Abstract
To date, the biological role of prokaryotic K+ channels remains unknown. Helicobacter pylori contains a gene encoding a putative K+ channel (HpKchA) of the two-transmembrane RCK (regulation of K+ conductance) domain family, but lacks known bacterial K+ uptake systems. A H. pylori ΔhpKchA mutant presented a strong growth defect at low K+ concentration, which was compensated by KCl addition. The role of the separate RCK domain was investigated in H. pylori by mutagenesis of its internal start codon, which led to a K+-dependent intermediate growth phenotype, consistent with RCK activating channel function. Tagging HpKchA C-terminally, we detected a 1:1 stoichiometry of the full-length HpKchA and the separate RCK domain. We constructed single amino-acid exchanges within the unusual selectivity filter of HpKchA (ATGFGA) in H. pylori and observed complete loss (G74A), a slight defect (G76A or F75G) or wild-type (A77D) channel function. HpKchA was essential for colonization of the murine stomach. These data show, for the first time, a biological function for a prokaryotic K+ channel, as a K+ uptake system, essential for the persistence of H. pylori in the gastric environment.
Keywords: gastric adaptation, mouse model, RCK domain, selectivity filter
Introduction
Potassium is the most abundant ion in the cytoplasm of all living cells. Cytoplasmic K+ is essential for many bacterial processes like regulation of cell turgor pressure, growth, cytoplasmic pH homeostasis and protein synthesis. In Escherichia coli, K+ uptake occurs through several systems, the constitutive and low-affinity transporters, Trk and Kup, and the high-affinity, inducible transport system, KdpFABC complex (Epstein et al, 1993). At low millimolar K+ concentrations, the constitutively expressed low-affinity transporters are sufficient for K+ acquisition under fluctuating osmotic pressure. Upon depletion of K+, the KdpFABC complex (P-type ATPase) is induced and takes over the function of Trk and Kup. Some other bacteria contain a fourth type of K+ uptake system, the Na+-dependent Ktr system (Nakamura et al, 1998).
Helicobacter pylori is involved in several gastric diseases, including chronic gastritis, gastric and duodenal ulcers and cancer (Ernst and Gold, 2000). Upon oral uptake, the pathogen has to pass the extreme acidic gastric lumen in order to reach the mucosa, its sole niche for proliferation, where the pH is close to neutrality (Schreiber et al, 2000). The concentration of K+ in the gastric juice is estimated to be around 12 mM (Greger and Windhorst, 1996). At the mucosa, the abundance of K+ is likely to be lower, as it is assumed to resemble that of blood (3.5–5 mM) (Devlin, 1997).
The systems involved in K+ uptake of H. pylori are not known. Its genome lacks genes encoding homologues of Kdp, Trk, Kup or Ktr. Instead, it contains a gene coding for a putative K+ channel (Tomb et al, 1997). This open reading frame (hp0490, called here hpkchA) codes for a protein with homology to the family of two-transmembrane (2TM) K+ channels, which are found in most eukaryotes and prokaryotes. This family includes KcsA from Streptomyces lividans, the bacterial Kirbac1.1 from Burkholderia pseudomallei and the inwardly rectifying MthK from the archaeon Methanobacterium thermoautotrophicum, for which crystal structures are available (Doyle et al, 1998; Jiang et al, 2002; Kuo et al, 2003). Functional K+ channels are homo-tetramers with the single subunits arranged in a four-fold symmetry around a water-filled core. In the 2TM K+ channels, the 2TM helices of each subunit are separated by a P-loop carrying the selectivity filter signature sequence, which contributes to the high specificity of K+ (Doyle et al, 1998). The canonical amino-acid sequence of the selectivity filter, TXGYGD, presents little variation among K+ channels. Frequently, the tyrosine is substituted by a phenylalanine, as in HpKchA. However, HpKchA exhibits further differences, as it presents the unusual selectivity filter sequence, ATGFGA. Strikingly, the negatively charged aspartate protruding at the external site of the channel is substituted by a neutral alanine. A non-conservative modification at this position is rare among K+ channels, as the aspartate is supposed to stabilize the filter structure (Chapman et al, 2001) and/or to be involved in the accumulation of the K+ ion close to the filter, increasing the rate of ion entry into the pore (Haug et al, 2004).
Like MthK, its close homologue, HpKchA contains a regulatory RCK (regulation of K+ conductance) domain of poorly defined function (Jiang et al, 2002). The MthK gene has its own internal start codon, which leads to the separate expression of the RCK domain in E. coli. From its crystal structure and from biochemical studies, a model for functional MthK was proposed, in which four full-length MthK subunits constitute the pore complex to which a tetramer of separate RCK domains is attached from the cytoplasmic side (Jiang et al, 2002).
So far, the biological role of K+ channels in prokaryotes is unknown (Kuo et al, 2005). On deleting the six-transmembrane K+ channel kch gene from E. coli, no ‘loss-of-function' phenotype was obtained. Further work suggests that Kch is an inwardly rectifying K+ channel, functioning as a voltage valve for the cells under yet unknown conditions (Kuo et al, 2003). Two K+ channels from the archaeon Methanococcus janaschii may play a role in K+ uptake, as they restored growth at low K+ concentrations of an E. coli mutant deficient in K+ transport (Hellmer and Zeilinger, 2003). However, as M. janaschii possesses an additional Trk system (Bult et al, 1996), it remains unclear whether its K+ channels play a role in K+ uptake or fulfill a different function in this organism.
As H. pylori presents no other commonly found K+ transport system, we chose HpKchA as a model protein to study the function of a prokaryotic K+ channel in its in vivo context. In this study, we show that HpKchA is essential for K+ uptake in H. pylori at low external K+ concentrations and for colonization of the gastric environment, corroborating its role as a unique uptake system for K+ in the gastric pathogen. In addition, we demonstrate that the expression of a separate RCK domain in addition to the full-length HpKchA is necessary for wild-type function and we discuss modifications of its unusual selectivity filter analyzed by site-directed mutagenesis.
Results
Features of the HpKchA primary sequence
HpKchA, which presents a calculated molecular mass of 43 kDa, exhibits significant homology with a number of K+ channel proteins or protein domains (Figure 1). The closest homologues of HpKchA are found in Helicobacter hepaticus (Suerbaum et al, 2003) and Wolinella succinogenes (Baar et al, 2003), which are members of the Helicobacteraceae. Its next best hits are with the two 2TM-K+ channels MjK1 and MjK2 from the archaeon M. jannaschii (Bult et al, 1996) (25 and 26% identity, respectively; Figure 1, lines 3 and 4, respectively). These proteins closely resemble the inwardly rectifying K+ channel MthK from M. thermoautotrophicum (Jiang et al, 2002) (Figure 1, line 1) and consist of a 2TM-K+ channel domain and a C-terminal regulatory RCK domain.
Figure 1.

Alignment of HpKchA with RCK-domain-containing K+ channel proteins. 2TM and 6TM, two-transmembrane and six-transmembrane K+ channel, respectively. Line 1: HpKchA (O25233); line 2: MthK, inwardly rectifying K+ channel of M. thermoautotrophicum (O27564); lines 3 and 4: MjK1 (Q57604) and MjK2 (Q58752), two K+ channels of M. jannaschii; line 5: partial EcKch, K+ channel of E. coli (P31069). Secondary structure assignment is based on the crystal structures of KcsA (outer helix, P-loop, inner helix) (Doyle et al, 1998) and MthK (RCK domain) (Jiang et al, 2002). α, α-helix; β, β-strand. The internal start codon of the RCK domain (M127) in HpKchA and the selectivity core glycine residues are highlighted in white on a black background. Residues identical to the corresponding amino acids in HpKchA are highlighted by a gray background.
Growth defect of the non-polar H. pylori ΔhpKchA mutant
To characterize the biological function of HpKchA, we constructed an isogenic deletion mutant of hp0490 in H. pylori strain X47-2AL by insertion of a non-polar kanamycin cassette. In brucella broth supplemented with 10% FCS, the hpKchA-deficient mutant had a strong growth defect (Figure 2A, open circles). The K+ concentration of this medium was determined by flame spectroscopy and amounts to 7.3±2.0 mM K+ (five independent measurements). Growth of the mutant was restored stepwise by addition of KCl. At K+ concentrations of around 22 mM, the growth defect of the ΔhpkchA strain was fully compensated to that of the wild-type level (Figure 2A and B).
Figure 2.
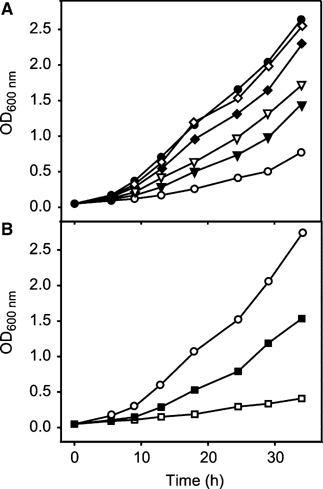
Growth of H. pylori ΔhpKchA strain is impaired at low K+ concentrations. H. pylori wild-type (X47-2AL) and its ΔhpKchA derivative were grown in brucella broth supplemented with 10% FCS. (A) ΔhpKchA strain at different K+ concentrations; ○, 7.3 mM K+ (basal K+ concentration of the medium); ▾, 10.3 mM KCl; ▿, 12.3 mM KCl; ♦, 17.3 mM KCl; ⋄, 22.3 mM KCl; •, 37.3 mM KCl. (B) growth of H. pylori wild-type in the presence or absence of the K+ channel blocker CsCl; ○, no addition (7.3 mM K+); □, 10 mM CsCl (and a basal level of 7.3 mM K+); ▪, 10 mM CsCl and 37.3 mM KCl.
Growth of wild-type H. pylori in the complex medium was completely inhibited in the presence of 10 mM CsCl, a well-known inhibitor of both eukaryotic inwardly rectifying K+ channels (Gay and Standfield, 1977) and bacterial KcsA (LeMasurier et al, 2001) (Figure 2B, open squares). This growth inhibition was only partially restored by addition of 30 mM KCl (Figure 2B, closed squares) suggesting that CsCl had also additional effects on cell growth, as growth at high K+ concentrations was independent of HpKchA (Figure 2A).
The K+ content of H. pylori cells was about 95 nmol/ml and the cytoplasmic water space about 0.5 μl/ml cell culture of OD600 nm=1 (∼5 × 108 CFU/ml). This gives a calculated cytoplasmic K+ concentration of 192±23 mM (seven independent measurements), which is similar to that of other bacteria (Walderhaug et al, 1987). It indicates that H. pylori accumulated K+ up to at least 30-fold from the liquid growth medium.
HpKchA is not required for pH homeostasis under acidic conditions
K+ transport processes are crucial for pH homeostasis of the cytoplasm at different values of medium pH in most bacteria (Booth, 1985). Therefore, the capacity to maintain pH homeostasis was compared between H. pylori ΔhpkchA and the wild-type. Urease activity was used to monitor cytoplasmic pH, as acidification of the cytoplasm has been shown to result in loss of activity of this enzyme and strongly correlates with survival of the bacterium (Rektorschek et al, 2000). Survival during 60 min of wild-type H. pylori at pH 2 was similar in brucella broth with a level of around 7 mM K+ and in citrate–phosphate buffer without KCl (Stingl et al, 2001), indicating that survival at acidity was independent of the presence of potassium. Moreover, urease activity of the ΔhpKchA mutant and wild-type strain in citrate–phosphate buffer in the presence of urea at pH 2 was comparable, with 12.5±3.6 μmol urea/(min × mg) protein for ΔhpKchA as compared to 16.4±3.9 μmol urea/(min × mg) protein for the wild-type (five independent measurements). Hence, HpKchA was dispensable for pH homeostasis of H. pylori at acidic pH in the presence of urea.
Purified His-tagged HpKchA formed oligomers
HpKchA was C-terminally fused to six histidine residues and the gene was overexpressed in E. coli C43 (Miroux and Walker, 1996). A total of 90% of the protein was solubilized from membranes with a 5:1 detergent mixture of N-dodecylmaltoside and SDS (Figure 3A and B, lanes 2–4). After binding to Ni2+-agarose, the eluted fraction (Figure 3A and B, lane 6) consisted of the 40 kDa HpKchA protein, sub-stoichiometric amounts of the 28 kDa RCK domain from HpKchA (both identified by N-terminal sequencing) and minor contaminants (partially HpKchA degradation products). As with MthK from M. thermoautotrophicum (Jiang et al, 2002) and MjK2 from M. jannaschii (Hellmer and Zeilinger, 2003), the conserved amino-acid residue M127 in HpKchA (Figures 1 and 3C) appears to serve as a start codon in E. coli, giving rise to the separate translation of the HpKchA-RCK domain. In a second purification step, the 40 kDa protein was electroeluted from the gel and re-chromatographed by SDS–PAGE (Figure 3D). At high SDS concentrations and in the presence of β-mercaptoethanol, we observed that a minor fraction of the electroeluted monomer formed a dimer at 80 kDa (Figure 3D, lane 1). In the absence of reducing agent and at low concentrations of SDS, the same electroeluted monomer formed two distinct bands at 40 and 35 kDa and several oligomerization states up to one tetramer (Figure 3D, lane 2). In further experiments, we verified that this migration difference was caused by the presence or absence of reducing agent and not by the concentration of SDS. We concluded that monomeric HpKchA exhibits two conformations under non-reducing conditions and that these monomers spontaneously formed dimers, trimers and tetramers, the latter being the functional unit of HpKchA.
Figure 3.
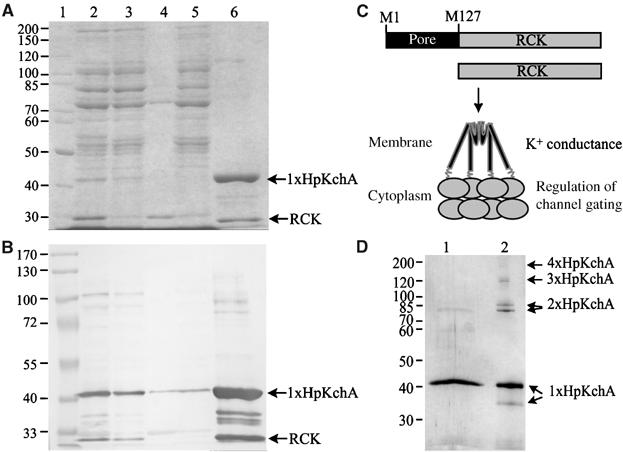
Purification of recombinant HpKchA. (A) Coomassie blue-stained 10% SDS–polyacrylamide gel. (B) Immunoblot using anti-penta-His antibodies. In panels A and B: lane 1, MW standard; lane 2, membranes before solubilization; lane 3, solubilized material; lane 4, membranes after solubilization; lane 5, flowthrough of the Ni2+-NTA resin; lane 6, eluted protein using 500 mM imidazol. Protein samples were dissolved in buffer with 2.7% SDS and 0.7% β-mercaptoethanol, heated at 50°C for 30 min and loaded on the gel. The relative amounts with respect to total cell protein in lanes 2–5 are comparable. Molecular weights are given in kDa. The 40 kDa HpKchA monomer (1xHpKchA) and its separate 28 kDa RCK domain are indicated with arrows; they were identified by N-terminal Edman sequencing. (C) Cartoon of the hpKchA gene illustrating its two start codons, one for the full-length protein and one for a separate RCK domain; the structural model modified after Jiang et al (2002) is depicted below in which a tetramer of the full-length channel interacts at the cytoplasmic site with an additional tetramer of the separate RCK domain. (D) Electroeluted monomeric HpKchA was mixed with loading buffer containing 0.7% SDS without reducing agent. After 1 h incubation at room temperature, the sample was divided into two equal aliquots. One aliquot received an additional 2% SDS and 0.7% β-mercaptoethanol and was heated for 30 min at 50°C. Both aliquots were run on a 10% SDS–polyacrylamide gel and stained with silver. Lane 1, HpKchA at 2.7% SDS plus 0.7% β-mercaptoethanol; Lane 2, HpKchA at 0.7% SDS. The oligomerization states are indicated on the right; the molecular weight is given on the left.
Detection of the full-length channel and a separate RCK domain in H. pylori
In order to test the expression of a separate RCK domain in H. pylori, we exchanged the methionine codon at the N-terminus of the predicted RCK domain (M127; Figure 1) in hpKchA by leucine. An M127L version of hpKchA was introduced into strain X47-2AL-Δhp490KmR. Subsequently, we tagged the wild-type hpKchA and the M127L-hpKchA mutant gene C-terminally with a tandem affinity purification (TAP) tag that we recently adapted for utilization in H. pylori (manuscript in preparation). As the TAP tag carries the antibody-binding domain of Protein A, its fusion to any H. pylori protein leads to specific detection of the target protein and its expression level by immunoblot analysis. We detected two separate protein bands in stochiometrically similar amounts in the hpKchA-TAP strain, corresponding to the full-length HpKchA and the separate RCK domain (Figure 4, lane 1). In the M127L-hpKchA-TAP strain, only the full-length HpKchA was detected (Figure 4, lane 2), indicating that the RCK domain was indeed expressed separately from the second methionine in position 127 in vivo. This shows that in H. pylori, HpKchA is expressed as a full-length protein plus a separate RCK domain in a 1:1 stoichiometry, which is in agreement with the proposed model of a tetrameric full-length pore complex associated with a tetrameric RCK domain complex (Jiang et al, 2002; Albright et al, 2006) (Figure 3C).
Figure 4.
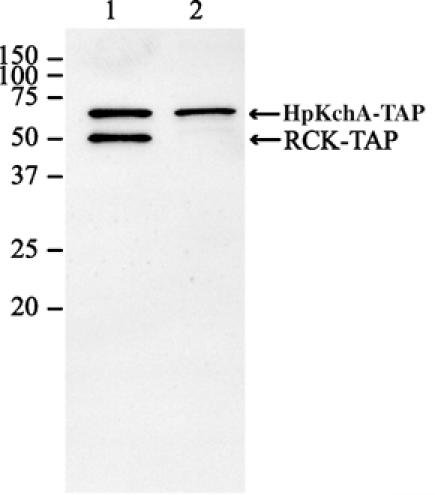
Immunoblot of TAP-tagged HpKchA in H. pylori. A 5 μg portion of H. pylori crude protein extracts was dissolved in loading buffer supplemented with 1% SDS and 50 mM DTT, separated on a 12.5% polyacrylamide gel and blotted to PVDF membrane. TAP-tagged protein was detected using the PAP antibody. Lane 1, HpKchA-TAP; lane 2, M127L-HpKchA-TAP.
In order to evaluate the impact of the expression of this separate RCK domain in the channel function of H. pylori, we studied growth of the M127L-hpKchA mutant in brucella broth. To accentuate the growth defect of HpKchA mutants, we supplemented the broth in all following experiments with β-cyclodextrin instead of FCS. This medium contained a lower and more reproducible concentration of 4.5±1 mM K+ (five different measurements). Growth of the wild-type H. pylori was independent of the concentration of additional KCl, whereas ΔhpKchA was strongly impaired in growth without supplementation of KCl (6% of OD600 nm at 30 h compared to the wild-type; Figure 5B, white bars, first two sets of columns), confirming and strengthening the data in FCS-supplemented medium (Figure 2). As expected, the hpKchA reinsertion mutant showed a similar growth behavior compared to the wild-type. Interestingly, the mutant carrying the single exchange of methionine 127 to leucine, lacking the expression of the separate RCK domain, exhibited a phenotype intermediate between that of the wild-type and the ΔhpKchA mutant (25% of OD600 nm at 30 h compared to the wild-type; Figure 5B, white bars, first and third sets of columns). This suggests that the separate RCK domain is essential for full function of HpKchA, and that the mutant channel conducts potassium less efficiently.
Figure 5.
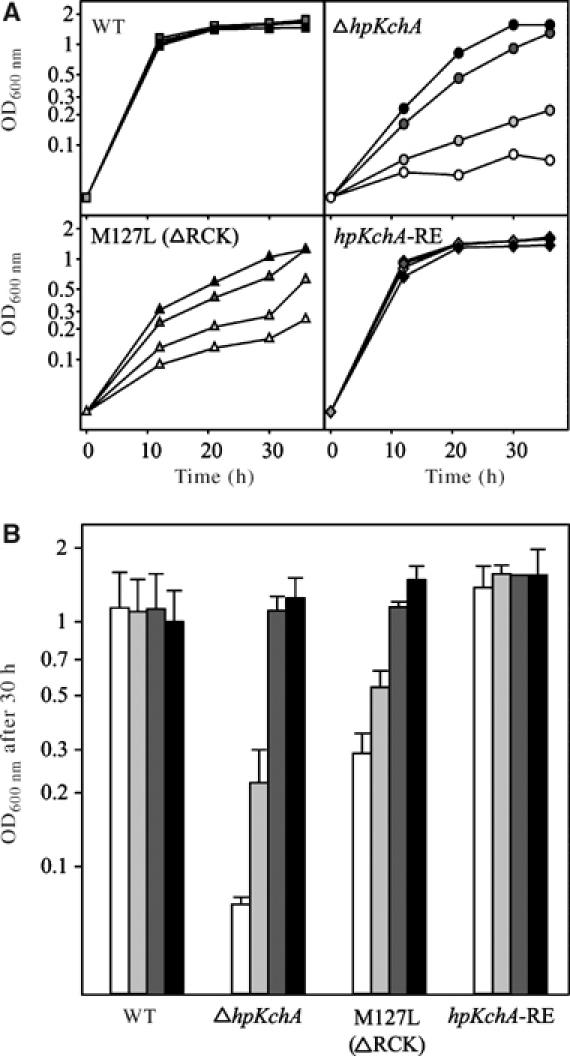
Growth phenotype of H. pylori lacking the separate RCK domain. Growth of H. pylori X47-2AL, its ΔhpKchA deletion mutant, the M127L-hpKchA mutant lacking the separate RCK domain and the hpKchA reinsertion mutant (hpKchA-RE) in brucella broth with β-cyclodextrin and different concentrations of K+ (added as KCl). (A) Representative growth curves. (B) Average OD600 nm after 30 h of growth (data from four independent growth curves for each strain); error bars correspond to standard deviation. Symbols are as follows: white, around 5 mM K+ (basal level of K+ in the medium); light gray, 10 mM K+; dark gray, 20 mM K+; black, 35 mM K+.
Selectivity filter mutants
The K+ filter sequence of HpKchA (ATGFGA; A72–A77; Figure 1) deviates from the canonical motif TXGYGD (T75–D80 in KcsA) (Figure 6, upper panel). Therefore, we studied the effect of mutations within this filter on the activity of the channel, monitored by the growth behavior of these H. pylori mutants at different K+ concentrations. Four different hpKchA versions, each carrying single codon exchange in the selectivity filter, were constructed and replaced the native gene on the chromosome of the H. pylori strain X47-2AL. The aspartate residue (of the canonical filter sequence) was shown to be important for accumulation of K+ before entering the pore (Haug et al, 2004) as well as for stabilization of the pocket structure of the filter (Chapman et al, 2001). An alanine is found at this position in HpKchA. When this alanine was exchanged to aspartate to fit the canonical sequence, we observed the same growth behavior on different K+ concentrations as for the wild-type (Figure 6B and C, first set of columns), indicating that at minimal K+ concentrations of around 5 mM, selectivity was not changed by altering the filter at this position. We then focused on the highly conserved canonical central motif, GYG, which, in wild-type HpKchA, is altered in a conservative manner to GFG. Exchanging the phenylalanine (F75) to a glycine (GFG → GGG) led only to a slight K+-dependent growth phenotype, indicating that K+ was still conducted by this mutant channel in a nearly as efficient way as by wild-type HpKchA (Figure 6B and C, second set of columns). Exchanging the first glycine (G74) to alanine (GFG → AFG) led to a complete loss of channel activity, as the growth behavior resembled that of the deletion mutant (Figures 5, 6B and C, third set of columns). In contrast, when the second glycine (G76) was substituted by alanine (GFG → GFA), growth of the mutant was only affected at the lowest concentration of K+ tested but reached wild-type level at 10 mM K+ (Figure 6B and C, fourth set of columns). The latter two results confirm the relative importance of these two glycine residues in coordination of K+ within the selectivity filter, as proposed from the crystal structure of KscA. As the first glycine is situated in the center of the filter pore, it is consistent with the model that modification to alanine leads to complete loss of activity. However, the second glycine was proposed to be located near the external site of the filter; thus, its minor change to alanine might be better tolerated.
Figure 6.
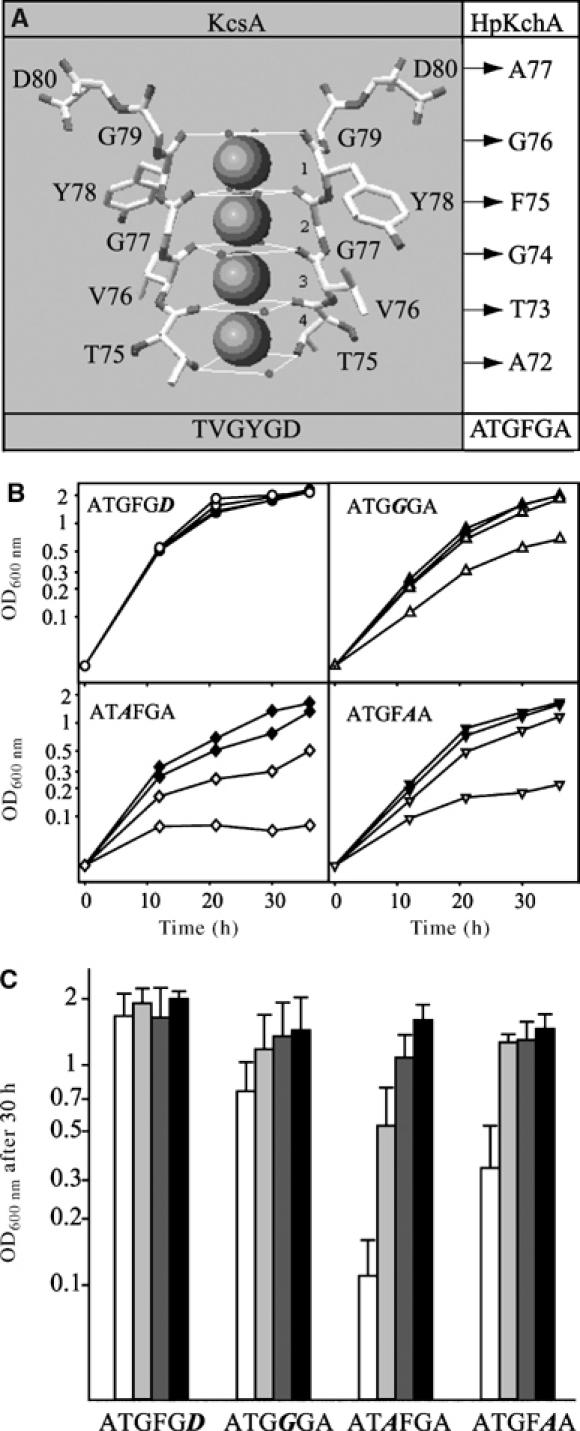
Modification of the selectivity filter in HpKchA. (A) Structural model of the selectivity filter of KcsA from S. lividans (modified by N Tholema according to the cover of Nature 414, 2001) and the corresponding amino-acid residues in HpKchA. Both hexameric filter sequences are summarized below. (B) Representative growth curves. (C) Average OD600 nm after 30 h of growth (data from 3 to 5 independent growth curves for each strain) of the selectivity filter mutants of H. pylori X47-2AL in brucella broth with β-cyclodextrin and different concentrations of K+ (added as KCl). Symbols are as follows: white, around 5 mM K+ (basal level of K+ in the medium); light gray, 10 mM K+; dark gray, 20 mM K+; black, 35 mM K+; error bars correspond to standard deviation.
Colonization of the gastric murine mucosa is dependent on HpKchA
We have demonstrated above that the K+ channel of H. pylori is essential for growth at low K+ concentrations in vitro. In order to test whether H. pylori encounters low K+ concentrations in its natural environment and whether HpKchA is essential for persisting in the host, we compared the capability of H. pylori wild-type and its isogenic hpKchA deletion mutant to colonize the gastric mucosa. For this purpose, we used the murine model for orogastric infection (Lee et al, 1997), counting the bacterial load at the gastric mucosa after 4 weeks of infection.
First, we used the X47-2AL-ΔhpKchA mutant resistant to kanamycin. In two independent experiments, the wild-type H. pylori X47-2AL colonized the murine gastric mucosa to a mean bacterial load of 105–106 CFU/g of stomach (Figure 7A and B). In contrast, none of the ΔhpKchA (XΔKmR) mutant cells was detected in the stomach of mice after 4 weeks of orogastric infection. In order to strengthen this result and use the same antibiotic resistance cassette for the complementation mutant, we constructed deletion mutants carrying a gentamycin resistance cassette in two different H. pylori strain backgrounds, the X47-2AL and the SS1 strain. These deletion mutants were then complemented by reinserting the native hpKchA into the same genetic locus using the kanamycin resistance cassette. In the X47-2AL genetic context, the gentamycin-resistant ΔhpKchA (XΔGmR) was not capable of colonizing the mouse mucosa (Figure 7C). For the SS1 genetic background, we detected only minor amounts of Δ-hpKchA(SΔGmR) in three out of seven mice (2–3 log less than median of wild-type; Figure 7D), leading to a geometrical median below the detection limit. For the reinsertion mutants of both strains, SS1 (S-REKmR) and the X47-2AL (X-REKmR), colonization of the gastric mouse mucosa (Figure 7C and D) was comparable to that of the wild-type, corroborating the result that the absence of HpKchA itself caused the dramatic decrease (for SS1) or even loss (X47-2AL) of capacity for colonization of the murine gastric environment. In conclusion, our results showed that HpKchA is essential for efficient colonization of the gastric environment, probably because of the presence of a concentration of potassium lower or equal to around 5 mM (as tested in vitro as minimal K+ concentration) at the site of bacterial proliferation.
Figure 7.
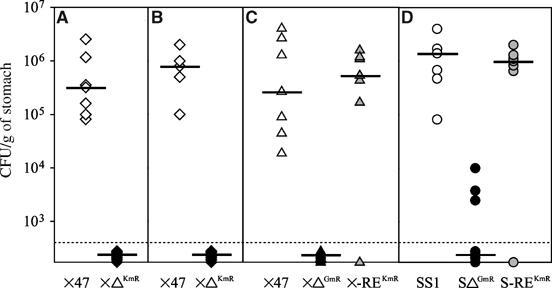
H. pylori ΔhpKchA fails to efficiently colonize the mouse gastric mucosa. The following H. pylori strains were tested for their capacity to colonize the murine stomach: two different wild-type H. pylori (X47-2AL and SS1), their isogenic ΔhpKchA deletion mutants carrying either a kanamycin or gentamycin resistance cassette (XΔKmR, XΔGmR and SΔGmR) as well as their hpKchA reinsertion strains (X-REKmR and S-REKmR). Each panel (A–D) corresponds to an independent experiment. Each data point represents the gastric colonization load of one mouse; horizontal bars, the geometric mean for each group of mice. The lower detection limit was 400 bacteria/g stomach (dashed line); points below this detection limit correspond to mice, in which no colonization of H. pylori was detected.
Discussion
Although a considerable amount of data are available on structural and mechanistic features of prokaryotic K+ channels, till now no clear biological function has been assigned to any of these systems (Kuo et al, 2005). In the present study, we identified for the first time an unequivocal biological role of a prokaryotic K+ channel, by demonstrating that HpkchA from H. pylori serves as bulk K+ uptake system for this gastric pathogen at low K+ concentrations. Moreover, HpKchA was essential for the efficient colonization of the mouse gastric mucosa (Figure 7), indicating that HpKchA serves as sole uptake system in this gastric pathogen.
The primary sequence shows that HpKchA belongs to the K+ channels of the 2TM RCK domain type family (Figure 1). Purified His-tagged monomeric HpKchA oligomerized spontaneously (Figure 3D), which is consistent with a K+ channel function, as the active form of K+ channels is a homotetramer (Doyle et al, 1998).
Unlike KcsA, HpKchA contains a regulatory RCK domain at its C-terminus. As for other related proteins, a potential internal second start codon (M127; Figure 1) for the RCK domain is found in HpKchA, and like MthK and MjK2, expression of HpKchA in E. coli leads to an additional 28 kDa RCK domain (Figure 3A and B; Jiang et al, 2002; Hellmer and Zeilinger, 2003; Parfenova et al, 2006). Till now, it was not known whether the separate RCK domain is expressed in vivo, and whether it complexes in a 1:1 stoichiometry with the complete channel subunit, forming a hetero-octamer as the K+-conducting channel entity (Jiang et al, 2002; Figure 3C). Using a tag fused to wild-type HpKchA and M127L-HpKchA, we showed that in H. pylori a separate RCK domain is indeed expressed in a 1:1 stoichiometry with the full-length HpKchA protein (Figure 4, lane 1). Moreover, loss of this separate RCK domain led to an intermediate growth phenotype at defined external K+ concentrations (Figure 5), indicating that the separate RCK domain is essential for full HpKchA activity. This could be explained by assuming that the open probability in the M127L-HpKchA variant is lower, and that the role of the separate RCK domain is activation of channel activity.
A highly conserved hexapeptide sequence within the pore-forming P-region determines the cation selectivity of K+ channels. HpKchA exhibits the unusual selectivity filter motif ATGFGA instead of the commonly found TXGYGD (Kuo et al, 2005). The sequence GYG is the critical determinant for K+ selectivity and conductance in KcsA (Heginbotham et al, 1992, 1994) and is conservatively modified to GFG in HpKchA. When we exchanged the phenylalanine to glycine (F75G), we observed a nearly wild-type growth phenotype of this H. pylori mutant (Figure 6). For the E. coli Kch channel, exchange of the corresponding tyrosine residue to anything else than phenylalanine led to loss of function (Kuo et al, 2003). However, in a eukaryotic 4TM K+ channel, the aromatic amino acid is replaced by leucine in one of the two P-loop motifs, which does not affect K+ selectivity (Lesage et al, 1996). As only the negatively charged carbonyl oxygens of the main polypeptide chain are involved in direct coordination of K+ in the filter pore, tolerance of amino-acid variations is supposed to be determined by the overall constitution of the filter and not only by the GYG core motif.
The two glycines in the filter core region are conserved in nearly all K+ channels, with the first glycine being the most important residue for K+ channel activity (Lu and Miller, 1995). The KcsA crystal structure shows that this residue (G77) is in a left-handed helical conformation, normally observed for D-amino acids, and that its essentiality at this position was explained by being the only natural amino acid that can exhibit this unusual conformation (Valiyaveetil et al, 2004). Accordingly, exchange of the corresponding glycine to alanine (G74A) in HpKchA led to complete loss of function of the K+ channel (Figure 6). However, when the second glycine (G76) was changed to alanine, we observed a growth defect at 5 mM K+, which was nearly completely compensated at 10 mM K+ (Figure 6). This only slight decrease in channel function is in agreement with glycine 76 being situated near to the external site of the filter pore, where an exchange to alanine might better be tolerated.
Changing the last alanine (A77) in the filter motif of HpKchA to the commonly used aspartate had no effect on channel function as observed under our conditions. Ionizable side chains in the vicinity of the filter pore are thought to affect the energetics of ion permeation (Ranatunga et al, 2001). At pH 7, aspartate is likely to be mainly deprotonated and therefore negatively charged. This might not be the case at slightly acidic pH, found in the natural environment of the pathogen. Thus, we speculate that under these conditions, the aspartate at the entry of the pore does not confer a major advantage for the conductance of K+ through the filter and was evolutionarily lost (i.e. exchanged to an alanine).
It has been predicted that the presence of a single K+ uptake channel might not be sufficient for K+ acquisition by H. pylori, and that yet unidentified transporters have to be present in this pathogen (Van Vliet et al, 2001). Deletion of all K+ transport systems in E. coli generates an E. coli mutant, requiring ⩾15 mM K+ for growth (Stumpe and Bakker, 1997), similar to that necessary to support growth of the H. pylori ΔhpKchA. For both organisms, it is unclear how potassium at high concentrations reaches the cytoplasm but it is likely that it enters less specifically through other ion transporters. At K+ concentrations lower than 20 mM, HpKchA is the sole K+ uptake system of this gastric bacterium. Assuming a membrane potential of around −180 mV at pH 7, a bacterial cell is capable of accumulating K+ about 1000-fold via such a K+-specific diffusion pathway. In most bacteria, the cytoplasmic K+ concentration amounts to 200 mM. Thus, a K+ channel would be sufficient for the cells to capture K+ down to a K+ concentration in the medium of 0.2 mM. In H. pylori, we measured a comparable internal K+ concentration (192±23 mM) and its membrane potential at pH 7 was previously determined to be at least −150 mV (Stingl et al, 2002), suggesting that H. pylori probably can grow down to K+ concentrations of at least 0.4 mM. The colonization site of H. pylori is the human gastric mucosa, where concentrations of K+ are expected to be 3.5–5 mM, which is far above the calculated minimal K+ concentration. Thus, under such conditions, HpKchA activity must be sufficient to satisfy the cells' need for K+. Consistently, HpKchA was essential for efficient colonization of the mouse gastric mucosa (Figure 7). This observation can be explained by assuming that at this site, the K+ concentration is indeed as low as 3.5–5 mM, conditions under which the ΔhpkchA strain is not able to sufficiently grow.
A role for a bacterial K+ channel as uptake system for bulk K+ might be an adaptation of the human pathogen to its K+-rich, competitor-free gastric environment, which is its sole proliferation niche. There are probably few other bacteria having only a K+ channel, like Ureaplama urealyticum, which inhabits the human urogenital tract, a site at which potassium is excreted. In contrast, it is hypothesized that most bacterial K+ channels affect the membrane potential and respond to changes in the environment (Kuo et al, 2005). The distribution of K+ transporters and K+ channels in closely related ɛ-proteobacteria in relation to their lifestyle led us to propose the following model (Figure 8). Bacteria such as H. pylori dwelling in one single, K+-rich niche have only a K+ channel for bulk K+ import. Campylobacter jejuni (Parkhill et al, 2000), being restricted to the mammalian or avian intestine, in which it faces high bacterial load and low external K+ concentrations, possesses only efficient K+ transporters for K+ acquisition. Furthermore, H. hepaticus (Suerbaum et al, 2003) and W. succinogenes (Baar et al, 2003) both possess a homologue of HpKchA in addition to other K+ transporters. H. hepaticus lives in the murine intestine and is found in liver tissue, whereas the commensal, W. succinogenes, occurs in the bovine rumen and has the potential for free living (Baar et al, 2003). In these two latter organisms, K+ transporters are proposed to accumulate K+, whereas their K+ channels can be utilized to respond to environmental changes. Consistently, the recently sequenced free-living ɛ-proteobacterium Thiomicrospira denitrificans also possesses a K+ transporter in addition to its K+ channel (Copeland, 2005, EMBL gene databank).
Figure 8.
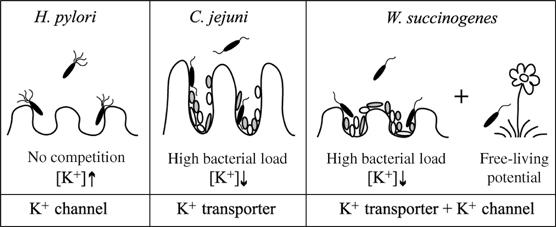
Function of prokaryotic potassium channels in the context of the lifestyle of three representative species of ɛ-proteobacteria. This model proposes the existence of a relation between the K+ content of single or multiple environmental niches and the need for a K+ channel, a K+ transport system or both. H. pylori, inhabiting a single, K+-rich niche, uses a channel for bulk potassium import; C. jejuni, inhabiting the intestine of different species and encountering high bacterial loads, faces low K+ concentrations. It uses efficient transport systems for K+ acquisition and no K+ channel; W. succinogenes, colonizing the rumen of cattle and presenting the potential for free-living as proposed by Baar et al (2003), has both, a K+ transporter for bulk K+ import and a K+ channel in order to adapt metabolic pathways upon environmental changes.
In conclusion, HpKchA is an excellent model protein, as it allows to link structural characteristics of a prokaryotic K+ channel to its biological function. Electrophysiological studies on HpKchA are under way to characterize original properties of this K+ channel, which might be a reflexion of adaptation to the gastric environment and might contribute to the development of specific inhibitors appropriate for anti-Helicobacter therapy.
Materials and methods
Bacterial strains and culture conditions
H. pylori strains DSM4867 (Deutsche Stammsammlung für Mikroorganismen, Braunschweig), X47-2AL (Ermak et al, 1998) and SS1 (Lee et al, 1997) were grown either on blood agar plates (Oxoid) supplemented with 10% defibrillated horse blood (bioMérieux) or in liquid culture using brucella broth with 10% fetal bovine serum (GibcoBRL) or with 0.2% β-cyclodextrin (Sigma). An antibiotic/anti-fungal cocktail was used at the following final concentrations per ml: 12.5 μg vancomycin, 3.1 μg polymyxin B, 6.25 μg trimethoprim and 2.5 μg amphotericin B. When appropriate, 20 μg/ml kanamycin, 4–8 μg/ml chloramphenicol or 50 μg/ml apramycin was used. Selection and precultures were performed on blood agar plates and liquid medium supplemented with 15 or 30 mM KCl, respectively. Plates were incubated at 37°C under a microaerobic atmosphere in jars using CampyGen (Oxoid). Liquid cultures were shaken at 140 r.p.m.
E. coli strains DH5α (Woodcock et al, 1989), C43 (Miroux and Walker, 1996) and MC1061 (Casadaban and Cohen, 1980) were grown at 37°C in Luria broth (Miller, 1992) or KML medium (10 g/l KCl, 10 g/l bactotryptone and 5 g/l yeast extract) with 100 μg/ml ampicillin or carbenicillin, 100 μg/ml spectinomycin or 20 μg/ml kanamycin, when appropriate.
Construction and purification of the His-tagged HpKchA protein
The homologue of the open reading frame, hp0490, from H. pylori 26695 was amplified from chromosomal DNA of strain H. pylori DSM4867, which was purified using the DNEasy Tissue Kit (Qiagen). The oligonucleotides Hp490L and Hp490R (Supplementary Table S1) were used to amplify the 1.1 kb hpkchA gene and to fuse six histidine codons to its 3′-end. Using these primers, the original TTG start codon of hp0490 was converted into ATG and an additional glycine was inserted as the second codon in order to create an NcoI restriction site. The PCR was performed using the high fidelity kit (Roche) as described by the manufacturer and the obtained sequence was checked. The fragment was ligated with plasmid pET16b opened with NcoI and BamHI. E. coli C43 was transformed with the ligation product, giving strain C43 with plasmid phpKchA. The purification of the His-tagged HpKchA was performed over Ni2+-NTA. For details see Supplementary data.
Construction of H. pylori strains carrying hpKchA mutations or C-terminal TAP fusions
Deletions, point mutations, fusions or reinsertions were introduced in H. pylori strains X47-2AL and/or SS1 by allelic exchange using plasmids in which H. pylori DNA regions are flanking antibiotic resistance cassettes, either the non-polar kanamycin cassette (Skouloubris et al, 1998) or the gentamycin resistance cassette (Bury-Moné et al, 2003). For the C-terminal TAP fusion (TAP tag composed of Protein A and a calmodulin-binding domain both separated by a TEV cleavage site; Rigaut et al, 1999), we used a plasmid, pILL851-C, in which the tag was fused to the non-polar kanamycin cassette (manuscript in preparation). Construction of the plasmids used is detailed in Supplementary data, with the corresponding PCR primers described in Supplementary Table S1. H. pylori mutants were obtained by natural transformation as previously described (Bury-Moné et al, 2001). Genomic DNA of the recombinant H. pylori mutants was isolated using the QIAamp DNA Mini Kit (Qiagen). All insertions into the chromosome were verified by PCR and sequenced with an ABI 310 automated DNA sequencer (Perkin-Elmer). Site-directed mutagenesis of the hpKchA was performed using the divergent PCR strategy described by Bury-Moné et al (2001) using phpKchA as template and the primers detailed in Supplementary Table S2.
Immunoblot analysis and N-terminal sequencing
For the analysis of TAP-tagged protein, H. pylori cells were sonicated twice for 30 s, debris was removed by centrifugation at 20 000 g for 5 min and 5 μl of protein extract was separated by 10 or 12.5% SDS–PAGE. Proteins were blotted onto nitrocellulose (Biometra, Göttingen, Germany) or polyvinylidene difluoride membrane (PVDF, Millipore). TAP-tagged protein was detected using a peroxidase-coupled anti-peroxidase antibody (Sigma) and the ECL reagent (Amersham) according to the manufacturer's instructions. His-tagged proteins were detected using an anti-penta-His-antibody (Qiagen, Hilden, Germany) at a concentration of 0.2 μg/ml and a second goat anti-mouse antibody (Pierce Biotechnology Inc., IL, USA) coupled to alkaline phosphatase.
For N-terminal sequencing, protein was blotted onto a PVDF membrane, from which the protein was N-terminally sequenced by Edman degradation (Applied Biosystems, 473 A apparatus).
Electroelution of monomeric HpKchA
Ni2+-NTA purified proteins were dissolved in the low SDS sample buffer without reducing agent, immediately separated by SDS–PAGE on a 10% gel and visualized by negative staining with 3 M KCl (Prussak et al, 1989). Gel pieces containing the 40 kDa HpKchA monomer were excised and transferred to an electroeluter (Bio-Rad Laboratories Inc., CA, USA). Protein was eluted into 500 μl of Laemmli buffer (0.1% SDS, 192 mM glycine and 25 mM Tris) by applying 10 mA per tube for 3 h at room temperature.
Determination of K+ concentrations
The volume of the cell pellet was determined as described previously (Stingl et al, 2002). For the determination of the cytoplasmic K+ concentration, the cell pellet was added to 1 ml of 5% TCA, frozen at −20°C overnight and boiled for 10 min. After addition of 3 ml of 3.7 mM CsCl, the K+ concentration was determined using an ELEX 6361 flame photometer (Eppendorf, Germany) and KCl as a standard. The K+ concentration of liquid medium was determined accordingly.
Determination of urease activity
H. pylori cells were harvested in logarithmic phase (at OD600 nm∼0.3) by centrifugation at 5000 g for 10 min. After resuspension in 1/10 volume of 0.9% NaCl, the cells were 10-fold diluted in 100 mM citric acid (pH 2) supplemented with 100 mM NaCl and 20 mM urea. Urease activity was determined by measuring the depletion of urea in the medium according to Rahmatullah and Boyde (1980).
Mouse model of colonization
H. pylori was grown on blood agar plates for 24 h and suspended in peptone broth at a concentration of 109 bacteria/ml. For the mutants, three independently selected clones were mixed before inoculation of the mice. The precise infection dose was determined by CFU enumeration. Female NMRI mice (4 weeks old, Iffa-Credo) were inoculated orogastrically with aliquots of 100 μl bacteria (108 bacteria) as described previously (Ferrero et al, 1998). Four mice were inoculated with peptone broth as a negative control. After 4 weeks, the mice were killed and the gastric tissue was homogenized in peptone broth. Bacterial counts were obtained by plating serial dilutions on blood agar plates containing per ml 200 μg bacitracine and 10 μg nalidixic acid in addition to the antibiotic/anti-fungal cocktail described before.
Supplementary Material
Supplementary Information and Tables
Acknowledgments
We thank C Weber-Sparenberg for construction of the selectivity filter mutant plasmids, E Limpinsel and J-M Thiberge for technical assistance and IG Boneca, R Guy and T Nakamura for helpful discussions. This work was supported by subsequent postdoctoral fellowships of the German Academic Exchange Servive (DAAD) and the Fondation pour la Recherche Médicale (FRM) to KS, by the Deutsche Forschungsgemeinschaft (SFB 431) and by the Fonds der Chemischen Industrie. CZ was supported by the Helmholtz-Gemeinschaft, Institut für Biologische Strukturforschung, VIBS (VH-VI-013).
References
- Albright RA, Ibar J-LV, Kim CU, Gruner SM, Morais-Cabral JH (2006) The RCK domain of the KtrAB K+ transporter: multiple conformations of an octameric ring. Cell 126: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Baar C, Eppinger M, Raddatz G, Simon J, Lanz C, Klimmek O, Nandakumar R, Gross R, Rosinus A, Keller H, Jagtap P, Linke B, Meyer F, Lederer H, Schuster SC (2003) Complete genome sequence and analysis of Wolinella succinogenes. Proc Natl Acad Sci USA 100: 11690–11695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth IR (1985) Regulation of cytoplasmic pH in bacteria. Microbiol Rev 49: 359–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb JF, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A, Scott JL, Geoghagen NS, Venter JC (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273: 1058–1073 [DOI] [PubMed] [Google Scholar]
- Bury-Moné S, Skouloubris S, Dauga C, Thiberge J-M, Dailidiene D, Berg DE, Labigne A, De Reuse H (2003) Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect Immun 71: 5613–5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Moné S, Skouloubris S, Labigne A, De Reuse H (2001) The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol Microbiol 42: 1021–1034 [DOI] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN (1980) Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol 138: 179–207 [DOI] [PubMed] [Google Scholar]
- Chapman ML, Krovetz HS, VanDongen AM (2001) GYGD pore motifs in neighbouring potassium channel subunits interact to determine ion selectivity. J Physiol 530: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin TM (ed.) (1997) Textbook of Biochemistry with Clinical Correlations. New York: Wiley-Liss [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280: 69–77 [DOI] [PubMed] [Google Scholar]
- Epstein W, Buurman E, McLaggan D, Naprstek J (1993) Multiple mechanisms, roles and controls of K+ transport in Escherichia coli. Biochem Soc Trans 21: 1006–1010 [DOI] [PubMed] [Google Scholar]
- Ermak TH, Giannasca PJ, Nichols R, Myers GA, Nedrud J, Weltzin R, Lee CK, Kleanthous H, Monath TP (1998) Immunization of mice with urease vaccine affords protection against Helicobacter pylori infection in the absence of antibodies and is mediated by MHC class II-restricted responses. J Exp Med 188: 2277–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst PB, Gold BD (2000) The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol 54: 615–640 [DOI] [PubMed] [Google Scholar]
- Ferrero RL, Thiberge JM, Huerre M, Labigne A (1998) Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect Immun 66: 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay LA, Standfield PR (1977) Cs+ causes a voltage-dependent block of inward K+ currents in resting skeletal muscle fibres. Nature 267: 169–170 [DOI] [PubMed] [Google Scholar]
- Greger R, Windhorst U (eds) (1996) Comprehensive Human Physiology: From Cellular Mechanisms to Integration. Berlin, Heidelberg: Springer-Verlag [Google Scholar]
- Haug T, Sigg D, Ciani S, Toro L, Stefani E, Olcese R (2004) Regulation of K+ flow by a ring of negative charges in the outer pore of BKCa channels. Part I: aspartate 292 modulates K+ conduction by external surface charge effect. J Gen Physiol 124: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heginbotham L, Abramson T, MacKinnon R (1992) A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science 258: 1152–1155 [DOI] [PubMed] [Google Scholar]
- Heginbotham L, Lu Z, Abramson T, MacKinnon R (1994) Mutations in the K+ channel signature sequence. Biophys J 66: 1061–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmer J, Zeilinger C (2003) MjK1, a K+ channel from M. jannaschii, mediates K+ uptake and K+ sensitivity in E. coli. FEBS Lett 547: 165–169 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R (2002) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417: 515–522 [DOI] [PubMed] [Google Scholar]
- Kuo MM, Haynes WJ, Loukin SH, Kung C, Saimi Y (2005) Prokaryotic K(+) channels: from crystal structures to diversity. FEMS Microbiol Rev 29: 961–985 [DOI] [PubMed] [Google Scholar]
- Kuo MM, Saimi Y, Kung C (2003) Gain-of-function mutations indicate that Escherichia coli Kch forms a functional K+ conduit in vivo. EMBO J 22: 4049–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF (1997) A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112: 1386–1397 [DOI] [PubMed] [Google Scholar]
- LeMasurier M, Heginbotham L, Miller C (2001) KcsA: it's a potassium channel. J Gen Physiol 118: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage F, Guillemare E, Fink M, Duprat F, Lazdunski M, Romey G, Barhanin J (1996) TWIK-1, a ubiquitous human weakly inward rectifying K+ channel with a novel structure. EMBO J 15: 1004–1011 [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Miller C (1995) Silver as a probe of pore-forming residues in a potassium channel. Science 268: 304–307 [DOI] [PubMed] [Google Scholar]
- Miller JF (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia Coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Miroux B, Walker JE (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260: 289–298 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yuda R, Unemoto T, Bakker EP (1998) KtrAB, a new type of bacterial K+-uptake system from Vibrio alginolyticus. J Bacteriol 180: 3491–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfenova LV, Crane BM, Rothberg BS (2006) Modulation of MthK potassium channel activity at the intracellular entrance to the pore. J Biol Chem 281: 21131–21138 [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403: 665–668 [DOI] [PubMed] [Google Scholar]
- Prussak CE, Almazan MT, Tseng BY (1989) Peptide production from proteins separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis. Anal Biochem 178: 233–238 [DOI] [PubMed] [Google Scholar]
- Rahmatullah M, Boyde TR (1980) Improvements in the determination of urea using diacetyl monoxime; methods with and without deproteinisation. Clin Chim Acta 107: 3–9 [DOI] [PubMed] [Google Scholar]
- Ranatunga KM, Shrivastava IH, Smith GR, Sansom MS (2001) Side-chain ionization states in a potassium channel. Biophys J 80: 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rektorschek M, Buhmann A, Weeks D, Schwan D, Bensch KW, Eskandari S, Scott D, Sachs G, Melchers K (2000) Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol Microbiol 36: 141–152 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Schreiber S, Nguyen TH, Stüben M, Scheid P (2000) Demonstration of a pH gradient in the gastric gland of the acid-secreting guinea pig mucosa. Am J Physiol Gastrointest Liver Physiol 279: G597–G604 [DOI] [PubMed] [Google Scholar]
- Skouloubris S, Thiberge JM, Labigne A, De Reuse H (1998) The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun 66: 4517–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl K, Uhlemann EM, Deckers-Hebestreit G, Schmid R, Bakker EP, Altendorf K (2001) Prolonged survival and cytoplasmic pH homeostasis of Helicobacter pylori at pH 1. Infect Immun 69: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl K, Uhlemann EM, Schmid R, Altendorf K, Bakker EP (2002) Energetics of Helicobacter pylori and its implications for the mechanism of urease-dependent acid tolerance at pH 1. J Bacteriol 184: 3053–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe S, Bakker EP (1997) Requirement of a large K+-uptake capacity and of extracytoplasmic protease activity for protamine resistance of Escherichia coli. Arch Microbiol 167: 126–136 [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, Bell M, Droge M, Fartmann B, Fischer HP, Ge Z, Horster A, Holland R, Klein K, Konig J, Macko L, Mendz GL, Nyakatura G, Schauer DB, Shen Z, Weber J, Frosch M, Fox JG (2003) The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc Natl Acad Sci USA 100: 7901–7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Venter JC (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388: 539–547 [DOI] [PubMed] [Google Scholar]
- Valiyaveetil FI, Sekedat M, Mackinnon R, Muir TW (2004) Glycine as a D-amino acid surrogate in the K(+)-selectivity filter. Proc Natl Acad Sci USA 101: 17045–17049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vliet AHM, Bereswill S, Kusters JG (2001) Ion metabolism and transport. In Helicobacter Pylori Physiology and Genetics, Mobley HLT, Mendz GL, Hazell SL (eds) pp 193–206. Washington, DC: ASM Press [PubMed] [Google Scholar]
- Walderhaug MO, Dosch DC, Epstein W (1987) Potassium transport in bacteria. In Ion Transport in Prokaryotes, Rosen BP, Silver S (eds) pp 85–130. New York: Academic Press [Google Scholar]
- Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17: 3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information and Tables


