tal-Singer R, Klos A. More than just innate immunity. Comparative analysis of Chlamydophila pneumoniae and Chlamydia trachomatis effects on host-cell gene regulation. Cell Microbiol. 2003;5:785–95. doi: 10.1046/j.1462-5822.2003.00319.x. [DOI] [PubMed] [Google Scholar]
- 19.Kaufmann SH. How can immunology contribute to the control of tuberculosis? Nat Rev Immunol. 2001;1:20–30. doi: 10.1038/35095558. [DOI] [PubMed] [Google Scholar]
- 20.de la Barrera Aleman M, Musella R, Schierloh P, Pasquinelli V, Garcia V, Abbate E, Sasiain MdelC. IL-10 down-regulates costimulatory molecules on Mycobacterium tuberculosis-pulsed macrophages and impairs the lytic activity of CD4 and CD8 CTL in tuberculosis patients. Clin Exp Immunol. 2004;138:128–38. doi: 10.1111/j.1365-2249.2004.02577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadek MI, Sada E, Toossi Z, Schwander SK, Rich EA. Chemokines induced by infection of mononuclear phagocytes with mycobacteria and present in lung alveoli during active pulmonary tuberculosis. Am J Respir Cell Mol Biol. 1998;19:513–21. doi: 10.1165/ajrcmb.19.3.2815. [DOI] [PubMed] [Google Scholar]
- 22.Denis M. Proinflammatory cytokines in hypersensitivity pneumonitis. Am J Respir Crit Care Med. 1995;151:164–9. doi: 10.1164/ajrccm.151.1.7812548. [DOI] [PubMed] [Google Scholar]
- 23.Standiford TJ, Rolfe MW, Kunkel SL, et al. Macrophage inflammatory protein-1 alpha expression in interstitial lung disease. J Immunol. 1993;151:2852–63. [PubMed] [Google Scholar]
- 24.Zhang YM, Broser H, Cohen M, Bodkin K, Law J, Reibman Rom WN. Enhanced interleukin-8 release and gene expression in macrophages after exposure to Mycobacterium tuberculosis and its components. J Clin Invest. 1995;95:586–92. doi: 10.1172/JCI117702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White JR, Lee JM, Young PR, et al. Identification of a potent, selective non-peptide CXCR2 antagonist that inhibits interleukin-8-induced neutrophil migration. J Biol Chem. 1998;273:10095–8. doi: 10.1074/jbc.273.17.10095. [DOI] [PubMed] [Google Scholar]
- 26.Bergeron A, Bonay M, Kambouchner M, Lecossier D, Riquet M, Soler P, Hance A, Tazi A. Cytokine patterns in tuberculous and sarcoid granulomas: correlations with histopathologic features of the granulomatous response. J Immunol. 1997;159:3034–43. [PubMed] [Google Scholar]
- 27.Chantry D, Romagnani P, Raport CJ, Wood CL, Epp A, Romagnani S, Gray PW. Macrophage-derived chemokine is localized to thymic medullary epithelial cells and is a chemoattractant for CD3(+), CD4(+), CD8(low) thymocytes. Blood. 1999;94:1890–8. [PubMed] [Google Scholar]
- 28.Yamashita U, Kuroda E. Regulation of macrophage-derived chemokine (MDC, CCL22) production. Crit Rev Immunol. 2002;22:105–14. [PubMed] [Google Scholar]
- 29.Matsukawa A, Hogaboam CM, Lukacs NW, Lincoln PM, Evanoff HL, Kunkel SL. Pivotal role of the CC chemokine, macrophage-derived chemokine, in the innate immune response. J Immunol. 2000;164:5362–8. doi: 10.4049/jimmunol.164.10.5362. [DOI] [PubMed] [Google Scholar]
- 30.Theus SA, Cave MD, Eisenach KD. Intracellular macrophage growth rates and cytokine profiles of Mycobacterium tuberculosis strains with different transmission dynamics. J Infect Dis. 2005;191:453–60. doi: 10.1086/425936. [DOI] [PubMed] [Google Scholar]
- 31.Bogdan C, Vodovotz Y, Nathan C. Macrophage deactivation by interleukin-10. J Exp Med. 1991;174:1549–55. doi: 10.1084/jem.174.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore KW, O'Garra A, de Waal Malefyt R, Vieira P, Mosmann TR. Interleukin-10. Annu Rev Immunol. 1993;11:165–90. doi: 10.1146/annurev.iy.11.040193.001121. [DOI] [PubMed] [Google Scholar]
- 33.Fulton SA, Cross JV, Toossi ZT, Boom WH. Regulation of interleukin-12 by interleukin-10, transforming growth factor-beta, tumor necrosis factor-alpha, and interferon-gamma in human monocytes infected with Mycobacterium tuberculosis H37Ra. J Infect Dis. 1998;178:1105–14. doi: 10.1086/515698. [DOI] [PubMed] [Google Scholar]
- 34.Gessani S, Belardelli F. IFN-gamma expression in macrophages and its possible biological significance. Cytokine Growth Factor Rev. 1998;9:117–23. doi: 10.1016/s1359-6101(98)00007-0. [DOI] [PubMed] [Google Scholar]
- 35.Fenton MJ, Vermeulen MW, Kim S, Burdick M, Strieter RM, Kornfeld H. Induction of gamma interferon production in human alveolar macrophages by Mycobacterium tuberculosis. Infect Immun. 1997;65:5149–56. doi: 10.1128/iai.65.12.5149-5156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cappelli G, Volpe P, Sanduzzi A, Sacchi A, Colizzi V, Mariani F. Human macrophage gamma interferon decreases gene expression but not replication of Mycobacterium tuberculosis: analysis of the host-pathogen reciprocal influence on transcription in a comparison of strains H37Rv and CMT97. Infect Immun. 2001;69:7262–70. doi: 10.1128/IAI.69.12.7262-7270.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortune SM, Solache A, Jaeger A, Hill PJ, Belisle JT, Bloom BR, Rubin EJ, Ernst JD. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J Immunol. 2004;172:6272–80. doi: 10.4049/jimmunol.172.10.6272. [DOI] [PubMed] [Google Scholar]
- 38.Greenwell-Wild T, Vazquez N, Sim D, Schito M, Chatterjee D, Orenstein JM, Wahl SM. Mycobacterium avium infection and modulation of human macrophage gene expression. J Immunol. 2002;169:6286–97. doi: 10.4049/jimmunol.169.11.6286. [DOI] [PubMed] [Google Scholar]
- 39.Burke B, Giannoudis A, Corke KP, Gill D, Wells M, Ziegler-Heitbrock L, Lewis CE. Hypoxia-induced gene expression in human macrophages. Implications for ischemic tissues and hypoxia-regulated gene therapy. Am J Pathol. 2003;163:1233–43. doi: 10.1016/S0002-9440(10)63483-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wayne LG, Lin KY. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immul. 1982;37:1042–9. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc Natl Acad Sci USA. 2001;13:7534–9. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desjardin LE, Hayes LG, Sohaskey CD, Wayne LG, Eisenach KD. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J Bacteriol. 2001;18:5311–16. doi: 10.1128/JB.183.18.5311-5316.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen F, Pufe T, Conradi L, Varoga D, Tsokos M, Papendieck J, Petersen W. Antimicrobial peptides are expressed and produced in healthy and inflamed human synovial membranes. J Pathol. 2002;198:369–77. doi: 10.1002/path.1224. [DOI] [PubMed] [Google Scholar]
- 44.Wu S, Howard ST, Lakey DL, et al. The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis in vivo. Mol Microbiol. 2004;51:1551–62. doi: 10.1111/j.1365-2958.2003.03922.x. [DOI] [PubMed] [Google Scholar]
- 45.Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 2004;14:628–38. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Guleria I, Pollard JW. Aberrant macrophage and neutrophil population dynamics and impaired Th1 response to Listeria monocytogenes in colony-stimulating factor 1-deficient mice. Infect Immun. 2001;69:1795–807. doi: 10.1128/IAI.69.3.1795-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowers K, Pitcher C, Marsh M, Int J. CD4. A co-receptor in the immune response and HIV infection. Biochem Cell Biol. 1997;29:871–5. doi: 10.1016/s1357-2725(96)00154-9. [DOI] [PubMed] [Google Scholar]
- 48.Center DM, Kornfeld H, Cruickshank WW. Interleukin 16 and its function as a CD4 ligand. Immunol Today. 1996;17:476–81. doi: 10.1016/0167-5699(96)10052-i. [DOI] [PubMed] [Google Scholar]
- 49.Hermann E, Darcissac E, Idziorek T, Capron A, Bahr M. Recombinant interleukin-16 selectively modulates surface receptor expression and cytokine release in macrophages and dendritic cells. Immunology. 1999;97:241–8. doi: 10.1046/j.1365-2567.1999.00786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharov V, Kwong KY, Frank B, et al. The limit of log-ratios. BMC Biotechnol. 2004;4:1–6. doi: 10.1186/1472-6750-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]