Abstract
Peritoneal B cells and their omental precursors play an important role in the immune response of the peritoneal cavity and mucosal surfaces in mice. We have previously shown that peritoneal and mucosal B lineage cells are unlikely to be significantly linked in humans. However, the status of the omentum remains unknown. Here, using immunohistochemistry, we observed that sparse, quiescent B cells and occasional clusters of B cells were present in the omentum and that plasma cells, predominantly with cytoplasmic immunoglobulin G (IgG), were present. We analysed sequences of immunoglobulin genes amplified using reverse transcriptase–polymerase chain reaction (RT-PCR) from the normal human greater omentum, and describe the characteristics of variable region genes used by IgG, IgA and IgM. We focused on the properties of IgVH4 and IgVH5 families to allow comparisons of like with like between different Ig isotypes and cells from different immune compartments. We observed that the IgM genes were derived from a mixed population with mutated and unmutated immunoglobulin sequences. All IgVH4 and IgVH5 genes used by IgA and IgG from omental cells showed evidence of somatic hypermutation but the load of mutations was not significantly different to that seen in either the systemic or the mucosal compartments. The trends observed, including the dominance of IgG plasma cells, the IgA1/IgA2 ratio being biased towards IgA1, JH1 usage, and a moderate level of somatic mutations, link omental B lineage cells with the systemic compartment. These observations reinforce previous studies highlighting the difference between human and murine B-cell compartments and their relationship to the mucosal immune system.
Keywords: human, omentum, immunoglobulin gene, B cell
Introduction
The human omentum is a double-layered sheet of fatty tissue in the peritoneum. There are two omenta in humans. The ‘fatty apron’-like greater omentum, made up of four layers of peritoneum, is attached from the greater curvature of the stomach to the transverse colon and hangs as an apron from the lower part of the transverse colon. The greater omentum contains numerous adipocytes and lymph nodes. The lesser omentum is attached to the top edge of the stomach, and extends to the undersurface of the liver. These omenta cover and support blood vessels and various abdominal organs; for example, they attach the stomach to the body wall. The human omentum has been considered as a lymphoid organ because of the presence of clusters of leucocytes, termed ‘milky spots’, which are most abundant in childhood.1,2 The fetal omentum is a site of lymphopoiesis.3 It has been speculated that the human omentum may be analogous to thymic tissue or may be a source of peritoneal macrophages.4–7 It has also been suggested that the omentum may be involved in immune responses to gut pathogens.4 Interest in the latter was increased recently with the report that B cells in the murine omentum are involved in the seeding of the peritoneal cavity with B1 cells.8 Murine B1 cells in the peritoneal cavity are considered to be a potential source of the immunoglobulin A (IgA) plasma cells that populate the intestinal lamina propria. Although we and others9,10 have previously shown that human peritoneal B cells are not related to the mucosal B-cell response, the status of the omentum in this context is not known. The aim of this study, therefore, was to characterize the cells of the B lineage in the adult human greater omentum.
Materials and methods
Origin of formalin-fixed, paraffin-embedded blocks of human omentum
Specimens of the human omentum were obtained from gynaecological surgery with the informed consent of the patients. Only patients with omentum without pathological changes, according to the histopathology report, were included in this study. Formalin-fixed, paraffin-embedded blocks of human omentum were studied from 16 patients. In addition, omental specimens from two female patients were obtained fresh (within 1 hr of surgery). They were snap-frozen and stored in liquid nitrogen until required. The first patient was a 68-year-old with clear cell carcinoma of the right ovary. The second patient was a 50-year-old with pelvic pain, irregular vaginal bleeding and an ovarian cyst, which, on histological examination, proved to be benign.
Immunohistochemical staining
All immunological reagents were purchased from DakoCytomation Ltd (Eli, UK), unless otherwise stated. In order to study the lymphocytes in the human omentum, the following primary antibodies were used on formalin-fixed, paraffin-embedded tissue sections: rabbit anti-human IgA, rabbit anti-human IgG, mouse monoclonal antibody (mAb) anti-human CD20cy (clone L26), rabbit anti-human CD3, and mouse mAb anti-human Ki-67 antigen (clone MIB-1). For single stain procedures, the immunoreactivity of the primary antibodies was demonstrated using an indirect immunoperoxidase method with a peroxidase-conjugated, swine anti-rabbit or rabbit anti-mouse immunoglobulin, as appropriate, and a 3,3′-diaminobenzidine tetrahydrochloride (DAB) substrate. For double stain procedures (e.g. CD20/MIB-1), the reactivity of the second immunostain was visualized using a biotinylated F(ab′)2 fragment of rabbit anti-mouse immunoglobulin followed by an Extravidin® alkaline phosphatase conjugate (Sigma-Aldrich Company Ltd, Gillingham, UK). Binding was visualized using fast blue, which contrasts with the brown DAB precipitate. Slides were washed in tap water without prior counterstain and then mounted in Aquamount (VWR International Ltd, Lutterworth, UK). Furthermore, tissue sections of human tonsil or appendix were processed in parallel to check the sensitivity of all our staining protocols.
Total RNA extraction and RT-PCR
Total RNA was extracted from frozen specimens of omentum, which had first been split into two fragments to look for evidence of related cells in a small environment. Total RNA extraction was carried out using 1 ml of TRIzol® reagent (Invitrogen Ltd, Paisley, UK) per sample according to the manufacturer's protocol.
Synthesis of first-strand complementary DNA (cDNA) was carried out as described elsewhere.9 Functional IgVH4 and IgVH5DJH-Cµ, -Cα and -Cγ transcripts were amplified using a semi-nested PCR strategy.
The sequences of all oligonucleotides used as sense PCR primers for the amplification of functional IgVH4 and IgVH5DJH-CH transcripts were taken from references.11–13 Both first- and second-round PCRs were carried out as described elsewhere.9 cDNA prepared from human tonsil was used as a template in positive control RT-PCR. For all negative control RT-PCRs, the murine Moloney leukaemia virus (MMLV) reverse transcriptase was omitted in the first-strand cDNA synthesis.
T-cloning of IgVH5DJH-CH functional transcripts and DNA sequencing
PCR products were size-fractionated by electrophoresis in a 3·3% MicroSieve® 3 : 1 agarose gel (FMC BioProducts, Flowgen, Lichfield, UK) under 1 × Tris-borate-ethylenediaminetetraacetic acid (EDTA) running buffer and stained with ethidium bromide. IgVH4 and IgVH5DJH-CH PCR products were cloned into the pGEM-T Vector using the pGEM®-T Vector System (Promega UK Ltd, Southampton, UK) and Escherichia coli JM109 competent cells. Cloned inserts were sequenced by contracted DNA sequencing (Lark Technologies, Inc., Takeley, UK).
Nucleotide sequences of IgVHDJH-Cµ, -Cα and -Cγ transcripts were analysed with GeneJockey II software (by Philip L. Taylor, distributed by Biosoft®, Cambridge, UK) using the V BASE database,14 which contains all known human germline immunoglobulin heavy- and light-chain gene segments. IgVH and JH gene segments were assigned manually. The third complementarity-determining regions of the immunoglobulin heavy chain (H-CDR3) were defined as starting at the first nucleotides after the end of IgVH gene segments and stopped just before the ‘TGGGG’ motif in the rearranged JH gene segment. Analysis of the H-CDR3 nucleotide sequences provided evidence of clonal interrelatedness, whereas analysis of the 5′ end of the CH exons provided evidence of immunoglobulin class switch DNA recombination. For each immunoglobulin transcript, the frequency of somatic hypermutation was computed as follows: (number of base substitutions identified on the IgVH gene segment when compared against the unrearrranged, germline IgVH gene sequence/total number of bases analysed along the IgVH gene) × 100. Analysed immunoglobulin sequences are accessible through the GenBank/EMBL/DDBJ nucleotide sequence databases (accession numbers AM233752–840).
Statistical methods
Comparisons of the frequencies of point mutations on the IgVH gene segments and comparison of H-CDR3 lengths were carried out using a Mann–Whitney U-test. Comparisons of the JH gene segment usages were carried out using χ2 tests in a Microsoft Excel spreadsheet. Observed differences were considered statistically significant at P ≤ 0·05.
Results
Immunohistochemical study
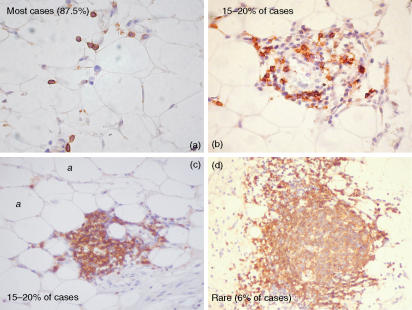
Blocks of formalin-fixed, paraffin-embedded omental tissues from 16 women were used for immunohistochemical analysis. We found that 14 cases (87·5%) showed isolated CD20+ and CD3+ cells only and three cases (18·8%) showed clusters containing both CD20+ and CD3+ cells but with no identifiable zonation and no germinal centres in the same or in serial tissue sections. A germinal centre-like structure with a small associated T-cell zone was observed in one case only. The range of appearance of CD20+ B cell-containing structures is illustrated in Fig. 1.
Figure 1.
Single immunostain using a mouse anti-human CD20 monoclonal antibody on formalin-fixed, paraffin-embedded tissue sections of the human greater omentum, illustrating the variety of microanatomical locations in which B cells were observed. CD20, a pan B-cell marker, appears as brown surface staining. a, adipocyte.
In order to test whether isolated cells and cells in small clusters of lymphoid tissues in human omentum were proliferating, sections of human omentum from five cases (four non-serial tissue sections per case) were single stained for MIB-1 or double stained for MIB-1 and CD20. Another set of 12 tissue sections from the same three cases was subjected to MIB-1/CD3 double staining. In these five cases, expression of MIB-1 antigen could not be detected, either alone or in association with CD20 or with CD3 expression. We therefore concluded that scattered lymphocytes in the human greater omentum were non-proliferative.
Plasma cells expressing IgG, IgA (Fig. 2) and IgM were present in the omental tissue sections. The plasma cells were commonly seen in confined spaces between adipocytes where they appeared ‘squashed’ into the triangular junctions between adipocytes. The resulting triangular appearance of the plasma cells is not typical. Normally, plasma cells are observed in diffuse stromal connective tissue such as that in the intestinal lamina propria, the bone marrow or the medullary cords of lymph nodes where they have a characteristic oval shape. IgG-expressing plasma cells were most abundant. By extrapolation, having counted plasma cells in known tissue volumes in tissue sections and consulted the histopathology reports providing us with the sizes of the omentum, we estimated that approximately 3 × 106 IgG-containing cells are present in the adult greater omentum.
Figure 2.
Single immunostain using a rabbit anti-human immunoglobulin A (IgA) polyclonal antibody on formalin-fixed, paraffin-embedded tissue sections of the human greater omentum. Note the adipocytes (labelled ‘a’) and their distended vacuoles encompassing lipids. The antibody-secreting cells, with intracytoplasmic IgA (arrowed) and an unusual triangular shape, appear compressed at the junction of the adipocytes.
Analysis of IgVHDJH-CH transcript DNA sequences amplified by RT-PCR from the human omentum
The immunoglobulin gene sequences were analysed to determine whether the plasma cells showed the hallmark of effector cells from mucosal or peripheral lymphoid tissues. A total of 124 immunoglobulin sequences were obtained and analysed, of which 89 were unique IgVHDJH gene segment rearrangements. We analysed 37 immunoglobulin transcripts from patient 1 and 52 transcripts from patient 2. Further details of the numbers of different immunoglobulin transcripts analysed per patient are shown in Table 1. Immunoglobulin transcripts PCR-amplified from human duodenal mucosa and splenic red pulp were retrieved from a previous study15 and were analysed here in order to provide an element of comparison for the mucosal and systemic immune compartments, respectively.
Table 1.
Number and origin of IgVH4 and IgVH5DJH-CH transcripts analysed
| Number of potentially functional IgVHDJH-CH transcripts analysed | |||||||
|---|---|---|---|---|---|---|---|
| Patient number | Age (years) | VH4-Cµ | VH4-Cα | VH4-Cγ | VH5-Cµ | VH5-Cα | VH5-Cγ |
| 1 | 68 | 7* | 9 | 7 | 3 | 3 | 8 |
| 2 | 50 | 10 | 18 | 10 | 4 | 5 | 5 |
| Total | 17 | 27 | 17 | 7 | 8 | 13 | |
Values represent the number of different IgVHDJH-CH transcripts analyzed, as determined by analysis of H-CDR3 nucleotide sequences.
Frequency of somatic hypermutation in IgVHDJH-CH transcripts amplified by RT-PCR from the human omentum
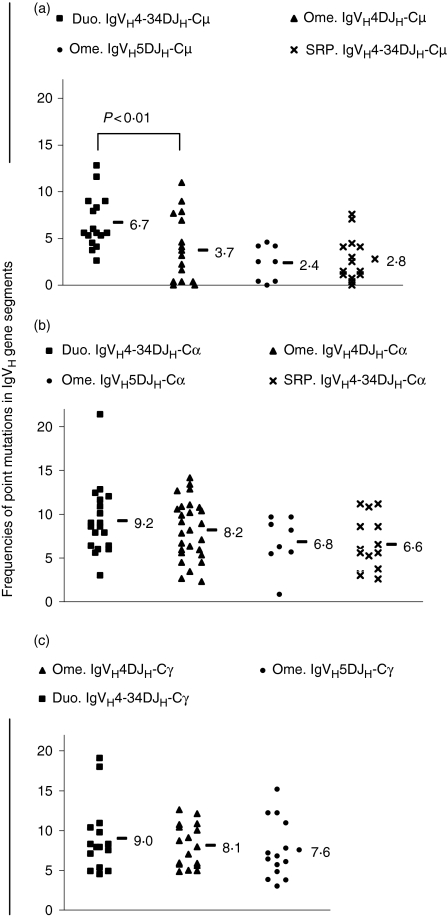
Figure 3 shows the frequencies of base substitutions in each IgVH gene segment for the IgVHDJH-Cµ transcripts (Fig. 3a), IgVHDJH-Cα transcripts (Fig. 3b) and IgVHDJH-Cγ transcripts (Fig. 3c). When data from the omentum were compared, within immunoglobulin isotypes, with a highly homologous group of sequences from the duodenum or splenic red pulp, no significant differences in the frequency of somatic hypermutation were found for any comparison.
Figure 3.
Frequencies of base substitutions observed in the rearranged IgVH gene segment of human IgVH4 and IgVH5DJH-CH transcripts isolated from the human adult greater omentum (Ome.), duodenal mucosa (Duo.) and splenic red pulp (SRP.). Each point on the scatter plot represents the number or mean number of point mutations for an individual IgVHDJH-CH transcript. When more than one nucleotide sequence was obtained for a given immunoglobulin (Ig) transcript (as determined by analysis of the CDR3 sequence), the average number was plotted as a single point to avoid data distortion. For each set of data, the average or mean number of point mutations is shown as appropriate. The numbers of base misinsertions resulting from Taq DNA polymerase errors were not subtracted from the values. For IgVH4DJH-Cµ transcripts, statistically significant differences in the frequencies of point mutations between sets of data obtained from the duodenal mucosa and the omentum are shown. (a) IgVHDJH-Cµ transcripts; (b) IgVHDJH-Cα transcripts; (c) IgVHDJH-Cγ transcripts.
The IgVH4DJH-Cµ transcripts were a mixed population of unmutated and mutated sequences, presumably representing the presence of naïve and post-germinal centre cells in the omentum. Therefore, although there was a significant difference between the frequency of mutation in the omental sequences and that in other populations of sequences (P < 0·01), this may not be of biological significance as it is probably not a comparison of like with like in terms of the composition of the population sampled.
Lengths of H-CDR3 in IgVHDJH-CH transcripts amplified by RT-PCR from the human omentum
The mean H-CDR3 lengths were 40·4, 40·2 and 34·8 nucleotides, respectively, for the VH4DJH-Cµ, -Cα and -Cγ transcripts and 41·4, 40·5 and 37·2 nucleotides, respectively, for the VH5DJH-Cµ, -Cα and -Cγ transcripts. No significant differences in H-CDR3 length were identified either within the data sets from the omentum or when they were compared with immunoglobulin sequences from the duodenal mucosa or splenic red pulp.
JH gene segment usage
The JH gene segment usage in immunoglobulin transcripts amplified by RT-PCR from the human omenta is illustrated in Table 2. Out of a total of 90 immunoglobulin transcripts analysed, the JH gene segment usage found can be sorted as follow: JH4 > JH5 > JH6 > JH3 > JH1 > JH2. Two potentially functional immunoglobulin transcripts encompassing a JH1 gene segment were identified: one IGHV4-34DJH1-Cα2 transcript and one IGHV4-31DJH1-Cγ2 transcript. Out of 324 different immunoglobulin rearrangements amplified from normal intestinal mucosa in previous studies from our own laboratory and others,16 the JH1 gene segment was not observed. Therefore, the JH1 gene segment was more frequently rearranged in the human omenta than in the intestinal mucosa (P = 0·007 by χ2 test). In contrast, when the JH1 gene segment usage was compared between the human omentum and the peripheral compartment [peripheral blood mononuclear cells (PBMC) and spleen], compiled from seven studies, nine sequences out of 568 examples of JH1 usage were observed, which is not significantly different from the two examples in 90 sequences observed here.16
Table 2.
JH gene segment usage in IgVH4 and IgVH5DJH-Cµ, -Cα and -Cγ transcripts amplified by RT-PCR from the human adult greater omenta
| Human omenta | ||||
|---|---|---|---|---|
| JH gene segment | IgVH4 + 5-IgM | IgVH4 + 5-IgA | IgVH4 + 5-IgG | Total |
| JH1 | 0 | 1 | 1 | 2* |
| JH2 | 0 | 0 | 0 | 0 |
| JH3 | 2 | 6 | 7 | 15 |
| JH4 | 9 | 13 | 13 | 35 |
| JH5 | 10 | 6 | 4 | 20 |
| JH6 | 3 | 10 | 5 | 18 |
| Total number ofimmunoglobulintranscripts | 24 | 36 | 30 | 90 |
JH1 gene segment usage in the omentum (two of 90) compared with the intestinal mucosa and parotid salivary gland (zero of 324) from Thoree et al.16. P = 0·007 (χ2 test). There was no significant difference in JH1 usage when compared to PBMC and spleen.
Identification of clonally related immunoglobulin transcripts based on their unique H-CDR3 DNA sequences
Examples of two clonally related immunoglobulin transcripts from the omentum are illustrated in Fig. 4. Both immunoglobulin transcripts carry the imprint of somatic hypermutation. When the pattern of point mutations was further analysed, 13 mutations were found to be shared between these two immunoglobulin transcripts (10 replacement mutations and three silent mutations), whereas two and three mutations were unique to these sequences. Of note, when the 99 bases of the IgCH exon between the splice site and the primer were analysed, one transcript displayed a 99% match with the Cγ2 exon while the other transcript displayed a complete match with the Cγ1 exon. There are seven base differences between the Cγ1 and Cγ2 exons over this sequence.
Figure 4.
(a) Nucleotide sequences and (b) deduced amino acid sequences of clonally related, productive IGHV4–4DJH6B-Cγ2 (labelled IgG2) and -Cγ1 functional transcripts (labelled IgG1). Sequences are shown aligned with the germline, unmutated IGHV4-4 and JH6B gene segments14 and with the 5′ end of the Cγ2 or Cγ1 exon. The location of the third complementarity-determining region of the immunoglobulin heavy chain (H-CDR3) is indicated. In (a), replacement point mutations are shown by uppercase letters, and silent point mutations by lowercase letters. In the IgVH4 gene segment, all base substitutions are shared by both immunoglobulin (Ig) transcripts sequences, except five mutations which are located in the FR1 domain. Dashes indicate identity with the above located nucleotide sequence. RNA splice sites (5′ end of Cγ exons) are represented by asterisks. Annealing sites of the sense VH5 FR1 and antisense Cγ PCR primers have been italicized. Differences in the nucleotide sequences between the human Cγ1 and Cγ2 exons have been underlined. Identical nucleotide sequences of the H-CDR3 and the partially shared pattern (72·2%) of base substitutions illustrate two clonally related omental cells of the B lineage which have diversified by class switch recombination event after somatic hypermutation.
Discussion
Peritoneal B cells and their omental precursors play an important role in the immune response of the peritoneal cavity and mucosal surfaces in mice. We have previously shown that peritoneal and mucosal B lineage cells are unlikely to be significantly linked in humans. In this study, we questioned whether the human omentum could harbour a putative reservoir of intestinal lamina propria plasma cell precursors analogous to the developmental lineage in the peritoneum/omentum in mice. B cells were sparse in the human omentum and occurred predominantly as isolated cells or in small, unstructured clusters. We found no evidence that omental lymphocytes were in cell cycle. Based on both these observations, we propose that the human omentum in adults is unlikely to be a significant source of B-cell or plasma cell precursors for export.
We found that the human greater omentum contains an IgG-containing plasma cell population. We estimated the size of this population at around 3 × 106 IgG-containing cells. In comparison, the size of the immunoglobulin-positive cell population in the human bone marrow has been estimated at 2·3 × 1010 cells,17 of which 51–66% have been estimated to contain IgG.18,19 The splenic immunoglobulin-positive cell population has been estimated at 4·7 × 108 cells,17 of which 90% have been estimated to be IgG positive.20 Therefore, our estimate demonstrates that the human omental IgG-containing cell population is minor, being only approximately 0·02% of the size of the bone marrow IgG-containing plasma cell population and 0·7% of the size of the splenic IgG-containing cell population.
The frequencies of point mutations acquired in the VH segment of IgVH4DJH-Cµ transcripts were significantly lower in the omentum than in the normal small intestinal mucosa (P < 0·01).12,15,21 However, this comparison is biased by the contribution of IgM+ cells harbouring rearranged but unmutated IgVH4 gene segments, presumably from naïve B cells. However, when other immunoglobulin isotypes were compared, differences in the frequencies of point mutations between IgVH genes from the omental and the small intestinal plasma cells were no longer apparent. The frequencies of point mutations in the IgVH4 segment from omental B cells class-switched to IgA or IgG were not significantly different when compared with those observed in the small intestinal plasma cell population or splenic red pulp, the average frequencies of point mutations being intermediate between those two sites. Similarly, analysis of the H-CDR3 lengths did not yield any statistically significant indication of whether the omental immunoglobulin sequences were from cells of mucosal or peripheral origin.
The VH-Cα1/VH-Cα2 ratio differs between peripheral blood (90% IgA1) and the intestinal mucosa (60% IgA2).22 In the present study, 68·6% of the VHDJH-Cα transcript sequences were spliced to the Cα1 exon, whereas 31·4% were spliced to the Cα2 exon. Therefore, as previously reported in human splenic red pulp15 and peritoneal B cells,9 the ratio of IgA subclasses was biased towards IgA1 in the human omentum. Analysis of the JH gene segments identified usage of the JH1 gene segment, which is associated with the peripheral B-cell repertoire. Out of a total of 90 potentially functional, mutated IgVHDJH-CH rearrangements, two examples of JH1 were observed in the omentum. This frequency was significantly higher than the JH1 gene segment usage reported in the parotid salivary gland and the intestinal mucosa but not significantly different from that in PBMC and the spleen.16
The group of clonally related IgVH4DJH6-Cγ transcripts, which includes IgG1- and IgG2-switched variants, is of interest. These sequences showed a similar pattern of point mutations on their productively rearranged IGHV4-4 gene segment. The 13 shared point mutations shown in Fig. 4 were accumulated either before any immunoglobulin class-switch DNA recombination, or at the IgVH4-Cγ1 stage, as the Cγ1 exon is located upstream of the Cγ2 exon in the human constant IgH chain locus.23 The identification of these immunoglobulin sequences supports the concept that clonally related cells of the B lineage can be disseminated within the omentum. There are two possible, non-mutually exclusive explanations for the presence of clonally related cells within the omentum. Either these cells might be derived from a distant site and arrive through the omental blood vessels and/or clonally related cells might be derived from the omental lymphoid aggregates or milky spots. Although such organized structures are rare and were not sampled by the biopsies taken from the omentum of patient 2, this does not preclude their existence. Such groups of clonally related DNA sequences are commonly observed at mucosal sites over large distances, although they are not exclusive to this compartment. Families of related immunoglobulin sequences have been observed in the following human tissues: the tonsil,24 spleen,25 subcarinae,26 parotid salivary gland,15 colonic mucosa,27 paediatric and adult duodenal mucosa12 and nasal mucosa.28
In conclusion, the human adult omental cells of the B lineage analysed in this study included quiescent, isolated and clustered B cells and isolated plasma cells, the IgG isotype being the more abundant. The mixed population of naïve B cells and plasma cells was reflected in mutated and unmutated IgVH4DJH-Cµ sequences. However, IgA and IgG sequences were somatically mutated. Although the omental IgVH4DJH-Cα transcripts used to determine the IgA1:IgA2 ratio were not significantly different from their mucosal and systemic counterparts, we observed a bias towards IgA1 in omental cells of the B lineage, as reported in human splenic red pulp and peritoneal B cells, and examples of JH1 rearrangement that are rare in the intestinal mucosal were seen. Therefore, it is unlikely that human omental cells of the B lineage are significant components of the intestinal immune system, either as a source of effector cells or as a significant site of localization of effector cells. Rather, the adult human omentum is likely to be a minor site of antibody production generated by immune responses predominantly in the systemic compartment.
Acknowledgments
This work was financially supported by a Biotechnology and Biological Sciences Research Council project grant to JS.
Abbreviations
- H-CDR3
third complementarity-determining region of the immunoglobulin heavy chain
- IgVHDJH-CH
immunoglobulin heavy chain productive/functional transcript
References
- 1.Krist LF, Eestermans IL, Steenbergen JJ, Hoefsmit EC, Cuesta MA, Meyer S, Beelen RH. Cellular composition of milky spots in the human greater omentum: an immunochemical and ultrastructural study. Anat Rec. 1995;241:163–74. doi: 10.1002/ar.1092410204. [DOI] [PubMed] [Google Scholar]
- 2.Shimotsuma M, Takahashi T, Kawata M, Dux K. Cellular subsets of the milky spots in the human greater omentum. Cell Tissue Res. 1991;264:599–601. doi: 10.1007/BF00319049. [DOI] [PubMed] [Google Scholar]
- 3.Solvason N, Kearney JF. The human fetal omentum: a site of B cell generation. J Exp Med. 1992;175:397–404. doi: 10.1084/jem.175.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koten JW, den Otter W. Are omental milky spots an intestinal thymus? Lancet. 1991;338:1189–90. doi: 10.1016/0140-6736(91)92043-2. [DOI] [PubMed] [Google Scholar]
- 5.Shimotsuma M, Simpson-Morgan M. Omental milky spots. Lancet. 1991;338:1596. doi: 10.1016/0140-6736(91)92419-3. [DOI] [PubMed] [Google Scholar]
- 6.Williams RJL, Davis AJS. Omental milky spots. Lancet. 1992;339:191–2. doi: 10.1016/0140-6736(92)90269-9. [DOI] [PubMed] [Google Scholar]
- 7.Beelen RHJ. Role of omental milky spots in the local immune response. Lancet. 1992;339:689. doi: 10.1016/0140-6736(92)90857-y. [DOI] [PubMed] [Google Scholar]
- 8.Ansel KM, Harris RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16:67–76. doi: 10.1016/s1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- 9.Boursier L, Farstad IN, Mellembakken JR, Brandtzaeg P, Spencer J. IgVH gene analysis suggests that peritoneal B cells do not contribute to the gut immune system in man. Eur J Immunol. 2002;32:2427–36. doi: 10.1002/1521-4141(200209)32:9<2427::AID-IMMU2427>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 10.Donze HH, Lue C, Julian BA, Kutteh WH, Kantele A, Mestecky J. Human peritoneal B-1 cells and the influence of continuous ambulatory peritoneal dialysis on peritoneal and peripheral blood mononuclear cell (PBMC) compostition and immunoglobulin levels. Clin Exp Immunol. 1997;109:356–61. doi: 10.1046/j.1365-2249.1997.4541352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn-Walters DK, Isaacson PG, Spencer J. Analysis of mutations in immunoglobulin heavy chain variable region genes of microdissected marginal zone (MGZ) B cells suggests that the MGZ of human spleen is a reservoir of memory B cells. J Exp Med. 1995;182:559–66. doi: 10.1084/jem.182.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boursier L, Dunn-Walters DK, Spencer J. Characteristics of IgVH genes used by human intestinal plasma cells from childhood. Immunology. 1999;97:558–64. doi: 10.1046/j.1365-2567.1999.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brezinschek H-P, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 14.Tomlinson IM, Williams SC, Ignatovich O, Corbett SJ, Winter GV. BASE Sequence Directory. Cambridge, UK: Medical Research Council Center for Protein Engineering; 1998. http://vbase.mrc-cpe.cam.ac.uk. [Google Scholar]
- 15.Dunn-Walters DK, Hackett M, Boursier L, Ciclitira PJ, Morgan P, Challacombe SJ, Spencer J. Characteristics of human IgA and IgM genes used by plasma cells in the salivary gland resemble those used in duodenum but not those used in the spleen. J Immunol. 2000;164:1595–601. doi: 10.4049/jimmunol.164.3.1595. [DOI] [PubMed] [Google Scholar]
- 16.Thoree VC, Golby SJ, Boursier L, Hackett M, Dunn-Walters DK, Sanderson JD, Spencer J. Related gA1 and IgG producing cells in blood and diseased mucosa in ulcerative colitis. Gut. 2002;51:44–50. doi: 10.1136/gut.51.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turesson I. Distribution of immunoglobulin-containing cells in human bone marrow and lymphoid tissues. Acta Med Scand. 1976;199:293–304. doi: 10.1111/j.0954-6820.1976.tb06735.x. [DOI] [PubMed] [Google Scholar]
- 18.Douglas AP, Crabbé PA, Hobbs JR. Immunochemical studies of the serum, intestinal secretions and intestinal mucosa in patients with adult celiac disease and other forms of the celiac syndrome. Gastroenterology. 1970;59:414–25. [PubMed] [Google Scholar]
- 19.Hijmans W, Schuit HR, Hulsing-Hesselink E. An immunofluorescence study on intracellular immunoglobulins in human bone marrow cells. Ann NY Acad Sci. 1971;177:290–305. doi: 10.1111/j.1749-6632.1971.tb35059.x. [DOI] [PubMed] [Google Scholar]
- 20.Chiappino G, Pernis B. Demonstration with immunofluorescence of 19S macroglobulins and 7S gamma globulins in different cells of the human spleen. Pathol Microbiol. 1964;27:8–15. doi: 10.1159/000161448. [DOI] [PubMed] [Google Scholar]
- 21.Fischer M, Küppers R. Human IgA- and IgM-secreting intestinal plasma cells carry heavily mutated VH region genes. Eur J Immunol. 1998;28:2971–7. doi: 10.1002/(SICI)1521-4141(199809)28:09<2971::AID-IMMU2971>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Brandtzaeg P, Halstensen TS, Kett K, Krajci P, Kvale D, Rognum TO, Scott H, Sollid LM. Immunobiology and immunopathology of human gut mucosa: humoral immunity and intraepithelial lymphocytes. Gastroenterology. 1989;97:1562–84. doi: 10.1016/0016-5085(89)90406-x. [DOI] [PubMed] [Google Scholar]
- 23.Hofker MH, Walter MA, Cox DW. Complete physical map of the human immunoglobulin heavy chain constant region gene complex. Proc Natl Acad Sci USA. 1989;86:5567–71. doi: 10.1073/pnas.86.14.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman CJ, Mockridge CI, Hamblin TJ, Stevenson FK. Tracking of the V4-34 (VH4-21) gene in human tonsil reveals clonal isotype switch events and a highly variable degree of somatic hypermutation. Clin Exp Immunol. 1996;105:360–8. doi: 10.1046/j.1365-2249.1996.d01-769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varade WS, Insel RA. Isolation of germinal center-like events from human spleen RNA. Somatic hypermutation of a clonally related VH6DJH rearrangement expressed with IgM, IgG, and IgA. J Clin Invest. 1993;91:1838–42. doi: 10.1172/JCI116397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snow RE, Djukanovic R, Stevenson FK. Analysis of immunoglobulin E VH transcripts in a bronchial biopsy of an asthmatic patient confirms bias towards VH5, and indicates local clonal expansion, somatic mutation and isotype switch events. Immunology. 1999;98:646–51. doi: 10.1046/j.1365-2567.1999.00910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holtmeier W, Hennemann A, Caspary WF. IgA and IgM VH repertoires in human colon: evidence for clonally expanded B cells that are widely disseminated. Gastroenterology. 2000;119:1253–68. doi: 10.1053/gast.2000.20219. [DOI] [PubMed] [Google Scholar]
- 28.Coker HA, Durham SR, Gould HJ. Local somatic hypermutation and class switch recombination in the nasal mucosa of allergic rhinitis patients. J Immunol. 2003;171:5602–10. doi: 10.4049/jimmunol.171.10.5602. [DOI] [PubMed] [Google Scholar]