Abstract
Antibody diversity is created by imprecise joining of the variability (V), diversity (D) and joining (J) gene segments of the heavy and light chain loci. Analysis of rearrangements is complicated by somatic hypermutations and uncertainty concerning the sources of gene segments and the precise way in which they recombine. It has been suggested that D genes with irregular recombination signal sequences (DIR) and chromosome 15 open reading frames (OR15) can replace conventional D genes, that two D genes or inverted D genes may be used and that the repertoire can be further diversified by heavy chain V gene (VH) replacement. Safe conclusions require large, well-defined sequence samples and algorithms minimizing stochastic assignment of segments. Two computer programs were developed for analysis of heavy chain joints. JointHMM is a profile hidden Markow model, while JointML is a maximum-likelihood-based method taking the lengths of the joint and the mutational status of the VH gene into account. The programs were applied to a set of 6329 clonally unrelated rearrangements. A conventional D gene was found in 80% of unmutated sequences and 64% of mutated sequences, while D-gene assignment was kept below 5% in artificial (randomly permutated) rearrangements. No evidence for the use of DIR, OR15, multiple D genes or VH replacements was found, while inverted D genes were used in less than 1‰ of the sequences. JointML was shown to have a higher predictive performance for D-gene assignment in mutated and unmutated sequences than four other publicly available programs. An online version 1·0 of JointML is available at http://www.cbs.dtu.dk/services/VDJsolver.
Keywords: antibodies, computer algorithm, gene rearrangements, human
Introduction
Diversity is a key feature of the adaptive immune system. Antibody diversity is partly created by combination of different heavy and light chains. Most of the diversity is, however, found within the complementarity determining region 3 (CDR3) which has a major influence on the binding of antigens.1 The heavy chain CDR3 is encoded by the junction of a variability (V), a diversity (D) and a joining (J) gene recombined during B-cell development in the bone marrow. Several copies of each gene segment exist and diversity can be created by sheer recombinatorial variation. However, the molecular processes linking the genes are imprecise and involve generation of palindromic (P) nucleotides,2–4 addition of non-templated (N) nucleotides by terminal deoxynucleotidyl transferase (TdT)4–6 and trimming of the gene ends,4 and therefore also play a major role in the generation of diversity.
Despite years of research which have provided detailed knowledge about the individual enzymes and complexes involved in the recombination process (e.g. references2,3,6–9), studying the actual repertoire is complicated by the imprecise nature of the recombination process, homology between the different genes and sometimes the presence of somatic hypermutations. Several approaches to evaluate the joint region have been employed, including simple alignment methods with mutations either permitted or not permitted,10–12 use of artificially generated sequences to define a threshold for D-gene lengths,13 a method of alignment taking the length of the joint region into account (joinsolver),14 a method incorporating the mutability of different codons,15 and a recent approach using a 3D dynamic programming alignment algorithm (SoDA)16. The methods have different advantages and drawbacks, and unfortunately the results of the various studies performed vary in a number of ways. The use of multiple D genes is an example of an area of controversy, as is the use of inverted D genes, the use of D genes with irregular recombination signal sequences (DIR) and the use of open reading frames encoded on chromosome 15 (OR15).11,13–15,17 These controversies need to be settled if algorithms of improved sensitivity and specificity for the D segment are to be developed, because algorithms that are too restrictive inevitably have low sensitivity and algorithms that are too relaxed lack specificity.
We sequenced the (to date) largest set of well-defined human heavy chain rearrangements totalling 6329 clonally unrelated sequences. To minimize misinterpretations in the V-gene assignment, in the analysis of trimming of heavy chain V gene (VH) and heavy chain J gene (JH) gene ends, and in the analysis of somatic hypermutations, we restricted the analysis to the most commonly used VH gene, IGHV3-23*01,10,18–20 and the two most commonly used JH genes, IGHJ4*02 and IGHJ6*02.10,21 Because of the experimental design, we also obtained some rearrangements using IGHJ5 and a few using IGHJ3. Furthermore, a set of 103 unrelated rearrangements using the IGHV3-h pseudogene was generated and used to confirm some of the findings. These sequences are well suited for this purpose because the rearrangements have not been selected for antigen binding because of a mutated translation initiation codon.22
The large set of sequences was used in the stepwise development and fine-tuning of algorithms of two programs for human heavy chain rearrangement analysis. JointML uses a maximum likelihood method taking both the length of the joint region and the mutational status of the VH gene into account. The other program, JointHMM, is a hidden Markov model superior at detecting insertions and deletions and with an approach that looks at the coding region as a whole. We were able to show that the final version of the JointML algorithm has a significantly better predictive performance for D-gene assignment than four publicly available programs.
Materials and methods
Sequences
A volume of 100 ml of peripheral blood was collected from 28 healthy adult volunteers after informed consent had been obtained. The protocol was approved by the regional ethics committee for Vejle and Funen counties. Before enrollment into the project, the genotypes for IGHV3-23 and IGHJ6 were determined by polymerase chain reaction (PCR) and direct sequencing as previously described.22 Only individuals homozygous for the most common genotypes, IGHV3-23*01 and IGHJ6*02, were included.
Peripheral blood mononuclear cells were isolated by density gradient centrifugation on Lymphoprep (Axis-Shield, Roskilde, Denmark) and enriched for memory B cells using the B Cell Isolation Kit II (Miltenyi, via Biotech Line, Slangerup, Denmark) followed by CD27 MicroBead separation (Miltenyi) according to the manufacturer's instructions. Memory B cells were chosen to enrich the material for mutated sequences. DNA was isolated from the enriched memory B-cell fraction by QIAmp Blood DNA Mini Kit (Qiagen via VWR, Albertslund, Denmark).
IGHV3-23-IGHD-IGHJ rearrangements were amplified in PCR reactions using VH3-23cn9.F (5′-CTGAGCTGGCTTTTTCTTGTG-3′) as the forward primer and a reverse primer binding downstream of either JH4 (5′-GCCGCTGTTGCCTCAGG-3′) or JH6 (5′-CCCACAGGCAGTAGCAGAA-3′). The forward primer was designed to be specific for the leader peptide sequence of IGHV3-23. However, it turned out also to bind in the leader sequence of IGHV3-h, leading to amplification of 150 IGHV3-h rearrangements. The PCR cycling conditions were 15 min at 95° and 38 cycles of 1 min at 94°, 1 min at 58°, and 30 seconds at 72°, followed by 10 min at 72°. The PCR products were cloned using the TOPO TA cloning kit (Invitrogen, Taastrup, Denmark) and carbenicillin (Sigma, Vallensbæk Strand, Denmark) and X-gal (Qbiogen via KemEnTec, Copenhagen, Denmark) selection. Plasmids were purified from overnight cultures of positive clones using the Wizard SV 9600 Plasmid Purification System (Promega via Ramcon, Birkerød, Denmark). Sequencing was performed with the BigDye Terminator kit (Applied Biosystems, Nærum, Denmark) and an ABI Prism 3100 genetic analyser (Applied Biosystems). Up to nine independent PCRs and cloning reactions were performed for each individual and 32–96 clones were sequenced per transfection.
Computer programs
Two suites of programs were developed to analyse the genetic rearrangement of immunoglobulin heavy chains. Both programs were generated in three versions (JointMLa, -b and -c and JointHMMa, -b and -c) successively reducing the accepted level of complexity in the genetic rearrangements. The initial a versions allow for all possible features described in the Introduction, including the use of multiple D genes, P nucleotides at VH, D and JH gene ends irrespective of trimming, inverted D genes, DIR, OR15 and conventional D genes. These programs were applied to analyse the use of P nucleotides. The b versions of the programs only allow for P nucleotides at untrimmed VH, D and JH gene ends. These programs were applied to analyse D-gene usage and to test the usage of multiple D genes, DIR and OR15 D genes, as well as the usage of inverted D genes. Finally, the c versions of the programs were constructed to allow only features that could be verified by statistical analysis, i.e. only one conventional D gene per joint and only P segments at untrimmed ends. Inversion of the D gene was allowed, but a penalty reducing illegitimate assignment of short or poorly matching inverted D segments was introduced.
The two program suites have fundamental differences in architecture and performance strategy. JointML was developed using Yabasic (http://www.yabasic.de). JointMLa uses the maximum likelihood method to obtain the best fit to the following model: VH-PVH-N1-PD1up-D1-PD1down-N2-PD2up-D2-PD2down-N3-PJH-JH, where Nx designates N and P indicates palindromic nucleotides. Any segment may be omitted except VH and JH. VH was compared with the IGHV3-23*01 germline gene (GenBank accession number M99660) while JH was compared with the germline JH gene with the highest identity score from codon 114 [the International Immunogenetics Information System (IMGT) nomenclature] through the splice site among all JH genes in the IMGT database (http://imgt.cines.fr/). The D segments were compared with any germline D gene available in the IMGT database including OR15 segments and DIR segments. P segments were defined as extensions, 2–8 nucleotides in length, from the VH, Dx or JH genes reverse-complementary to the corresponding germline sequence. Maximum likelihood was determined by running through all possible combinations of segments for a given rearrangement and finding the combination maximizing the likelihood score. The score was defined as the product of estimated probabilities for any event deviating from the germline sequences in question. Probabilities for substitutions in the terminal part of VH and in the Dx and JH segments were estimated to be the substitution prevalences in the VH region from codons 1 to 100. For unmutated sequences, the estimated Taq error rate was used. A given N nucleotide was attributed a probability equal to its frequency in all N segments. A dynamic probability for including a D segment was introduced dependent on the length of the part of the joint region not interpreted as being derived from the VH or JH gene. The probability was reduced by a factor dependent on the mutation rate of the VH region. Both parameters were fine-tuned to find a D segment in 5% of the sequences from a set of artificial rearrangements with 0–50 mutations in the VH region and a number of random bases reflecting the prevalences and lengths in the segment between VH and JH in real rearrangements. D segments were generally at least 8 nucleotides long.
JointHMM is a hidden Markov model (HMM),23 where each germline gene is encoded as a profile HMM. Palindrome segments were encoded as transitions from the germline gene to the corresponding position in the inverted gene. Multiple D-gene segments are encoded through a loop in the D-gene profile HMM. All D genes are selected with equal probability. Somatic hypermutations (SHMs) in the form of nucleotide transitions and transversions are encoded directly in the emission probabilities for the match states. Transition probabilities within the different profile HMMs are estimates from a small set of 200 immunoglobulin sequences. The probability for identifying a D gene was fitted to allow for identification of at most 5% D-gene hits among artificial (randomly permutated) rearrangements. Alignment of sequences to germline sequences in the JointHMM model was performed using the viterby algorithm.24
Statistical tests
To evaluate the significance of the identified features of the immunoglobulin rearrangements, we compared the results with values obtained in a random set of permutated immunoglobulin sequences (see below). We performed two types of comparisons based on number of observations and the length distribution of an observed phenomenon. In the first situation, we applied Fisher's exact test to determine if the observed phenomenon in the experimental sequences was statistically different from that found in the permutated sequences. In the other situation, we applied a Student's t-test for significantly different means.25 In both cases, we considered a P-value <0·05 as an indication of statistical significance. Other statistical tests were performed using the analyse-it addition to Microsoft Excel.
Permutated sequences
The permutated sequences were generated from the real sequences by permutation of the nucleotides lying between the appointed VH and JH gene segments. The VH and JH gene sequences were used with the degree of trimming and the mutations found in the real sequences. Thus, our reference sequences had the nucleotide composition found in real rearrangements. For analysis of P nucleotides around D genes, we made another set of permutated sequences where the D gene was also preserved so only N and P nucleotides were permutated. For these analyses, only sequences with exactly one assigned D gene segment were used.
Results
Validation of data and cluster definition
A sequence was entered into the database if it contained the first codon of the IGHV gene and the splice site of the IGHJ gene. Between 17 and 1091 (median 205) sequences from each of the 28 persons were entered into the database; in total, 9464 sequences. A smaller database of 150 sequences using the IGHV3-h pseudogene was also created.
A cluster analysis was performed to exclude sequences originating from the same cell or from cells derived from the same founder cell. Sequences were taken to be clustered if they had the same joint lengths (defined as codon 101 through the IGHJ gene splice site) and up to two nucleotide differences in the joint region. Sequences with the same joint lengths and three or four nucleotide differences were taken to be clustered only if they also shared at least seven mutations in the rest of the VH region. A total of 1704 IGHV3-23 and 27 IGHV3-h clusters containing two to 28 sequences were identified and only a single sequence (the first entered) from each cluster was used. A total of 3167 IGHV3-23 sequences and 41 IGHV3-h sequences were discarded as a result of clustering. The data were further scrutinized by identifying sequences with more than 10 shared mutations in the VH region or sequences that could be clustered by allowing a deletion or an insertion in the joint region. In this way, a further 32 IGHV3-23 and six IGHV3-h sequences were removed. The final database contained 6329 unique sequences using IGHV3-23[European Molecular Biology Laboratory (EMBL) accession numbers AM076988–AM083316] and 103 sequences using IGHV3-h (EMBL accession numbers AM282702–AM282804). Unless otherwise stated, the results in the following sections were generated for the IGHV3-23 rearrangements only.
Estimation of the Taq error rate
The Taq error rate was estimated to be 0·00048 mutations per nucleotide per sequence by PCR amplification, cloning and sequencing of an already sequenced rearrangement in the form of a plasmid. Eighty-four clones were sequenced, of which 13 had one substitution, two had two substitutions and one had three substitutions.
Of the 6329 sequences, 1495 were found to contain no substitutions, and, consistent with the presence of Taq errors, 458 sequences had one and 148 had two substitutions. This is compatible with a Poisson distribution yielding an estimated Taq error rate of 0·0015 mutations per nucleotide per sequence in our experimental system. The true error rate is probably somewhere between this result and the result obtained by amplification and sequencing of the plasmid. Sequences with three substitutions (123) were too numerous to be accounted for by Taq errors alone, suggesting a significant contribution from somatic hypermutations. On the basic of these results, we classified all sequences containing up to two substitutions as unmutated with respect to SHMs (2101 sequences), while sequences containing more than three substitutions were classified as mutated (4105 sequences). When the sequences were divided into mutated and unmutated sequences, sequences with exactly three substitutions were omitted.
P nucleotides
The presence of P nucleotides at trimmed and untrimmed gene ends was analysed in sequences with exactly one D gene using the a versions of the programs (Table 1). P nucleotide segments up to 7 bp in lengths were seen, but about 90% were 2 or 3 nucleotides long. Comparison of the numbers of P nucleotides found in the experimental sequences with those found in permutated sequences by Fisher's exact test showed with statistical significance that P nucleotides are present at the last position of VH, the first position of JH and the first and last positions of D – that is, at untrimmed gene ends. All other positions up to 10 bp from the heptamer were tested, but the presence of P segments after these positions did not differ significantly from that of random sequences.
Table 1.
Number of sequences with palindromic (P) segments of 2–8 bp in length downstream/upstream of the VH, diversity (D) and JH gene segments. The P segments were found using JointMLa, which allows P nucleotides to start at the end of the gene segment irrespective of the amount of trimming. P-values are calculated using Fisher's exact test. Similar results were obtained using JointHMMa (data not shown)
| Sequences | Permutated sequences* | ||||||
|---|---|---|---|---|---|---|---|
| Distance from heptamer to gene end† | No. of sequences | No. with P | % with P | No. of sequences | No. with P | % with P | P-value |
| VH gene | |||||||
| 1 | 1448 | 474 | 32·7 | 1635 | 103 | 6·3 | < 10−5 |
| 2 | 1027 | 48 | 4·7 | 1068 | 65 | 6·1 | 0·091 |
| 3 | 762 | 53 | 7·0 | 612 | 36 | 5·9 | 0·245 |
| JH gene | |||||||
| 1 | 324 | 60 | 18·5 | 350 | 23 | 6·6 | < 10−5 |
| 2 | 184 | 2 | 1·0 | 209 | 3 | 1·4 | 0·560 |
| 3 | 219 | 8 | 3·7 | 250 | 14 | 5·6 | 0·220 |
| 5′ end of D gene | |||||||
| 1 | 519 | 128 | 24·7 | 619 | 54 | 8·7 | < 10−5 |
| 2 | 343 | 31 | 9·0 | 347 | 26 | 7·5 | 0·275 |
| 3 | 474 | 25 | 5·3 | 454 | 17 | 3·7 | 0·168 |
| 3′ end of D gene | |||||||
| 1 | 616 | 86 | 14·0 | 684 | 58 | 8·5 | 0·001 |
| 2 | 266 | 30 | 11·3 | 276 | 24 | 8·7 | 0·195 |
| 3 | 460 | 5 | 1·1 | 485 | 9 | 1·9 | 0·241 |
See the Materials and methods section.
All positions up to 10 bp from the heptamer were analysed. No statistically significant differences were found except for at untrimmed ends.
On the basis of this observation, JointML and JointHMM were modified to only allow P nucleotides at untrimmed ends. Results obtained using these b versions confirm that P nucleotides can indeed be found after VH and D genes and before JH and D genes (P < 0·001). Significantly more P nucleotides were found after VH (34·3% of the sequences) and upstream of D (25·0%) than downstream of D (13·9%) and before JH (19·8%) (P < 0·05, pair-wise comparisons). The differences are not caused by selection as the fractions of sequences with and without P nucleotides at the four positions were the same in productive and non-productive rearrangements (data not shown). The frequency of VH ends with P nucleotides was similar in the IGHV3-h sequences (29·3%, P = 0·31 compared to IGHV3-23).
Thirty-three per cent of the first nucleotides of N1 (the N region between VH and D) at untrimmed VH ends (A as the last nucleotide) were found to be T. The frequency of T over the entire N1 segment for the same subset of sequences was only 22%, and this difference indicates that single-nucleotide P segments were indeed present. However, assuming an equal distribution of T over the entire N1 region, these numbers also indicate that accepting 1-nucleotide-long P segments would lead to approximately two out of three falsely identified 1 bp P segments created by N addition.
Identification of D genes
Identification of D-gene segments in rearrangements is complicated by at least two problems. (1) A wrong D gene may be assigned by chance because of accidental homology to a part of the joint sequence. Such a segment will usually be short and can be controlled for by comparing with the frequency at which similar matches are found in permutated joint sequences. (2) A wrong D gene may be assigned instead of a highly homologous (correct) one because of accidental identity with flanking bases or with bases changed by mutations in the original gene segment. This problem particularly affects identification of DIR, OR15 sequences and even inverted D genes as these genes all contain stretches homologous to parts of conventional D genes. Such wrong D-gene assignment will tend to be more commonly found in real sequences than in permutated ones and the wrong segment will (as a result of incorporation of flanking bases) tend to be slightly longer than the original D segment. Therefore, special analyses are required to clarify if these unconventional genes are used.
JointMLb found 4128 D segments in the 6329 sequences. Of these, 3923 (95%) were conventional D genes in normal orientation, while a minority of the sequences were interpreted to include DIR (41; 1·0%), inverted DIR (50; 1·2%), OR15 (42; 1·0%), inverted OR15 (eight; 0·2%) or inverted conventional D genes (64; 1·6%). Sixty-five sequences were assigned two D genes. The corresponding values for JointHMMb were similar (data not shown). Analysis of these assignments is discussed further in the following sections.
Use of DIR
In the sequences, we found 41 DIR and 50 inverted DIR segments. The median lengths were 12 and 11 bp, respectively, which are much shorter than conventional D segments (median 17 bp, P < 0·0001 in both cases), suggesting that assigned DIR and inverted DIR segments are random hits. For inverted DIR, this was confirmed by the fact that the average lengths were not different from the average length of DIR segments found in permuted sequences (11 bp; P = 0·57). Concerning DIR in normal orientation, analysis of the individual sequences was required. Germline DIR are very long (over 180 bp) and include a conventional D gene from family 1 at the 3′ end. Of the 41 DIR segments found, 12 could be explained as a conventional family 1 D gene extended upstream by 1 to 4 N nucleotides, resembling the flanking germline DIR sequence. When these sequences were removed from the calculations, the median length of the remaining DIR segments was 11 bp, which is not different from that of DIR in permutated sequences (11 bp; P = 0·20). Thus, we conclude that DIR are not used at a significant frequency in the normal memory repertoire.
Use of chromosome 15 open reading frames
The 42 OR15 segments found in the sequences appeared to be longer than D genes in the permutated sequences (median 14 bp compared with 10 bp). The OR15 genes all have very high identity to a conventional D gene and most of the sequence differences are in a hotspot for SHM (RGYW) in the conventional D genes (data not shown). To test if the assignment of OR15 could be a consequence of SHM in a conventional D gene leading to sequence mimicry with an OR15, we compared the assignment of OR15 in mutated and unmutated sequences (judged by mutations in the VH region). Of the 42 OR15 assignments, only five were in unmutated sequences (out of 2113 unmutated sequences in total) which is statistically different from the frequency in mutated sequences (37 OR15 in 4086 sequences) (χ2 = 8·29; P = 0·004). Furthermore, the OR15 found in unmutated sequences had a median length of 9 bp, which is not different from the median lengths of D segments found in permutated sequences (P = 0·18), and they are therefore probably accidental hits. Only eight inverted OR15 segments were found in the sequences. These had a median length not different from that of the D genes in permutated sequences (P = 0·91). Thus, hotspot mutations in conventional D genes are able to account for the OR15 that are not explainable as stochastic hits. We therefore conclude that OR15 are not used at a significant frequency in the memory B-cell repertoire, if they are used at all.
Use of multiple D genes
Using JointMLb, we found 4082 sequences with exactly one D-gene segment and 65 sequences with two D segments. For the permutated sequences the numbers were 324 and five sequences, respectively. The ratios between sequences with one and two D genes in experimental and permutated sequences were not statistically significantly different (χ2 = 0·03; P = 0·87). With JointHHMb the numbers were 4023, 77, 321 and six, respectively (χ2 = 0·0032; P = 1·0). Therefore, our data do not support the idea that rearrangements containing two D genes are used at a detectable frequency in the peripheral repertoire.
This conclusion was further strengthened by a detailed analysis of the sequences with two apparent D genes. Nine of the 65 sequences could be explained as one long D-gene segment by allowing a single nucleotide deletion that could have arisen by a Taq error or SHM. Further, eight D-gene combinations were not feasible by deletional rearrangement because of the mutual chromosomal location of the genes – that is, the D gene closest to the JH gene in the rearrangement had a chromosomal location upstream of the D gene closest to the VH gene. In 17 other sequences, one or both of the D genes found were DIR or OR15 genes, i.e. genes not likely to be used in vivo.
The average length of the shortest of the D-gene segments in sequences with two apparent D genes was 11·6 bp, which is equivalent to that of D-gene segments in permutated sequences (11·3 bp) (P = 0·25). The average length of the longest of the two D segments (18·6 bp) was not statistically different from the average length of normal D segments in the sequences (17·8 bp) (P > 0·05) but was significantly different from that of D segments in permutated sequences (P < 0·001).
Use of inverted conventional D genes
Sixty-four sequences were assigned an inverted conventional D gene. The median length of 11 bp was significantly shorter than that of D-gene segments in normal orientation (17 bp; P = 0·005). Nevertheless, the inverted segments were slightly longer than both inverted D segments in permutated sequences (10 bp; P = 0·02) and all D segments in permutated sequences (10 bp; P = 0·001). This may, at least in part, be explained by the fact that there is a certain sequence identity between conventional D genes in the inverted and normal directions. However, while no inverted D segments in the permutated sequences were longer than 15 bp, three particularly long inverted D segments (20, 20 and 22 bp, respectively) were found in the experimental sample, prompting us to carry out a more detailed evaluation. Using JointHMMb, 100 joint permutations of the entire data set were performed and reanalysed. In no case could an inverted D segment longer than 18 bp be detected by chance. We also performed a Bootstrap analysis25 by generating 100 data sets of 6329 experimental sequences and 6329 permutated sequences from the original sets with replacements. This analysis also supported the finding that inverted D segments longer than 15 bp were found significantly more frequently amongst the experimental sequences (data not shown). Therefore, we cannot exclude the use of long inverted D genes.
The final versions of the programs (JointMLc and JointHMMc) were made to accept inverted conventional D genes with a penalty reducing illegitimate assignment of short or poorly matching segments. Using these parameters, we found two inverted D genes (20 and 22 bp long, respectively). Running the algorithm with a penalty for inversion on the entire data set using inverted D gene templates, we recovered 2302 of 4420 (52%) of the assigned D genes as inverted inverted D genes (= normal reading direction) despite the penalty. For D genes 15 bp or longer, the recovery rate was 80%. It is therefore concluded that if inverted D genes were used with the same length distribution as D genes in the normal reading direction they would often be identified by our algorithm. As only two inverted D segments were found, they constitute less than 1‰ of the D-gene segments in the peripheral memory B-cell repertoire.
PCR and cloning artefacts
Zylstra et al. claim that the risk of generating PCR and cloning crossing-over artefacts when using a template of highly homologous sequences is so large that a distinction between non-productive and productive rearrangements cannot be made.26,27 To test if this was also the case with our material, we divided our sequences into non-productive (657 sequences) and productive (5672 sequences) rearrangements based on the joint region only. A non-productive rearrangement was defined as a sequence with a joint region with one or more premature stop codons presumed to have arisen at the time of rearrangement (i.e. prior to SHM) or a number of nucleotides that changed the reading frame of the JH gene. Afterwards, insertions and deletions in the VH region upstream of the joint area were analysed. Table 2 shows that insertions and deletions preserved the reading frame in more than 95% of the rearrangements that we have marked as productive based on their joint region, while this was the case in less than 45% of non-productive rearrangements. These prevalences were significantly different for both deletions and insertions (P < 0·0001), which was expected if crossing-over was rare because productive rearrangements should show signs of selection while non-productive rearrangements should not. On the basis of these observations, we find that non-productive and productive rearrangements can indeed be distinguished in our experimental system.
Table 2.
Insertions and deletions in the VH region of sequences classified as being productive or non-productive based on their joint region
| Insertions | Deletions | |||
|---|---|---|---|---|
| Consequence of insertion/deletion | Productive | Non-Productive | Productive | Non-Productive |
| RF maintained | 100 | 24 | 137 | 26 |
| RF not maintained | 3 | 29 | 7 | 78 |
Insertions and deletions are said to maintain the reading frame (RF) if their lengths are divisible by 3. Significantly more insertions (P < 0·0001) and deletions (P < 0·0001) maintaining the reading frame were found in productive rearrangements than in non-productive rearrangements. One-bp insertions and deletions are excluded because they are thought to be predominantly caused by Taq errors. However, inclusion of these insertions/deletions does not change the results (P < 0·0001 in both cases).
D gene usage and experimental bias
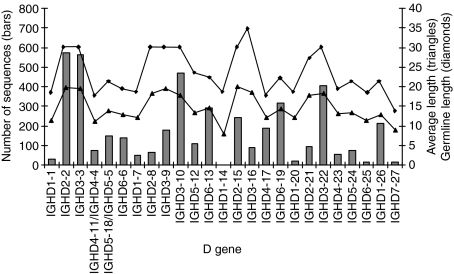
Not all D genes are used at equal frequency. Figure 1 shows the D-gene usage as well as the average lengths of each D-gene segment. The average length varied greatly from 8·9 bp for IGDH7-27 (median 9 bp) to 20·1 bp for IGHD2-15 (median 21 bp); however, as can be seen from Fig. 1, this is almost exclusively a result of differences in the length of the germline genes. IGHD2-2, IGHD3-3, IGHD3-10 and IGHD3-22 are the most commonly used D genes, while most genes from D gene families 1, 4 and 6 and IGDH7-27 are rarely used. The apparent, although not complete, correlation between germline length and D gene usage prompted us to investigate whether we systematically overlooked D genes with short germline lengths.
Figure 1.
Diversity (D)-gene usage and average lengths of the different D genes found in all rearrangements. IGHD4-4 and IGHD4-11 have the same coding sequences, and these two genes can thus not be distinguished. The same is the case for IGHD5-5 and IGHD5-11. If more than one allele of a given D gene exists, the numbers for all alleles have been added. The figure shows that the different D genes are used at very different frequencies, ranging from one sequence for IGHD1-14 to 572 for IGHD2-2. The average lengths of the used D genes correspond to the germline lengths, and the D genes with longer germline sequences tend to be used more often than the shorter D genes.
Two sets of 15 000 artificial sequences were generated using IGHV3-23*01 and the JH genes with the same frequency as found amongst our experimental sequences. Each of the 32 D-gene alleles was used at equal frequency. Other features of the joint region were constructed using the frequencies found in non-productive, unmutated sequences and one set of sequences were randomly mutated using a 5% substitution rate for each nucleotide and a 1 : 1 transitions:transversion rate. The sequences were analysed using JointMLc and the number of assigned D genes for each allele was counted and compared with that for the alleles used to generate the sequences. For both data sets, there was a strong correlation between the frequency of recovery of a D gene and the germline length of that gene. For the unmutated sequences, JointMLc identified 98–101% (median 100%) of the D genes with germline lengths of 28–38 bp (families 2 and 3). For germline lengths of 20–23 bp, 82–94% (median 84%) of the D genes were found, and for germline lengths of 16–19 bp, 52–85% (median 73%) were found, while only 21% of the very short IGHD7-27 (germline length 11 bp) were found. The corresponding numbers for the mutated sequences were: 28–38 bp, 96–103% (median 99%); 20–23 bp, 73–87% (median 79%); 16–19 bp, 46–79% (median 65%), and IGHD7-27, 15%. Thus, the shorter the germline length of a D gene, the greater the chance of JointMLc overlooking the D gene when it is trimmed. This effect was slightly increased in mutated sequences. Although this means that the D-gene usage of families 1, 4, 5, and 6 was probably underestimated by 20–54%, the effect was not able to explain the major differences (in some cases over 40-fold) in D-gene usage outlined in Fig. 1.
Thus, we can show that the skewed D-gene usage in the peripheral memory B-cell repertoire is a real biological phenomenon that is not just a consequence of differences in D-gene germline lengths. In support of this conclusion, a skewed D-gene usage with a similar pattern was also found in the sequences using IGHV3-h (data not shown).
Footprints of VH replacement
It has been suggested that autoreactive B cells can be rescued from apoptosis in the bone marrow by replacing the VH gene with another VH gene (for example, references28,29). The mechanism is thought to involve the use of a cryptic heptamer sequence found at the 3′ end of most VH genes,29 and a replacement reaction will leave behind a small 3–10-bp footprint sequence from the first VH gene. We searched for perfect matches of 4 to 8 nucleotides from footprints from other VH genes in N1 (the N region between VH and D) of our sequences. The numbers of matches of different lengths were compared with the numbers of matches of identical lengths found in a set of permutated sequences where only N1 was permutated. We used two different sets of footprint sequences. One contained the footprints of VH genes located centromeric to IGHV3-23, as only primary rearrangements using these genes would be expected to be able to make a VH replacement with IGHV3-23. The other set was a control set of footprint sequences from IGHV3-23 itself and all genes located telomeric to this gene as these genes cannot be replaced by IGHV3-23. We found many matches with both sets of footprint sequences (Table 3). The frequencies of matches, however, did not differ significantly from the frequencies found in the permutated sequences, and we therefore conclude that VH replacement does not significantly influence the repertoire. The same conclusion was reached for the sequences using IGHV3-h (data not shown). Another control experiment looking for footprint sequences in N2 (the N region between D and JH) gave equally high frequencies of matches (data not shown), also supporting the conclusion that the matches are random hits.
Table 3.
Number of sequences containing a match of the given length of any of the VH footprint sequences found in the experimental sequences (seq) and the permutated sequences (perm), respectively
| Footprint sequences centromeric to IGHV3-23 | Footprint sequences telomeric to IGHV3-23$ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Match length (bp) | Seq match | Seq No match | Perm match | Perm No match | P-value* | Seq match | Seq No match | Perm match | Perm No match | P-value |
| 4 | 1301 | 2318 | 1214 | 2405 | 0·017 | 1939 | 1680 | 1860 | 1759 | 0·033 |
| 5 | 377 | 2867 | 347 | 2897 | 0·126 | 564 | 2680 | 495 | 2749 | 0·011 |
| 6 | 66 | 2812 | 56 | 2822 | 0·205 | 100 | 2778 | 91 | 2787 | 0·270 |
| 7 | 8 | 2443 | 12 | 2439 | 0·251 | 16 | 2435 | 8 | 2443 | 0·075 |
| 8 | 0 | 2115 | 0 | 2115 | 1·000 | 0 | 2115 | 0 | 2115 | 1·000 |
Because of the many comparisons, differences are only considered to be significant if P < 0·01.
Footprint sequences from VH genes telomeric to IGHV3-23 were included as a negative control, as replacement of any of these sequences by IGHV3-23 would require the use of the homologous chromosome, which we believe is very unlikely.
Comparison between JointMLc and JointHMM
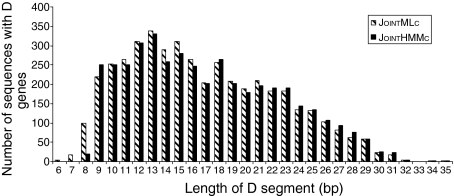
A detailed comparison of the performance of JointMLc and JointHMMc was made. Figure 2 demonstrates a high degree of consistency between the D genes identified by the two programs. However, JointMLc identified 4420 D genes, which is significantly more than JointHMMc (4285 D genes) (P = 0·01). The programs showed a strong agreement as to which sequences could be assigned a D gene and which could not (agreement for 94% of the sequences). Eighty-eight per cent of the identified D genes were identical, 9% were from different alleles of the same gene, 2% were from different genes from the same family, and only 1% were from different families. JointMLc found significantly more short D-gene segments (< 15 bp) than JointHMMc (P < 0·0001), whereas JointHMMc found significantly more long (> 20-bp) D-gene segments. However, when the D genes with insertions and deletions were omitted, the latter difference disappeared (P = 0·9).
Figure 2.
Length of the diversity (D) genes identified by the computer programs JointMLc and JointHMMc. JointMLc and JointHMMc identified 4420 and 4285 D genes, respectively, in the 6329 sequences. JointMLc identified significantly more short D genes (length < 15 bp) than JointHMMc (P < 0·0001).
The use of insertions and deletions in the D segments was not permitted in JointMLc. With JointHMMc, we identified 148 sequences with insertions or deletions in the identified D segment (3%); of these, 60 were in unmutated sequences and 88 were in mutated sequences. The ratio of D genes with insertions/deletions was thus similar in the mutated and unmutated sequences (P =0·90), suggesting that the insertions/deletions in D segments were either wrongly assigned by JointHMMc or had not arisen by SHM but perhaps by Taq errors.
It was concluded that the programs were in close agreement, but that JointMLc was slightly superior in recognizing short D segments in mutated sequences and therefore preferred in the following analysis.
Comparison with other publicly available programs
To test the performance of JointMLc against that of other publicly available programs, an unmutated and a mutated set each consisting of 1000 artificial test sequences using IGHV3-23*01 and JH6*02 were constructed. D-gene usage, CDR3 lengths, trimming of the gene ends, P nucleotide lengths and frequencies and N nucleotide lengths and compositions were constructed to resemble the frequencies found in unmutated productive sequences and mutated productive sequences, respectively. For the mutated data set, mutations in residues in the VH and JH genes were introduced independently of each other at the frequency at which they were found in productive, mutated sequences. In the sequence between the VH and JH genes, nucleotides were mutated at the frequency found for the same nucleotide when situated in the middle of a similar 5-bp motif in the VH region of productive, mutated sequences.
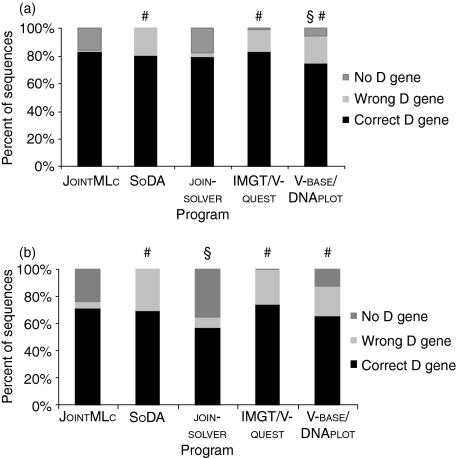
Both sets of sequences were tested using JointMLc and four publicly available programs: joinsolver14 (http://joinsolver.niams.nih.gov/) (current version January 2006), SoDA16 (http://dulci.org/soda/index.html) (current version January 2006), IMGT/V-quest12 (http://imgt.cines.fr/textes/vquest/) (current version January 2006) and V-base/DNAplot(http://vbase.mrc-cpe.cam.ac.uk/) (current version January 2006). The numbers of correctly and wrongly assigned D genes for each program were noted as a measure of the sensitivity (fraction of sequences with a correct D-gene assignment) and the specificity [1 − (fraction of sequences with a wrong D-gene assignment)] of the programs, respectively. A total of 1000 sequences were analysed by JointMLc and SoDA, which can perform bulk analysis, whereas only 200 sequences were analysed by the other programs. To be counted as correctly assigned D genes, the D genes found by joinsolver had to conform to the required minimum match length for the given VH–JH distance as indicated by the authors.14Figure 3 shows the percentage of correctly and wrongly assigned D genes as well as the percentage of sequences in which the programs could no find a D gene in the unmutated (Fig. 3a) and mutated (Fig. 3b) data sets, respectively. JointMLc found more correct D genes in the unmutated sequences than SoDA, joinsolver and V-quest; however, the sensitivity of JointMLc was not significantly better (P > 0·05 for all comparisons). JointMLc had a significantly higher sensitivity than V-base/DNAplot on unmutated sequences (P =0·006). When comparing the number of wrongly assigned D genes, JointMLc found significantly fewer false D genes than SoDA, IMGT/V-quest and V-base-DNAplot (P < 0·001), showing that JointMLc has a higher specificity. Also, because both IMGT/V-quest and V-base/DNAplot do not distinguish between mutated and unmutated sequences, more than half of the D genes correctly assigned by these two programs were too long because of accepted mutations in the flanks of the D-gene segments. No differences were found between the specificities of JointMLc and joinsolver (P = 0·5) on unmutated sequences.
Figure 3.
Comparison of the fraction of correctly and wrongly assigned D genes and sequences where a D gene could not be assigned by JointMLc, SoDA, joinsolver, IMGT/V-quest and V-base/DNAplot in a test set of (a) unmutated artificial sequences and (b) mutated artificial sequences. For JointMLc and SoDA the frequencies are based on the analysis of 1000 sequences from each data set, whereas only 200 sequences were analysed with the other programs.
§JointMLc has significantly higher sensitivity (more correctly assigned D genes) (P < 0·01).
#JointMLc has significantly higher specificity (fewer wrongly assigned D genes) (P < 0·001).
For mutated sequences, JointMLc also showed a significantly higher specificity than SoDA, IMGT/V-quest and V-base/DNAplot (P < 0·001) but its specificity was still not higher than that of joinsolver (P = 0·07). The sensitivity of JointMLc was significantly better than that of joinsolver (P < 0·001) but, despite finding more correct D genes, it was not significantly better than SoDA, IMGT/V-quest and V-base/DNAplot (P > 0·05). Thus, JointMLc has a higher predictive performance for D gene assignment in mutated and unmutated sequences than the four other algorithms.
When analysing the available sequences from the joinsolver material,14 JointMLc found more D genes in both the productive (66·5% compared with 64·4%) and the non-productive (78·4% compared with 71·4%) sequences, but the differences were not significant.
Discussion
Reliable analysis of V(D)J rearrangements requires computer algorithms with high specificity and sensitivity for the different components of the joint, even in the presence of somatic hypermutations. Development and optimization of such algorithms, however, demand a very detailed knowledge of the preferences of the rearrangement machinery and mutation machinery forming the antibody repertoire. Improvement of algorithms is therefore an iterative procedure in which improved definition of the joint components and improved algorithms go hand in hand. In order to improve the analysis of human heavy chain V(D)J rearrangements, we sequenced the (to date) largest sample of well-defined rearrangements from memory B cells. The data set contained 6329 non-clustered sequences using IGHV3-23*01, and our analyses showed that neither Taq errors nor recombination artefacts arising during PCR or cloning influenced our results significantly. We developed two algorithms employing different strategies to analyse the rearrangements. Using a stepwise strategy, both programs were initially very flexible but were gradually restricted to only accepting features that could be validated by rigorous statistical analyses. One of the algorithms, JointMLc, turned out to be slightly better than the other, and the data discussed in the following sections are based on this algorithm.
P nucleotides are found at untrimmed ends only and their frequency depends on the gene
P nucleotides arise when the Artemis–DNA–protein kinase catalytic subunit (PKcs) complex cuts the hairpin loop of the coding end at a position different from the position of loop formation. We found that P nucleotides were created only at untrimmed ends, which is consistent with previous results.14,30 It therefore appears that the recombination activated gene/high-mobility group I (RAG/HMG1) complex exclusively cuts the DNA strand exactly between the heptamer and the coding sequence in humans.
We chose not to accept P nucleotides 1 nucleotide in length because of the high likelihood of accidental matches to N nucleotides. However, our analysis showed that the N nucleotide immediately adjacent to untrimmed ends was more often than expected found to be the nucleotide that would be created as a 1-nucleotide P segment. The length of the accepted P nucleotides varied from 2 to 7 nucleotides, but 69% were 2 nucleotides long. This is in good agreement with a study showing that the Artemis–DNA–PKcs complex most often cuts at position +2 in vitro.3 This is likely to be a result of the single-stranded nature of the tip of the DNA loop, as the Artemis–DNA–PKcs complex is known to cut preferentially at transitions between single- and double-stranded DNA.31 We found that the frequency and the length of P nucleotides varied depending on the germline gene by which they were templated. This is consistent with an earlier report.30 VH (34·3% of untrimmed ends) and upstream ends of D genes (25·0%) had P nucleotides more often than JH genes (13·9%) and downstream ends of D genes (19·8%). There are two possible explanations for this: either the Artemis–DNA–PKcs complex acts differently on the two joints, perhaps because of differences in the sequences involved, or the degree of trimming varies during the formation of the two joints. If more trimming takes place during the formation of the D–JH joint, the P nucleotides formed are more likely to be trimmed off. In support of this hypothesis, we found that JH genes were trimmed to a significantly greater extent than VH genes (7·2 bp trimmed off the JH genes on average compared with 1·6 bp for VH genes). This was true even for non-productive rearrangements and therefore was not attributable to selection. However, no difference was found in the respective ends of the D genes (4·6 bp at the 3′ end compared with 4·7 bp at the 5′ end), which argues against this mechanism. Differences in trimming will be described in detail elsewhere (L. Ohm-Laursen et al., manuscript in preparation). For the individual JH or D genes, we found significant variations in the frequencies of P nucleotides (data not shown), suggesting that the local sequence plays a role in cutting and/or trimming. However, the data provide no clear picture regarding which sequences promote or inhibit the formation of P nucleotides.
DIR and OR15 play no significant role in the peripheral immunoglobulin repertoire, while inverted D genes are used infrequently
Thorough analysis of the length distribution and nucleotide composition of the DIR, OR15 and multiple D-gene segments showed that the presence of these genes could be explained by either N nucleotide additions to conventional genes, somatic hypermutations of conventional genes or hits by random chance. We therefore suggest that these phenomena do not play a significant role in the peripheral repertoire of memory B cells, which is consistent with the results of Corbett et al.13
Others have, however, reported that DIR, OR15 and multiple D genes can be found in the peripheral repertoire;11,14,15 the contrasting findings obtained here may be a result of our larger, better defined and more thoroughly analysed material.
The use of inverted D genes could not be rejected but was found to be a very rare event, present in less than 1‰ in the peripheral memory B cells. This finding is supported by in vitro studies showing that inversion is mechanistically possible but relatively rare (compared with deletional rearrangement resulting in D genes in the normal reading direction) as a result of the sequence of the D gene itself and the RSS.32,33
Skewed D-gene usage
The usage of the different D genes was strongly skewed, and the five most frequently used genes (IGHD2-2, IGHD3-3, IGHD3-10, IGHD3-22 and IGHD6-19) accounted for more than 50% of the D-gene usage. These D genes also dominated the repertoire in other studies13,14 and in the rearrangements using the IGHV3-h pseudogene, and it is therefore unlikely that the result is caused by VH-gene restriction in this study. The D genes belonging to families 2 and 3 are the longest of the D genes, making them more likely to be recognized by the algorithms even after extensive trimming. An analysis of artificially generated sequences showed that JointMLc recovered 100% of the long D genes and a decreasing fraction of the shorter genes correlating with decreasing germline length. This bias is probably an inherent feature of all algorithms for identification of D genes but has, to our knowledge, not been addressed by others. However, even if we corrected for the bias of the JointMLc algorithm towards D genes with longer germline lengths, the same genes still dominated the repertoire and genes belonging to families 1 and 7 were still rare. This further indicates that the skewed usage of D genes was not an artefact of the algorithm.
Sequences without a detectable D-gene segment
In 36% of the mutated and 20% of the unmutated sequences, the JointMLc algorithm could not assign a D gene. These rearrangements could have arisen in non-conventional recombination events directly between a VH gene and a JH gene. However, we consider it much more likely that they were conventional rearrangements where the D gene had been extensively trimmed and/or mutated so that it could not be assigned by our algorithms. The fact that we found more D genes in the unmutated sequences supports this notion. Also, the analysis of the artificial sequences showed that over 54% of the sequences, unmutated as well as mutated, for which JointMLc could not assign a D gene had a D gene that was shorter than 5 bp in length, making secure identification very difficult.
Footprints of VH replacement could not be found
In vitro studies of the human B-cell line EU1229 or the Abelson mouse pre-B-cell line34 and in vivo studies of mice made transgenic for an autoreactive rearrangement28 have provided compelling evidence that rearrangements can be edited and the VH-gene segment substituted by another VH gene by use of a cryptic heptamer sequence found at the 3′ end of most VH genes.29 For light chains, receptor editing deleting a whole non-productive rearrangement and replacing it with a new one has also been shown.35 Several studies claimed to find footprints of other VH genes in about 5% of N1 regions in human B cells, suggesting that VH replacement is an important contributor to the repertoire.15,29 We found footprint sequences in approximately 12% of our sequences when we looked for a 5-nucleotide match. However, this frequency is not significantly different from the frequency of footprint sequences found in permutated sequences. Similar results were obtained by analysis of sequences using IGHV3-h. Using the N2 region as a negative control, like Collins et al.,15 we found an equally high frequency of footprints in N2, supporting the possibility that VH footprints are indeed random hits. Also, we found an even higher frequency (approximately 17%) of illegitimate footprint sequences defined as footprints of VH genes with a chromosomal location telomeric to IGHV3-23. As we consider it very unlikely that replacements can use VH genes on the other chromosome 14 at this high frequency, we suggest that footprint sequences found in our sequences are a result of N nucleotides added by TdT and hence that VH replacement does not play an important role in the repertoire in humans. It is therefore likely that this process is restricted to cell lines or certain animals.
Comparison with other programs
JointMLc was compared with four publicly available programs using two sets of artificial rearrangements made to resemble productive unmutated or mutated sequences, respectively. JointMLc performed significantly better than SoDA, IMGT/V-quest and V-base/DNAplot in terms of specificity on both unmutated and mutated sequences. Compared with V-base/DNAplot it also had a better sensitivity on unmutated sequences. One explanation is that V-base/DNAplot does not take VH and JH matches into account when searching for a D gene, sometimes leading to an incorrect D gene assignment.
The joinsolver algorithm has been fine-tuned for high specificity,14 and joinsolver and JointMLc performed equally well on this parameter on both data sets. However, JointMLc had a significantly higher sensitivity than joinsolver on mutated sequences. JointMLc also found more D genes in the experimental sequences analysed by Souto-Carneiro et al.14 The differences were, however, not significant, which was possibly a result of the limited size of the data sets or the fact that many of the sequences in the set were unmutated.
The results of the comparisons clearly show that an algorithm must be fine-tuned for maximum sensitivity and a minimum of false positive hits (maximum specificity). For either of the two parameters, JointMLc performed significantly better than other publicly available programs. We have made an online program (VDJsolver, version 1·0) based on the JointMLc algorithm available at http://www.cbs.dtu.dk/services/VDJsolver. The program accepts bulk submissions and has been modified to be able to analyse rearrangements to all the VH genes from the IMGT database (http://imgt.cines.fr/), making it a user-friendly analysis tool.
Acknowledgments
We thank Nina Eggers for excellent technical assistance. This study was supported by grants from the Danish Medical Research Council (grant 22-01-0156), the Institute of Clinical Research, the University of Southern Denmark, the Toyota Foundation, and the Lands-Landsdelspuljen for the County of Funen.
Abbreviations
- DIR
D gene with irregular recombination signal sequence
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- HMM
hidden Markov model
- IGHJ
immunoglobulin heavy chain J gene
- IGHV
immunoglobulin heavy chain V gene
- JH
heavy chain J gene
- N nucleotide
non-templated nucleotide
- OR15
chromosome 15 open reading frame
- P nucleotide
palindromic nucleotide
- SHM
somatic hypermutation
- TdT
terminal deoxynucleotidyl transferase
- VH
heavy chain V gene
References
- 1.Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 2000;13:37–45. doi: 10.1016/s1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–26. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–94. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 4.Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–69. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Repasky JA, Corbett E, Boboila C, Schatz DG. Mutational analysis of terminal deoxynucleotidyltransferase-mediated N-nucleotide addition in V(D)J recombination. J Immunol. 2004;172:5478–88. doi: 10.4049/jimmunol.172.9.5478. [DOI] [PubMed] [Google Scholar]
- 6.Thai TH, Purugganan MM, Roth DB, Kearney JF. Distinct and opposite diversifying activities of terminal transferase splice variants. Nat Immunol. 2002;3:457–62. doi: 10.1038/ni788. [DOI] [PubMed] [Google Scholar]
- 7.O'Driscoll M, Cerosaletti KM, Girard PM, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–85. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 8.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–9. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 9.Bogue MA, Wang C, Zhu C, Roth DB. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 10.Brezinschek HP, Foster SJ, Brezinschek RI, Dorner T, Domiati-Saad R, Lipsky PE. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(–)/IgM+ B cells. J Clin Invest. 1997;99:2488–501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanz I. Multiple mechanisms participate in the generation of diversity of human H chain CDR3 regions. J Immunol. 1991;147:1720–9. [PubMed] [Google Scholar]
- 12.Giudicelli V, Chaume D, Lefranc MP. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 2004;32:W435–40. doi: 10.1093/nar/gkh412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett SJ, Tomlinson IM, Sonnhammer EL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, ‘minor’ D segments or D-D recombination. J Mol Biol. 1997;270:587–97. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- 14.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J Immunol. 2004;172:6790–802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 15.Collins AM, Ikutani M, Puiu D, Buck GA, Nadkarni A, Gaeta B. Partitioning of rearranged Ig genes by mutation analysis demonstrates D-D fusion and V gene replacement in the expressed human repertoire. J Immunol. 2004;172:340–8. doi: 10.4049/jimmunol.172.1.340. [DOI] [PubMed] [Google Scholar]
- 16.Volpe JM, Cowell LG, Kepler TB. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. 2006;22:438–44. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 17.Moore BB, Meek K. Recombination potential of the human DIR elements. J Immunol. 1995;154:2175–87. [PubMed] [Google Scholar]
- 18.Kraj P, Rao SP, Glas AM, Hardy RR, Milner EC, Silberstein LE. The human heavy chain Ig V region gene repertoire is biased at all stages of B cell ontogeny, including early pre-B cells. J Immunol. 1997;158:5824–32. [PubMed] [Google Scholar]
- 19.Suzuki I, Pfister L, Glas A, Nottenburg C, Milner EC. Representation of rearranged VH gene segments in the human adult antibody repertoire. J Immunol. 1995;154:3902–11. [PubMed] [Google Scholar]
- 20.Hufnagle WO, Huang SC, Suzuki I, Milner EC. A complete preimmune human VH3 repertoire. Ann N Y Acad Sci. 1995;764:293–5. doi: 10.1111/j.1749-6632.1995.tb55839.x. [DOI] [PubMed] [Google Scholar]
- 21.Wasserman R, Ito Y, Galili N, et al. The pattern of joining (JH) gene usage in the human IgH chain is established predominantly at the B precursor cell stage. J Immunol. 1992;149:511–6. [PubMed] [Google Scholar]
- 22.Ohm-Laursen L, Larsen SR, Barington T. Identification of two new alleles, IGHV3-23*04 and IGHJ6*04, and the complete sequence of the IGHV3-h pseudogene in the human immunoglobulin locus and their prevalences in Danish Caucasians. Immunogenetics. 2005;57:621–7. doi: 10.1007/s00251-005-0035-8. [DOI] [PubMed] [Google Scholar]
- 23.Krogh A, Brown M, Mian IS, Sjolander K, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J Mol Biol. 1994;235:1501–31. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 24.Durbin R, Eddy SR, Krogh A, Mitchison GJ. Biological Sequence AnalysisProbabilistic Models of Proteins and Nucleic Acids. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 25.Press WH, Flannery BP, Teukolsky SA, Vetteling WT. Numerical Recipes in C. The Art of Scientific Computing. Cambridge, UK: Cambridge University Press; 1992. [Google Scholar]
- 26.Blanden RV, Steele EJ. Misinterpretation of DNA sequence data generated by polymerase chain reactions. Mol Immunol. 2000;37:329. doi: 10.1016/s0161-5890(00)00051-1. [DOI] [PubMed] [Google Scholar]
- 27.Zylstra P, Rothenfluh HS, Weiller GF, Blanden RV, Steele EJ. PCR amplification of murine immunoglobulin germline V genes: strategies for minimization of recombination artefacts. Immunol Cell Biol. 1998;76:395–405. doi: 10.1046/j.1440-1711.1998.00772.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 1995;3:747–55. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Zemlin M, Wang YH, et al. Contribution of Vh gene replacement to the primary B cell repertoire. Immunity. 2003;19:21–31. doi: 10.1016/s1074-7613(03)00170-5. [DOI] [PubMed] [Google Scholar]
- 30.Meier JT, Lewis SM. P nucleotides in V(D)J recombination: a fine-structure analysis. Mol Cell Biol. 1993;13:1078–92. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y, Schwarz K, Lieber MR. The Artemis. DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–51. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Gauss GH, Lieber MR. The basis for the mechanistic bias for deletional over inversional V(D)J recombination. Genes Dev. 1992;6:1553–61. doi: 10.1101/gad.6.8.1553. [DOI] [PubMed] [Google Scholar]
- 33.Pan PY, Lieber MR, Teale JM. The role of recombination signal sequences in the preferential joining by deletion in DH-JH recombination and in the ordered rearrangement of the IgH locus. Int Immunol. 1997;9:515–22. doi: 10.1093/intimm/9.4.515. [DOI] [PubMed] [Google Scholar]
- 34.Reth M, Gehrmann P, Petrac E, Wiese P. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature. 1986;322:840–2. doi: 10.1038/322840a0. [DOI] [PubMed] [Google Scholar]
- 35.Casellas R, Shih TA, Kleinewietfeld M, et al. Contribution of receptor editing to the antibody repertoire. Science. 2001;291:1541–4. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]