Abstract
The mouse Pactolus gene is an evolutionary paralogue to the CD18/β2 integrin subunit and is preferentially expressed by neutrophils. When first identified, it was assumed Pactolus would function as an adhesion receptor similar to other β integrin subunits. The analysis of mice genetically deficient in Pactolus, however, did not define any lesion in neutrophil migration, adhesion or phagocytosis. Therefore a wider analysis of the Pactolus deficiency was initiated using transcriptional profiling during an inflammatory insult. This screen identified a single transcript, CXCL13, that was elevated in cells from a peritoneal lavage of the wild type animal compared to the Pactolus-deficient animal. Our analyses confirmed resident macrophages as being responsible for the chemokine using intracellular CXCL13 staining and additional cell markers to phenotypically characterize such cells. The resident CXCL13-expressing cells (which do not express Pactolus) are functionally distinct from the macrophages recruited into the peritoneal cavity following the inflammatory stimulation since the recruited macrophages do not express detectable levels of the chemokine. The numbers and expression patterns of these resident CXCL13-expressing cells do not vary in naïve animals of wild type or Pactolus-deficient origin. Additionally, Pactolus-deficient neutrophils do not preferentially kill (compared to wild type) CXC13-expressing macrophages. These data suggest that during an inflammatory response, Pactolus may help retain CXCL13-expressing cells within the peritoneal environment.
Keywords: neutrophils, adhesion receptor, chemokine, cell migration
Introduction
The mouse Pactolus protein is encoded by the itgb2l (integrin beta 2-like) gene which is a direct duplication of that encoding the integrin β2 (CD18) subunit.1,2 Although the genes for these two proteins reside on different chromosomes, they possess virtually identical exon/intron junctions and encode extremely similar proteins. The Pactolus protein, however, does vary from CD18 in a number of key regions as it lacks the metal ion dependent adhesion site (MIDAS) domain structure that is critical for a chain pairing and substrate recognition3–7 and the cytoplasmic and transmembrane domains that show little similarity to the same domains of any of the β integrin subunits. The Pactolus protein is more heavily modified by N-linked glycosylation and sialic acid additions than CD18 even though the glycosylation addition sites are largely conserved between the two proteins.8,9 Pactolus also varies from CD18 in its tissue specific expression profile. While CD18 is expressed by granulocytes, lymphocytes and macrophages, the transcription of the Pactolus gene in vivo is restricted to cells of the neutrophil lineage.8 The rat possesses a similar gene but it is not present within the human genome.
The Pactolus protein is expressed in the earliest stages of neutrophil development. The vast majority of the protein is shuttled into a set of dense granules where it remains until the cells are stimulated.8,9 Developing granulocytes in the marrow and circulating neutrophils in the blood possess minimal Pactolus protein on their cell surface. However, neutrophils that are recruited into the tissue via an inflammatory irritation (such as injection of thioglycollate into the peritoneal cavity) have translocated virtually all of the intracellular stores of Pactolus to the cell surface. Induction of apoptosis/necrosis of murine neutrophils also results in the translocation of Pactolus to the cell surface.
A Pactolus-deficient animal was generated and analysed; the animal did not show any gross abnormalities in any organ, or in any cell distribution in the spleen or bone marrow, and blood constituents were identical to wild type.9 Migration of neutrophils into the skin via a contact hypersensitivity assay and into the peritoneal cavity via thioglycollate injection did not show any alteration in timing or cell type indicating this protein does not function in cell migration as does the CD18 protein. The Pactolus-deficient animal also did not show any difference in susceptibility to gram positive or Gram-negative bacteria compared to the wild type animal.9
Defining the function of the Pactolus protein has been an experimental challenge. Its movement to the cell surface at the terminal stages of the neutrophil life cycle suggests it could serve as a phagocytic receptor, yet targeting complexes to the protein does not enhance their uptake (as opposed to targeting complexes to CD18, which is a very effective phagocytic receptor).10 Conversely, Pactolus may serve a ligand function for the recognition and uptake by macrophages of dead and dying neutrophils. Macrophages possess a number of receptors that bind to ligands on the surface of apoptotic and necrotic cells including scavenger receptors, the phosphatidyl serine receptor, integrins and lectins.11–14 The lectins presumably bind to sugars (either constitutively displayed or shuttled to the cell surface upon apoptosis) held by proteins or lipids. Pactolus could serve as a lectin ligand because of the extensive addition of sialic acid residues.
Based upon the models described above, Pactolus binding to a cognate receptor could have an inhibitory or activating response to the responding cell. Such a response could be expected to alter the function(s) of a cell including potentially altering its transcriptional profile. To test if this was the case, an experimental plan was devised using Pactolus-deficient mice to create an inflammatory response that would bring necrotic and apoptotic neutrophils together with macrophages recruited into the site. Transcriptional profiling of this event comparing the Pactolus-deficient animal with wild type animals identified a single gene product, CXCL13, that varied between the types of mice. The source of the CXCL13, however, was not from the recruited macrophages but from the resident, peritoneal cells present prior to the inflammatory insult, suggesting that the Pactolus deficiency alters the maintenance of such cells within the peritoneal cavity.
Materials and methods
Mice
BALB/c mice were obtained from the National Cancer Institute, National Institutes of Health (Bethesda, MD), and 129/sv mice were obtained from the Jackson Laboratory (Bar Harbor, ME). All animal experiments were reviewed and approved by the University of Utah Animal Use Committee. Previously described Pactolus-deficient mouse9 were back-crossed seven times to BALB/c and used for the described experiments. Genotyping of such animals by microsatellite analysis was carried out by the University of Utah DNA Sequencing core facility.
Affymetrix analysis
Total RNA was isolated from peritoneal lavage cells using CsCl guanidine.15 Equal amount of total RNA (3·33 µg) from five mice were pooled. Duplicate mouse 2.0 Affymetrix chips were hybridized per pool. All of the individual mouse RNA samples were then analysed with gene specific sequences and quantitative reverse transcription–polymerase chain reaction (RT–PCR) to verify the Affymetrix data.
RT–PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA was resuspended in water and quantified by A260 absorbance. cDNA was synthesized from 5 µg of RNA, 10 µl of 5× first strand buffer, 5 µl of 5 mm deoxynucleotide phosphate (dNTP), 5 µl of 0·1 m dithiothreitol, 2 µl of 1·25 mm random primers (Life Technologies, Inc., Gaithersburg, MD), 2 µl of Moloney murine leukaemia virus reverse transcriptase, and water to final volume of 50 µl. The reaction was incubated at 37° for 1 hr cDNA was cleaned using a PCR purification kit (Qiagen, Valencia, CA). cDNA levels were assessed using quantitative PCR on the LightCycler (Roche, Indianopolis, IN). Amplification was performed in a 10 µl final volume containing 3 mm MgCl2, 50 mm Tris (pH 8·3), 500 ng of bovine serum albumin/µl, 200 µm (each) dNTP, 1 : 30 000 dilution of SYBR Green I (Molecular Probes, Eugene, OR), 5 µm (each) primer, 0·05 U of Taq polymerase/µl, 11 ng of TaqStart antibody (BD Biosciences, San Diego, CA)/µl. Oligonucleotides used were Actin: #62; 5′-GTAACAATGCCATGTTCAAT-3′, and #339; 5′-CTCCATCGTGGGCCGCTCTAG-3′, CXCL13: #2499; 5′-AACTCCACCTCCAGGCAGAATG-3′, and #2500; 5′-TGTGTAATGGGCTTCCAGAATACC-3′, β-2 microglobulin#2723; 5′-AGACTGATACATACGCCTGCAG-3′, and #2724; 5′-GCAGGTTCAAATGAATCTTCAG-3′, F4/80#2715; 5′-TCCTCTTCTGGGGCTTCAG, and #2716; 5′-GAAAGTTGGTTTGTCCATTGC-3′.
Neutrophil isolation
Bone marrow neutrophils were isolated following the protocol previously described.16 Briefly, bone marrow tissue was flushed from murine femurs with cold phosphate-buffered saline (PBS). The red blood cells were lysed by treating the marrow with ACK lysing buffer (0·15 m NH4CL, 1 mm KHCO3, and 0·1 mm Na2-ethylenediaminetetra-acetic acid) for 4 min on ice. After two washes, the cells were resuspended in PBS and layered onto a Percoll (Amersham Pharmacia Biotech, Arlington Heights, IL) gradient consisting of equal volumes of the different densities of PBS and 1·080 g/ml percoll (lower layer). The gradients were then centrifuged at 1000 g for 20 min at 4°. Cells were visible as a pellet (most mature neutrophils) and at the interface of the two densities (mostly less mature neutrophils and B cells). The pellet was removed and centrifuged in PBS to remove the Percoll and were ready for use. Gradient dilutions were made following the protocol from Amersham Pharmacia Biotech using 0·15 m NaCl.
Thioglycollate recruitment
Cells were recruited to the peritoneal cavity after injecting 0·5 ml 3% thioglycollate (Difco, B. D. Diagnostics, Franklin Lakes, NJ). At specific time points a peritoneal lavage was performed using cold PBS. For isolating neutrophils from the peritoneal lavage, a Percoll density of 1·06 g/ml was used (see neutrophil isolation).
In vitro macrophage incubation
Peritoneal macrophages were harvested 4 days after 0·5 ml 3% thioglycollate was administered. Lavages from three individual mice were pooled and plated into six-well dishes (∼1 × 106/well) and incubated over night at 37°. Mature bone marrow neutrophils (∼5 × 106), and lipopolysaccharide (LPS, 5 ng/ml) was added. At each time point the wells were washed in PBS and the adherent cells were lysed in TRIzol for total RNA purification.
Fluorescence-activated cell sorting (FACS) analysis
After harvesting peritoneal cells, red blood cells were lysed with ACK (see neutrophil isolation). 1 × 106 Cells were resuspended in 100 ml staining buffer (PBS + 0·1% bovine serum albumin (BSA)) prior to antibody staining. Antibody used were Gr-1 (clone RB6-8C5), B220 (clone RA3-6B2), CXCR5 (clone 2G8) and I-A/I-E (clone 2G9) all from BD Bioscience (San Diego, CA). F4/80 (clone BM8) from Caltag (Burlingame, CA). CXCL13 (polyclonal goat) from R & D Systems (Minneapolis, MN).
Magnetic bead separation
After harvesting peritoneal cells, cells were stained and separated as recomendet by Miltenyi Biotech (Auburn, CA). Cells were labelled with B220-Microbeads from Miltenyi Biotech. or F4/80-biotin (clone BM8) from Caltag plus Streptavidin Microbeads from Miltenyi Biotech. Manufacturer's protocol was followed.
Results
Depressed CXCL13 expression in inflammatory responses of Pactolus-deficient animals
A Pactolus-deficient animal was created using standard gene knockout technologies, and analysed.9 Because the Pactolus gene is a direct duplication of that of CD18/β2 integrin2 phenotypes similar to those of animals lacking CD18 (adhesion, migration, phagocytosis) were anticipated.17,18 However, animals lacking Pactolus were normal for marrow and blood populations, for migration of inflammatory cells into sites of irritation, and for responses to infectious agents. Reasoning that any phenotype associated with a Pactolus deficiency would most likely be when the protein is translocated to the cell surface, we utilized a standard peritonitis model to recruit terminal neutrophils into the peritoneal cavity of the mouse. Neutrophils recruited into such a site do not re-circulate back into the animal but are removed by recruited macrophages.11 Our hypothesis was that the presence of Pactolus on the surface of recruited neutrophils would influence their interactions with other cell types in the peritoneal cavity (potentially both resident and recruited), and the absence of such interactions with the Pactolus-deficient animal might result in altered transcriptional responses.
The majority of the Pactolus protein is held within the maturing neutrophil in granules until such cells are induced to degranulate, either via necrosis and apoptosis, or direct activation. The vast majority of Pactolus is translocated to the surface of the neutrophil at the terminal stage of its life span.9 The Pactolus deficiency (originally derived from a 129/sv and C57BL/6 chimera) was bred upon the BALB/c background for seven generations. Micro-satellite mapping of the backcrossed animals indicated that the bulk of chromosome 16 (and the rest of the genome) was of BALB/c origin except for the most telomeric region of that chromosome which is where the Pactolus gene resides. This region of the chromosome was of 129/sv derivation consistent with the genetic background of the ES cell used to create the knockout animal (Table 1).
Table 1.
Genotype of chromosome 16 from Pactolus-deficient BALB/c mouse
| Locus | Distance (cM) | MGI (cM) | Strain |
|---|---|---|---|
| D16Mit165 | 10·9 | 10·3 | BALB |
| D16Mit146 | 17·5 | 16·9 | BALB |
| D16Mit136 | 26·2 | 28·2 | BALB |
| D16Mit157 | 29·5 | 34·1 | BALB |
| D16Mit49 | 39·3 | 53 | BALB |
| D16Mit118 | 44·8 | 57 | BALB |
| D16Mit52 | 51·4 | 66·8 | 129/sv |
| Pactolus | 69 |
Thioglycollate-induced peritonitis was induced in five wild type BALB/c and five Pactolus-deficient animals and total cells were isolated from the peritoneal cavity 16 hr after treatment. FACS analysis of each animal's peritoneal cells after harvest showed about 60% of the cell population were Gr-1 positive neutrophils and 25% were F4/80 positive macrophages (data not shown). This single time point was chosen because it represented the time at which neutrophil recruitment into the cavity was waning and the influx of macrophages was increasing. RNA was obtained from the total cell population of each treated animal, pooled and was used to screen the 2.0 version of the mouse Affymetrix gene array. Rank product analysis was performed on the samples and genes showing enhanced or depressed expression in the two sets of samples were ranked.
A number of genes displayed altered expression profiles in this screen, these genes were subdivided into three groups. The first group could not be reproduced by quantitative RT–PCR from each of the individual animal samples used to generate the pool thus the altered expression was caused by a single aberrant sample in the original pool. The second set mapped to genes on chromosome 16 closely linked to Pactolus, and were presumably of 129/sv origin. These genes showed expression differences in comparing 129/sv recruited cells versus those of BALB/c derivation, and were eliminated from our study. The third set consisted of a single gene, CXCL13, which was identified as having a 2·4-fold elevation of expression in the wild type animal compared to the knockout (the CXCL13 gene is on murine chromosome 5). Each of the samples used to create the pool were independently analysed: each wild type sample expressed a higher level of CXCL13 than any of the Pactolus-deficient samples (data not shown).
The altered expression of CXCL13 in the Pactolus-deficient animal, compared to wild type BALB/c animals, was analysed. Cells were obtained from test animals following thioglycollate injection 16 hr after induction and evaluated for CXCL13 expression. As shown in Fig. 1(a), there is a reduction in CXCL13 expression in the 16 hr recruited cells between the BALB/c sample versus that of the Pactolus-deficient animal (t = 0·06). A comparison of CXCL13 expression between the BALB/c recruited cells versus those from 129/sv lineage (Fig. 1b) did not demonstrate any significant difference. The influx of neutrophils into the peritoneal cavity 16 hr after the inflammatory irritation was the same between the BALB/c wild type and Pactolus-deficient animal (Fig. 1c). To determine if there were inherent differences in CXCL13 expression between the various strains of mice used in these analyses, we analysed untreated resident peritoneal cells for transcript anomalies. As shown in Fig. 1(d), the levels of CXCL13 expression in cells from a naïve, unfractionated peritoneal wash from 129/sv, BALB/c and the Pactolus-deficient animals were not significantly different. Note that the number of CXCL13 transcripts per 1000 β-actin transcripts is about 40 fold higher in this panel versus Fig. 1(a or b). This apparent reduction is caused, in part, by the influx of neutrophils and other cell types that possess transcripts for β-actin but lack those for CXCL13 (see below).
Figure 1.
Differential expression of CXCL13 in wild type and Pactolus-deficient animals. (a) Four BALB/c and four Pactolus-deficient mice (bred upon BALB/c background for seven generations) were induced with a peritoneal injection of thioglycollate, cells were isolated from the cavity 16 hr later. Quantitative RT–PCR was done on each sample independently comparing CXCL13 transcripts per 1 × 103β-actin transcripts. *t = 0·06. (b) Eight 129/sv and four BALB/c mice were induced with a peritoneal injection of thioglycollate and cells were isolated from the peritoneal cavity 16 hr later. Quantitative RT–PCR was done comparing CXCL13 transcripts per 1 × 103β-actin transcripts on each individual sample. (c) Percentage of GR-1 positive cells (granulocytes) in a peritoneal lavage 16 hr after thioglycollate treatment between the BALB/c (five mice examined and averaged) and Pactolus-deficient animals (five animals examined and averaged). (d) Cells were taken from a naïve peritoneal lavage from five 129/sv, five BALB/c and four Pactolus-deficient animals. RNA was obtained from the individual cell populations and analysed for CXCL13 transcripts per 1 × 103β-actin transcripts. Analysis of single experiments is shown but each were done multiple times and generated the same data.
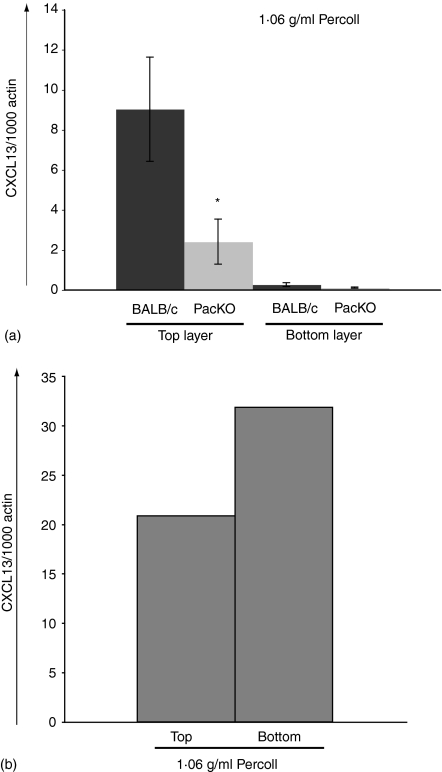
The previous analyses were performed on RNA obtained from the total cellular pool of recruited and resident cells. The expansion of cell numbers 16 hr after injection of thioglycollate is 30–40-fold (data not shown) and is primarily made up of neutrophils and macrophages. To help determine the source of the CXCL13 transcripts in such samples, thioglycollate recruited cells from BALB/c and Pactolus-deficient animals were separated by Percoll sedimentation into fractions enriched for neutrophils or macrophages. As shown in Fig. 2(a) the dense neutrophil-enriched population possessed very low levels of CXCL13 transcripts per β-actin transcripts. The top layer, that was enriched for macrophages and lymphocytes, showed a much higher level of expression of CXCL13 per β-actin equivalence, and a nearly fourfold increase of CXCL13 expression in the BALB/c sample versus the Pactolus-deficient animal. The difference in CXCL13 expression between wild type and knockout animal is more pronounced in the more homogeneous cell samples in this figure (versus that of Fig. 1) which would be expected as cells that possess β-actin transcripts but not those for CXCL13 were removed from the sample. The low level of expression of CXCL13 in the neutrophil-enriched sample was most likely caused by the very small percentage of cellular contamination inevitable with such a separation protocol. When naive, wild type peritoneal washes were examined in the same fashion, Fig. 2(b), CXCL13 transcripts were found in both fractions suggesting that alterations in the density of cells expressing the chemokine may occur during an inflammatory response. Increasing the Percoll step gradient to 1·08 g/ml still split the CXCL13-producing cells into the top and bottom fractions (data not shown).
Figure 2.
CXCL13 transcripts are found in the nongranulocyte cell population. Total peritoneal lavage samples 16 hr after thioglycollate treatment (four mice per set) (a) or from naïve mice (b) were obtained and separated, based upon density, in a Percoll gradient (1·06 g/ml) (see Methods) RNA was obtained from the samples and analysed for CXCL13 transcripts (per β-actin control) by quantitative PCR. Results from a single experiment are shown but is representative of multiple identical experiments. (*t = 0·03).
The CXCL13 chemokine is recognized by the CXCR5 receptor found on B and T cells.19 To determine if the depressed level of CXCL13 transcripts in the Pactolus-deficient animal following inflammatory irritation would result in a depression of migration of CXCR5-bearing cells, we obtained total peritoneal cells from animals 24 and 96 hr after thioglycollate injection, and analysed them for populations of CXCR5-possessing cells (Fig. 3). Two discrete populations of CXCR5 positive cells (B220+ B cells and B200– T cells) were obtained from such animals but there was no significant difference in either of these CXCR5+ populations between the wild type and Pactolus-deficient animals. The number of neutrophils in wild type and Pactolus-deficient mice drops dramatically after recruitment such that 96 hr after thioglycollate induction, only about 1% of the peritoneal cells are GR1+ neutrophils (data not shown). Such cells are removed by macrophage phagocytosis: we do not see any preferential phagocytosis of wild type or Pactolus-deficient neutrophils (data not shown).
Figure 3.
Recruitment of CXCR5 possessing cells following an inflammatory stimulus. Thioglycollate was injected into the peritoneal cavity of four BALB/c mice and four Pactolus-deficient (knockout) mice. After 24 and 96 hr, peritoneal cells were isolated from pools of two mice per time point, and analysed by FACS for expression of CXCR5 and the B-cell marker B220. Similar trends in cell recruitment were observed in other identical experiments.
Expression of CXCL13 following inflammatory insult is caused by resident cells not recruited macrophages
The preceding data in this report indicated that the primary source of CXCL13 products following thioglycollate recruitment was from low density cells such as dendritic cells or macrophages suggesting that differential interactions between the wild type and Pactolus-deficient neutrophils with either resident or recruited CXCL13-expressing cells were responsible for the difference in CXCL13 phenotype. We sought to duplicate these responses in vitro by incubating naïve and recruited peritoneal cellular populations (both wild type and Pactolus-deficient) with neutrophils isolated from the bone marrow of wild type and Pactolus-deficient animals. Incubating such purified neutrophil populations overnight induces the translocation of Pactolus to the cell surface.9
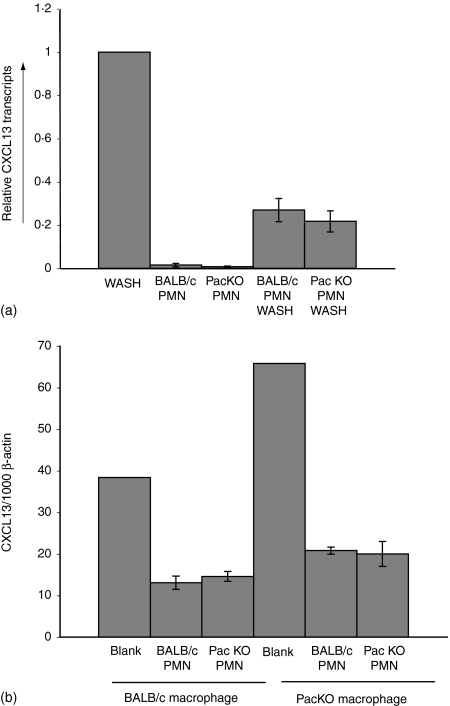
Peritoneal wash populations were isolated from naïve BALB/c animals, held alone, or with the addition of four to one bone-marrow derived neutrophils for 16 hr. The isolated bone marrow polymorphonuclear cells (PMNs) were also analysed without the addition of the peritoneal wash cells. As shown in Fig. 4(a), the addition of the bone marrow derived PMNs resulted in about a four- to fivefold drop in CXCL13 expression. However, the quantification of CXCL13 expression is based upon normalization to β-actin transcripts: the PMNs possessed β-actin mRNA but not CXCL13 products. Therefore, the four- to fivefold drop in CXCL13 transcripts with the addition of the knockout and wild type PMNs is wholly accounted for by the relative dilution of the CXCL13 to β-actin ratio. However, if the Pactolus-deficient neutrophils were more prone to kill the CXCL13 expressing cells within the peritoneal cavity, compared to the wild type PMNs, then a more pronounced depression of CXCL13 transcripts should have been evident. The PMN samples alone (wild type and knockout) did not possess any CXCL13 transcription products.
Figure 4.
Expression of CXCL13 in in vitro cultures. (a) Total peritoneal washes from naïve animals were incubated alone (WASH) or with bone marrow isolated neutrophils from wild type (BALB/c PMN WASH) or from Pactolus-deficient animals (PacKO PMN WASH), for 16 hr in tissue culture. Equivalent samples of the isolated neutrophils (BALB/c PMN or PacKO PMN) were also analysed in parallel. Relative levels of CXCL13 transcripts were obtained from identical experiments and normalized to a 100% value from the WASH sample. (b) Total peritoneal washes from wild type (BALB/c Macrophage) and Pactolus-deficient (PacKO Macrophage) animals 96 hr after thioglycollate treatment were incubated either alone (Blank) or with neutrophils isolated from the bone marrow of Pactolus-deficient (PacKO PMN) or wild type (BALB/c PMN) animals. The analysis of a single experiment is shown but is indicative of a number of independent analyses that provided the same result.
In a second analysis, peritoneal wash cells were derived from wild type and Pactolus-deficient animals 96 hr after thioglycollate injection and incubated with wild type or Pactolus-deficient bone marrow derived neutrophils (Fig. 4b). The apparent quantity of CXCL13 transcripts was again reduced due to the addition the β-actin transcripts contained within the PMNs. Again we saw no specific depression of CXCL13 gene products after incubation with the Pactolus-deficient neutrophils, compared to the wild type cells, suggesting that the preferential reduction in CXCL13 products seen during the peritoneal inflammatory response (see Figs 1 and 2) is not caused by preferential cell killing by the Pactolus-deficient neutrophils. Again, as shown previously, the quantity of CXCL13 products produced by the peritoneal cells of the Pactolus-deficient animal is comparable to those of wild type (except at the height of the inflammatory response, 16 hr after induction).
The preceding data left open the question as to whether the CXCL13 transcripts observed in the peritoneal washes 16 hr after induction were caused by freshly recruited macrophages, the resident peritoneal population, or both. This question was addressed, in part, by analysing relative levels of CXCL13 expression at various time points following thioglycollate injection. CXCL13 expression was quantified as copies per 1000 copies of β-actin. If the expression of the chemokine was due solely to the resident cells, then the addition of recruited cells not expressing the chemokine would serve to depress this ratio. On the other hand, if recruited macrophages were expressing CXCL13, then the relative level of the chemokine per β-actin transcripts should remain high with a maximal value at the apex of macrophage recruitment, 20–40 hr after thioglycollate induction. As shown in Fig. 5, as cells are recruited into the cavity following thioglycollate recruitment, the ratio of CXCL13 transcripts to β-actin transcripts falls dramatically over the 42 hr time period indicating that the resident peritoneal cells are largely responsible for the production of the chemokine, not the recruited cells. It is at the 16 hr time point of this assay that a significant difference in response within the wild type and Pactolus-deficient animals was observed (see Fig. 1).
Figure 5.
Enumeration of CXCL13 transcripts in peritoneal cell washed from naïve and thioglycollate treated animals. Total cell populations were obtained from naïve (0 hr, five BALB/c and four Pactolus-deficient animals) and thioglycollate treated animals 5, 16 and 42 hr after induction (four BALB/c and three Pactolus-deficient, four and four, and four and three, respectively) of the inflammatory response. Total RNA samples were analysed for CXCL13 expression per 1 × 103β-actin transcripts. Averages between the different mouse samples are shown. The same experiment was done repeatedly and provided the same result.
Characterization of the naïve peritoneal CXCL13-expressing cells and modulation during an inflammatory response
The expression of CXCL13 in the normal, uninflamed peritoneal cavity has been attributed to resident macrophages and cells found within the peritoneal omentum and peritoneal wall. The CXCL13-expressing peritoneal macrophages were characterized as CD11c+, CD11b+, F4/80+ and B220–.20,21 Radiation chimera analysis demonstrated that the peritoneal macrophage population was sensitive to radiation treatment but that the bulk of CXCL13-expressing cells within the omentum were not.21 Such macrophage-like cells within the omentum had been previously proposed to originate from local precursors and not to be dependent upon the bone marrow indicating that there are two distinct populations of CXCL13-expressing cells within the peritoneal environment. A number of groups have also described dendritic cells as significant sources of CXCL13 (dendritic cells adhere weakly to plastic plates).22–24 Additionally, the expression of CXCL13 by B cells has been documented even though CXCL13 is a strong B-cell chemoattractant and critical in establishing correct lymphoid structures.25–27
The preceding data indicated that the expression of the CXCL13 chemokine seen during an inflammatory response was linked to the resident cells of the peritoneal cavity and could not be attributed to the recruited macrophages. These CXCL13-expressing resident cells must differentially interact with the Pactolus-deficient or normal neutrophils in the first wave of inflammatory recruitment to give rise to the altered levels of CXCL13 transcripts.
We chose to more fully characterize the type of cells in the peritoneal cavity that express CXCL13 (Fig. 6a). The free peritoneal cells responsible for the CXCL13 expression were F4/80 positive and B220 negative, consistent with a macrophage-like phenotype. This analysis was extended using an antibody specific for CXCL13 for intracellular staining of such cells. As shown (Fig. 6b), when naïve peritoneal lavage cells analysed by FACS analysis using anti-F4/80 and anti-CXCL13 antibodies, a small subset, but not all, of the F4/80+ cells also stain for CXCL13. Peritoneal lavage cells from naïve mice were further analysed using double stains of anti-CXCL13 and anti-CD11b, anti-B220 or anti-major histocompatibility complex (MHC) II. Only a subset of the CD11bHI cells expressed CXCL13 while MHCIIHI cells (B cells) did not express the chemokine (Fig. 6c). As shown, the bulk of CXCL13 product was associated with B220– cells. Because we observed a depression in CXCL13 gene products 16 hr after thioglycollate recruitment and we can detect CXCL13-expressing cells by FACS analysis, we analysed a total peritoneal lavage wash for any differences in the numbers of CXCL13-expressing cells between the wild type and Pactolus knockout animals 16 hr after thioglycollate treatment. As shown in Fig. 6(d), we could not specifically detect CXCL13-expressing cells in the peritoneal wash of the wild type or the Pactolus-deficient animals. At this time point following thioglyocollate recruitment, the major influx of neutrophils is over and the entry of macrophages is robust such that the CXCL13-expressing cells observed in the quiescent peritoneal lavage would be lost in the population of recruited cells. Attempting to gate on subpopulations before analysis (such as CD11b, F4/80) did not reduce the background such that the CXCL13-expressing cells could be identified and quantified in either the wild type or Pactolus knockout samples. Alternatively, the ability to detect the protein in these cells could be lost following inflammatory activation.
Figure 6.
Expression of CXCL13 in naïve peritoneal cells colocalizes with expression of F4/80. (a) Total peritoneal cells were isolated naïve BALB/c animals and fractionated, via magnetic bead separation, into B220+ populations, and F4/80+ populations. Cells were then lysed, RNA obtained and analysed for CXCL13, CD19, and F4/80 transcripts per 1 × 103β-actin transcripts. Two BALB/c mouse samples were pooled for the analysis: data shown is percentage of total. (b) Total peritoneal cells were isolated from naïve BALB/c animals and stained for flow cytometry. Cells were either treated (+Bref) or not (–Bref) with Brefeldin A. Cells were labelled with antibodies against F4/80 in combination with isotype immunoglobulin (Ig) and CXCL13. The RI region of the side scatter versus forward scatter contained those cells positive for CXCL13 staining: the R1 region was used in the lower four panels. (c) Total peritoneal cells were isolated from naïve BALB/c animals and stained for flow cytometry. Cells from the R1 gate (not shown) were analysed. Cells were treated with Brefeldin A, and labelled with antibodies against CXCL13 in combination with CD11b, B220 and MHC class II. (d) Total peritoneal lavage cells 16 hr after thioglycollate recruitment from wild type (WT) and the Pactolus-deficient animals (KO) were analysed for F4/80 and CXCL13 protein expression. Cells were treated with Brefeldin A, permeabilized and fixed prior to staining. There was no difference noted in WT or KO cells for CXCL13 staining compared to the isotype control antibody (Ig).
Previous experiments in this report detail the differential loss of CXCL13 transcripts in wild type versus Pactolus-deficient animals during an inflammatory insult. The loss of CXCL13 gene products from these resident cells of the peritoneal cavity could be caused by a number of reasons: including preferential transcript degradation, silencing of gene transcription, differential migration out of the cavity, or apoptosis of resident cells expressing the product. Co-incubation of fresh, naïve peritoneal washes with neutrophils did not demonstrate a net loss of CXCL13 transcripts over a 16 hr period suggesting that transcription was not being quenched and CXCL13-expressing cells were not being preferentially killed by the Pactolus-deficient neutrophils (Fig. 4). These observations suggested that differential migration of such cells out of the peritoneal cavity was the most likely cause for the differences between the Pactolus-deficient and wild type animals.
The other cell type in the peritoneum that has been described for CXCL13 production is a macrophage-like cell embedded within the peritoneal wall, omentum mesentary and diaphragm.21 We reasoned that if alterations in numbers of the CXCL13-expressing cells in the peritoneal wall were obvious hours after an inflammatory stimulus, then it would be most likely due to a soluble product from responding cells such as neutrophils. Alternatively, if alterations were obvious in the free peritoneal cells but not apparent for the CXCL13-expressing cells in the peritoneal wall, then cell to cell contact between the wild type or Pactolus-deficient neutrophils might be causative. Accordingly, we first treated multiple BALB/c mice with PBS or thioglycollate injections and evaluated peritoneal lavage and peritoneal wall samples for CXCL13 and control products. As shown in Fig. 7(a), CXCL13 gene products were significantly reduced in the thioglycollate treated samples, compared to the β-actin controls. As discussed previously, the reduction in CXCL13/β-actin transcripts for the peritoneal lavage is largely caused by the migration into the cavity of other cells possessing β-actin but not CXCL13 gene products. The same would not be true for the peritoneal wall analysis in that the structure of the wall was not remarkably influenced and the total number of cells within it (expressing β-actin) should stay relatively uniform. Only a very small minority of cells in the peritoneal wall express CXCL13.21 As internal controls for this experiment, we analysed transcripts for F4/80 and β-2 microglobulin (again expressed as transcripts per 1000 β-actin control). The former showed loss for the marker within the peritoneal wall (consistent with migration out of that site), while the latter was not significantly altered between PBS and thioglycollate treatment. The most likely interpretation of these data is that the CXCL13-expressing cells migrate out of the peritoneal wall.
Figure 7.
Modulation of cells expressing CXCL13 in peritoneal wall samples following inflammatory activation. Three BALB/c animals were mock treated (PBS) or injected with thioglycollate (Thio) for 16 hr. The peritoneal cavity constituents were first isolated (peritoneal cavity). Anatomically identical sites of the peritoneal wall were excised and lysed (Peritoneal wall). Total RNA was obtained and analysed for (a) CXCL13, (b) F4/80 and (c) β-2 microglobulin expression, by quantitative RT–PCR. This experiment is one of multiple that showed the same trend.
We then sought to determine if there was any difference in the loss of CXCL13 transcripts with the peritoneal wall sample between the wild type and Pactolus-deficient animal following a 16 hr inflammatory stimulus. Physically identical samples of the peritoneal wall and omentum were compared. As shown (Fig. 8), a significant reduction in CXCL13 expression in the Pactolus-deficient animal is observed with both the peritoneal wall and peritoneal wash samples while that for the omentum showed equivalent transcript levels between the two sets of animals. These data suggest that the CXCl13-expressing cells in the peritoneal wall are preferentially lost in that anatomical site of the Pactolus-deficient animal during an inflammatory response.
Figure 8.
Differential loss of CXCL13 transcripts from the peritoneal wall between Pactolus-deficient and BALB/c mice. Three BALB/c and three Pactolus-deficient (knockout) animals were injected with thioglycollate and samples taken 16 hr later. Total peritoneal cells were obtained from the cavity, as well as anatomically identical samples of the peritoneal wall and the greater omentum. Total RNA was extracted and analysed for CXCL13 transcripts by quantitative RT–PCR. Averages are shown from the sets of three mice. Other similar experiments showed the same trend. #t = 0·065: *t = 0·027.
Discussion
In this report we have analysed the novel finding that the level of expression of CXCL13 found in peritoneal cells after a thioglycollate recruitment is depressed in animals lacking the Pactolus protein compared to wild type controls. This observation was made at the time point at which we had hypothesized the effect of a Pactolus deficiency might be most evident when interactions between recruited neutrophils and macrophages would be maximal. Pactolus is shuttled to the surface of the activated neutrophil prior to that cells entry into apoptosis and/or necrosis, presumably to either interact with another neutrophil ligand, or to bind to a cognate receptor on the surface of macrophages. The difference in CXCL13 transcripts between the wild type and Pactolus-deficient animals was not evident 4 hr after induction (primarily during the neutrophil influx) nor 48 hr after induction (at which point most of the neutrophils had been eliminated) but instead 16 hr post induction when the ratio of neutrophil to macrophages is about 3 : 1.
One concern about our observations between the wild type (BALB/c) and Pactolus-deficient animal (extensively bred upon the BALB/c background) was that the differential level of CXCL13 expression by resident cells was preset prior to the inflammatory induction. Thus a 129/sv allele of a gene (that was functionally different from that of BALB/c lineage) linked to the Pactolus knockout insertion could differentially influence the level of expression (either by manipulating total cell number or the level of transcription) of CXCL13. However, all of our analyses have shown that the quantity of CXCL13 transcripts within various tissue samples was the same in the BALB/c, 129/sv controls and the Pactolus-deficient animal prior to the inflammatory response. Therefore, the absence of the Pactolus protein during the inflammatory response must be causative for this phenomenon.
Our experimental hypothesis was that the greatest effect of the Pactolus deficiency would have been linked to the function of macrophages recruited into the peritoneal cavity. Therefore, we were surprised to find that virtually all of the CXCL13 response (which was differential between the wild type and Pactolus knockout animal) was from the resident cells of the peritoneal cavity. Previous reports have identified resident macrophages (CD11bHI/F4/80HI) as the primary CXCL13-expressing cells within the quiescent peritoneal cavity, the omentum and peritoneal wall.20,21 CXCL13 is a B-cell homing chemokine critical for lymphocyte homing to lymphoid tissues and localization within the peritoneal cavity. Animals deficient in CXCL13 show a lack of B1 cell localization in the peritoneal cavity.21
The macrophages recruited into the peritoneal cavity (both wild type and Pactolus-deficient) following the thioglycollate treatment were CD11bHI/F4/80HI, but were not expressing the CXCL13 chemokine. The induction of an inflammatory response such as that created with a peritonitis model is characterized by a immediate migration of neutrophils into the site followed by a macrophages and, still later, T and B cells. We assumed that CXCL13 would have been expressed by the recruited macrophages to induce the migration of CXCR5-bearing lymphocytes; however, that was not the case. This observation suggests that the resident peritoneal CXCL13 expressing macrophages are fundamentally different cells than those macrophages recruited into the body cavity following an inflammatory stimulus. Previous reports have suggested CXCL13 expression by dendritic cells (CD11bHI/CD11cHI), and, in one report, the cell surface markers of the peritoneal CXCL13-expressing cells do express both CD11b and CD11c.22 Therefore, at this time, it is unclear whether or not the CXCL13 expressing cells in the peritoneal cavity represent a specialized class of macrophages or dendritic cells.
The central question left unresolved by this work is why the recruitment of a Pactolus-deficient neutrophil would promote the loss of CXCL13-expressing cells from the peritoneal cavity during an inflammatory response. The in vitro coculture of such cells with neutrophils did not induce their death nor was there evidence of depression of CXCL13 transcription leaving the preferential migration of such cells out of the peritoneal cavity as the likely reason for the Pactolus-deficient phenotype. Such a model suggests the differential release of a neutrophil product that would serve to retain (wild type) or exclude (Pactolus deficiency) CXCL13-expressing peritoneal cells during the inflammatory response. Determining whether or not this model is correct will require additional inquiry into differential product release from wild type and Pactolus-deficient neutrophils.
Acknowledgments
The authors would like to thank Hilary Crandall for her assistance in the Affymetrix analysis, to thank the members of the Weis labs for their critical analysis of the work, and to acknowledge the support of the Research Cores at the University of Utah. This research was supported by NIH grants AI-42032 (J.H.W.), and NIH grants AI-32223 and AR-43521 (J.J.W.).
Abbreviations
- MIDAS
metal ion dependent adhesion site
- PCR
polymerase chain reaction
- RT
reverse transcription
- PBS
phosphate-buffered saline
- LPS
lipopolysaccharide
- SNP
single nucleotide polymorphism
- FACS
fluorescence activated cell sorting
References
- 1.Chen Y, Garrison S, Weis JJ, Weis JH. Identification of pactolus, an integrin beta subunit-like cell-surface protein preferentially expressed by cells of the bone marrow. J Biol Chem. 1998;273:8711–8. doi: 10.1074/jbc.273.15.8711. [DOI] [PubMed] [Google Scholar]
- 2.Margraf RL, Chen Y, Garrison S, Weis JJ, Weis JH. Genomic organization, chromosomal localization, and transcriptional variants of the murine Pactolus gene. Mamm Genome. 1999;10:1075–81. doi: 10.1007/s003359901164. [DOI] [PubMed] [Google Scholar]
- 3.Bajt ML, Loftus JC. Mutation of a ligand binding domain of beta 3 integrin. Integral role of oxygenated residues in alpha IIb beta 3 (GPIIb-IIIa) receptor function. J Biol Chem. 1994;269:20913–9. [PubMed] [Google Scholar]
- 4.Wardlaw AJ, Hibbs ML, Stacker SA, Springer TA. Distinct mutations in two patients with leukocyte adhesion deficiency and their functional correlates. J Exp Med. 1990;172:335–45. doi: 10.1084/jem.172.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michishita M, Videm V, Arnaout MA. A novel divalent cation-binding site in the A domain of the beta 2 integrin CR3 (CD11b/CD18) is essential for ligand binding. Cell. 1993;72:857–67. doi: 10.1016/0092-8674(93)90575-b. [DOI] [PubMed] [Google Scholar]
- 6.Ueda T, Rieu P, Brayer J, Arnaout MA. Identification of the complement iC3b binding site in the beta 2 integrin CR3 (CD11b/CD18) Proc Natl Acad Sci USA. 1994;91(22):10680–4. doi: 10.1073/pnas.91.22.10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rieu P, Ueda T, Haruta I, Sharma CP, Arnaout MA. The A-domain of beta 2 integrin CR3 (CD11b/CD18) is a receptor for the hookworm-derived neutrophil adhesion inhibitor NIF. J Cell Biol. 1994;127:2081–91. doi: 10.1083/jcb.127.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrison S, Hojgaard A, Patillo D, Weis JJ, Weis JH. Functional characterization of Pactolus, a beta-integrin-like protein preferentially expressed by neutrophils. J Biol Chem. 2001;276:35500–11. doi: 10.1074/jbc.M104369200. [DOI] [PubMed] [Google Scholar]
- 9.Garrison S, Hojgaard A, Margraf R, Weis JJ, Weis JH. Surface translocation of pactolus is induced by cell activation and death, but is not required for neutrophil migration and function. J Immunol. 2003;171:6795–806. doi: 10.4049/jimmunol.171.12.6795. [DOI] [PubMed] [Google Scholar]
- 10.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–52. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 11.Cox G, Crossley J, Xing Z. Macrophage engulfment of apoptotic neutrophils contributes to the resolution of acute pulmonary inflammation in vivo. Am J Respir Cell Mol Biol. 1995;12:232–7. doi: 10.1165/ajrcmb.12.2.7865221. [DOI] [PubMed] [Google Scholar]
- 12.Fadok VA, Savill JS, Haslett C, Bratton DL, Doherty DE, Campbell PA, Henson PM. Different populations of macrophages use either the vitronectin receptor or the phosphatidylserine receptor to recognize and remove apoptotic cells. J Immunol. 1992;149:4029–35. [PubMed] [Google Scholar]
- 13.Fadok VA, Bratton DL, Frasch SC, Warner ML, Henson PM. The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ. 1998;5:551–62. doi: 10.1038/sj.cdd.4400404. [DOI] [PubMed] [Google Scholar]
- 14.Ramprasad MP, Terpstra V, Kondratenko N, Quehenberger O, Steinberg D. Cell surface expression of mouse macrosialin and human CD68 and their role as macrophage receptors for oxidized low density lipoprotein. Proc Natl Acad Sci USA. 1996;93:14833–8. doi: 10.1073/pnas.93.25.14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–9. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 16.Kjeldsen L, Sengelov H, Borregaard N. Subcellular fractionation of human neutrophils on Percoll density gradients. J Immunol Methods. 1999;232:131–43. doi: 10.1016/s0022-1759(99)00171-4. [DOI] [PubMed] [Google Scholar]
- 17.Mizgerd JP, Kubo H, Kutkoski GJ, Bhagwan SD, Scharffetter-Kochanek K, Beaudet AL, Doerschuk CM. Neutrophil emigration in the skin, lungs, and peritoneum: different requirements for CD11/CD18 revealed by CD18-deficient mice. J Exp Med. 1997;186:1357–64. doi: 10.1084/jem.186.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walzog B, Scharffetter-Kochanek K, Gaehtgens P. Impairment of neutrophil emigration in CD18-null mice. Am J Physiol. 1999;276:G1125–30. doi: 10.1152/ajpgi.1999.276.5.G1125. [DOI] [PubMed] [Google Scholar]
- 19.Legler DF, Loetscher M, Roos RS, Clark-Lewis I, Baggiolini M, Moser B. B cell-attracting chemokine 1, a human CXC chemokine expressed in lymphoid tissues, selectively attracts B lymphocytes via BLR1/CXCR5. J Exp Med. 1998;187:655–60. doi: 10.1084/jem.187.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Ishikawa S, Sato T, et al. Defective B1 cell homing to the peritoneal cavity and preferential recruitment of B1 cells in the target organs in a murine model for systemic lupus erythematosus. J Immunol. 2004;172:3628–34. doi: 10.4049/jimmunol.172.6.3628. [DOI] [PubMed] [Google Scholar]
- 21.Ansel KM, Harris RB, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16:67–76. doi: 10.1016/s1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa S, Sato T, Abe M, et al. Aberrant high expression of B lymphocyte chemokine (BLC/CXCL13) by C11b+ CD11c+ dendritic cells in murine lupus and preferential chemotaxis of B1 cells towards BLC. J Exp Med. 2001;193:1393–402. doi: 10.1084/jem.193.12.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa S, Nagai S, Sato T, Akadegawa K, Yoneyama H, Zhang YY, Onai N, Matsushima K. Increased circulating CD11b+ CD11c+ dendritic cells (DC) in aged BWF1 mice which can be matured by TNF-alpha into BLC/CXCL13-producing DC. Eur J Immunol. 2002;32(7):1881–7. doi: 10.1002/1521-4141(200207)32:7<1881::AID-IMMU1881>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 24.Vissers JL, Hartgers FC, Lindhout E, Figdor CG, Adema GJ. BLC (CXCL13) is expressed by different dendritic cell subsets in vitro and in vivo. Eur J Immunol. 2001;31:1544–9. doi: 10.1002/1521-4141(200105)31:5<1544::AID-IMMU1544>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 25.Steinmetz OM, Panzer U, Kneissler U, Harendza S, Lipp M, Helmchen U, Stahl RA. BCA-1/CXCL13 expression is associated with CXCR5-positive B-cell cluster formation in acute renal transplant rejection. Kidney Int. 2005;67:1616–21. doi: 10.1111/j.1523-1755.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 26.Falkenhagen KM, Braziel RM, Fraunfelder FW, Smith JR. B-Cells in ocular adnexal lymphoproliferative lesions express B-cell attracting chemokine 1 (CXCL13) Am J Ophthalmol. 2005;140:335–7. doi: 10.1016/j.ajo.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 27.Ansel KM, Ngo VN, Hyman PL, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–14. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]