Abstract
Nonsense-mediated mRNA decay (NMD) is an mRNA surveillance pathway that recognizes and degrades aberrant mRNAs containing premature stop codons. A critical protein in NMD is Upf1p, which belongs to the helicase super family 1 (SF1), and is thought to utilize the energy of ATP hydrolysis to promote transitions in the structure of RNA or RNA–protein complexes. The crystal structure of the catalytic core of human Upf1p determined in three states (phosphate-, AMPPNP- and ADP-bound forms) reveals an overall structure containing two RecA-like domains with two additional domains protruding from the N-terminal RecA-like domain. Structural comparison combined with mutational analysis identifies a likely single-stranded RNA (ssRNA)-binding channel, and a cycle of conformational change coupled to ATP binding and hydrolysis. These conformational changes alter the likely ssRNA-binding channel in a manner that can explain how ATP binding destabilizes ssRNA binding to Upf1p.
Keywords: mRNA decay, nonsense-mediated mRNA decay, RNA helicase, Upf1, X-ray crystallography
Introduction
Nonsense-mediated mRNA decay (NMD) is an evolutionarily conserved mRNA surveillance pathway that recognizes and eliminates aberrant mRNAs harboring premature termination codons (PTC), thereby preventing the accumulation of nonfunctional or potentially deleterious truncated proteins in the cells (Baker and Parker, 2004; Conti and Izaurralde, 2005; Lejeune and Maquat, 2005). In addition to mRNAs with PTCs, NMD degrades a variety of naturally occurring transcripts to suppress genomic noise (He et al, 2003; Mendell et al, 2004). An important step in NMD is the translation-dependent recognition of transcripts with aberrant termination events and then targeting those mRNAs for destruction.
The three Upf proteins, Upf1, Upf2 and Upf3, appear to constitute the core NMD machinery as they are conserved and required for NMD in Saccharomyces cerevisiae (Cui et al, 1995; He et al, 1997), Caenorhabditis elegans (Hodgkin et al, 1989), Drosophila melanogaster (Gatfield et al, 2003) and in mammalian cells (Lykke-Andersen et al, 2000; Serin et al, 2001). A key member of this protein set is Upf1, which appears to recognize aberrant translation termination events (Amrani et al, 2004) and, then in a subsequent step, interacts with Upf2 and Upf3 to trigger degradation of mRNA (Kashima et al, 2006; Sheth and Parker, 2006).
Upf1p also regulates mRNAs in a manner independent of NMD. For example, in a process referred to as Staufen-mediated decay, human Upf1 (hUpf1) is recruited downstream of a termination codon by the RNA-binding protein Staufen to degrade some mRNAs (Kim et al, 2005). hUpf1 also functions in the regulated degradation of histone mRNAs in mammalian cells (Kaygun and Marzluff, 2005). Moreover, hUpf1 has been shown to play distinct roles in NMD and nonsense-mediated altered splicing (NAS) (Mendell et al, 2002). Finally, Upf1 has been proposed to be involved in the cellular response to genotoxic stress, as exposure of cells to ionizing radiation results in its phosphorylation (Brumbaugh et al, 2004).
Sequence analysis indicates that Upf1 contains two conserved functional regions, an N-terminal Cys-rich domain required for binding to Upf2 and a C-terminal helicase domain (Applequist et al, 1997; Culbertson and Leeds, 2003). The conserved helicase domain of Upf1 belongs to superfamily 1 (SF1) of DNA/RNA helicases, and contains all seven motifs common to the SF1 and SF2 helicases (Applequist et al, 1997). RNA helicases are generally thought to use the energy of ATP hydrolysis to promote transitions in RNA or RNP structure (Tanner and Linder, 2001). Consistent with this notion, S. cerevisiae and human Upf1 proteins exhibit RNA binding, RNA-dependent ATP hydrolysis and 5′–3′ ATP-dependent RNA helicase activities. Moreover, these activities are essential for NMD (Czaplinski et al, 1995; Weng et al, 1996; Bhattacharya et al, 2000). To understand how this ATP hydrolysis is coupled to Upf1 function, it will be important to understand the structure of Upf1 and how it changes during the cycle of ATP binding and hydrolysis.
Crystal structures of the related SF1 DNA helicases PcrA and Escherichia coli Rep protein in complex with DNA substrates and nucleotide have been solved (Korolev et al, 1997; Velankar et al, 1999). These structures revealed monomeric enzymes consisting of two domains, with each domain comprising two subdomains (1A, 1B and 2A, 2B). In each case, two RecA-like domains (1A and 2A) have been proposed to act as a ‘motor' to translocate on the bound nucleic acid using the energy of ATP hydrolysis, with subdomains 1B and 2B acting as a ‘wedge' to destabilize the double-stranded nucleic acid (Soultanas and Wigley, 2001). Similarly, the recent crystal structure of the Drosophila DEAD-box protein Vasa, an SF2 RNA helicase, in complex with a single-stranded RNA (ssRNA) and AMPPNP suggests that the RNA is bound with a tight turn in the backbone, which would be expected to destabilize an RNA duplex (Sengoku et al, 2006).
To gain insight into the molecular function of Upf1 in NMD, we determined the crystal structure of the helicase domain of hUpf1 in AMPPNP-, ADP- and phosphate-bound forms. These results revealed that the hUpf1 helicase core domain is composed of four domains, two of which have a RecA-like fold. Structural comparison with other members of the SF1 helicases, combined with mutagenesis provides a view of the important functional states of hUpf1, and suggests a mechanism by which ATP binding/hydrolysis could be coupled with RNA-binding activity.
Results
Structure determination
The helicase domain of hUpf1 (residues 295–914, designated as hUpf1hd; Supplementary Figure S1) was expressed in E. coli and purified to homogeneity. Gel filtration showed that the protein is monomeric in solution (Supplementary Figure S2). The structure of hUpf1hd with bound AMPPNP (hUpf1hd-AMPPNP) was solved by the single-wavelength anomalous dispersion (SAD) method at a resolution of 2.6 Å. Difference Fourier maps clearly showed the bound AMPPNP and Mg2+. Residues 584–587 are disordered in the structure. The final model has an Rfree of 27.1% with good stereochemistry.
The ADP-bound complex (hUpf1hd-ADP) was solved at 2.4 Å resolution by molecular replacement using the structure of hUpf1hd-AMPPNP without bound nucleotide as a search model. In the structure of hUpf1hd-ADP, an ADP molecule plus a phosphate ion occupy the nucleotide-binding pocket for both molecules in the asymmetric unit with Mg2+ situated in a position similar to that in the AMPPNP structure. Residues 349–355, 583–586, and 844–849 for both molecules are disordered. As no substantial differences are observed between two molecules (r.m.s.d. of 0.6 Å for all the equivalent Cα atoms), and molecule A is more complete than B, all subsequent analyses are based on molecule A.
The structure of nucleotide-free hUpf1hd was solved by SAD phasing combined with molecular replacement at a resolution of 2.8 Å. As a phosphate ion is situated in a position corresponding to that occupied by the γ-phosphate of AMPPNP in the hUpf1hd–AMPPNP complex, this structure is designated as hUpf1hd-phosphate. Residues 337–338, 367–373, 401–405 and 584–587 are disordered in the hUpf1hd-phosphate structure. Details of all the structure determinations and refinements are summarized in Table I (see Materials and methods).
Table 1.
Data collection and refinement statistics
| hUpf1hd-AMPPNP | hUpf1hd-phosphate | hUpf1hd-ADP | |
|---|---|---|---|
| Data collection | |||
| Derivative | SeMet | — | — |
| Wavelength (Å) | 0.9793 | 0.9793 | 0.9793 |
| Resolution limit (Å) | 2.6 | 2.8 | 2.4 |
| Space group | P43212 | P43212 | P1 |
| Cell parameters | |||
| a/b/c (Å) | 189.0/189.0/44.9 | 195.2/195.2/45.5 | 63.3/67.8/87.3 |
| α/β/γ (deg) | 90/90/90 | 90/90/90 | 114.4/90.1/110.2 |
| Unique reflections (N) | 27 959 | 248 96 | 43 423 |
| I/σ | 6.0 (2.1) | 4.5 (2.3) | 8.8 (3.1) |
| Completeness (%) | 97.2 (98.9) | 100 (100) | 90.8 (90.8) |
| Rmergea | 0.094 (0.379) | 0.102 (0.395) | 0.059 (0.24) |
| Number of Se sites | 13 | ||
| Refinement statistics | |||
| Data range (Å) | 2.0–2.6 | 2.0–2.8 | 2.0–2.4 |
| Used reflections (N) | 24 065 | 21 121 | 41 163 |
| Nonhydrogen atoms (water) | 4877 (110) | 4720 (63) | 9555 (243) |
| Rworkb/Rfreec (%) | 24.5/27.1 | 25.8/29.8 | 23.5/27.2 |
| R.m.s. deviation | |||
| Bond length (Å) | 0.006 | 0.012 | 0.008 |
| Bond angles (deg) | 1.048 | 1.294 | 1.011 |
| Average B factors (Å2) | |||
| 1A/1B/1C/2A | 27.4/60.4/48.3/48.7 | 57.2/92.1/51.9/77.1 | 25.5/41.8/42.6/39.3 |
| Ramachandran plot | |||
| Most favored region (%) | 83.9 | 85.2 | 87.6 |
| Number of outliers | 1 | 1 | 0 |
| Values in parentheses indicate the specific values in the highest resolution shell. | |||
| Rmerge=∑∣Ij−〈I〉∣/∑Ij, where Ij is the intensity of an individual reflection, and 〈I〉 is the average intensity of that reflection. | |||
| Rwork=∑∣∣Fo∣−∣Fc∣∣/∑∣Fc∣, where Fo denotes the observed structure factor amplitude, and Fc denotes the structure factor amplitude calculated from the model. | |||
| Rfree is as for Rcryst, but calculated with 5.0% of randomly chosen reflections omitted from the refinement. | |||
Description of overall structure
In the AMPPNP-, ADP- and phosphate-bound forms, hUpf1hd is composed of two domains (domains 1 and 2) (Figure 1A). Both of these two domains contain a ‘RecA-like' α/β domain designated as 1A and 2A, respectively. In addition to subdomain 1A, domain 1 also contains two insertions, each of which forms a distinct subdomain, denoted as domains 1B and 1C, respectively. Domain 1A consists of residues 295–324, 415–555 and 610–700 and forms a parallel seven-stranded β-sheet surrounded by seven helices on one side and three helices on the other. Domain 2A contains a parallel six-stranded β-sheet flanked by three helices on both sides. The seven classical sequence motifs of SF1/SF2 helicases are located in these two domains with motifs I, Ia, II and III in domain 1A and motifs IV, V and VI in domain 2A. The nucleotide-binding site is located in a deep cleft separating domains 1A and 2A. The core RecA-like architecture of hUpf1hd resembles that found in SF1 DNA helicase PcrA (Velankar et al, 1999) and SF2 RNA helicase Vasa (Sengoku et al, 2006). Superposition of these two domains (1A and 2A) with those of PcrA (Velankar et al, 1999) with bound ADPNP and double-stranded DNA (dsDNA) and Vasa with bound ssRNA and AMPPNP gives an r.m.s.d. of 1.5 and 1.9 Å, for 212 and 149 equivalent Cα atoms, respectively (Figure 1B and C). This suggests that the relative orientations of domains 1A and 2A are largely conserved among these helicases with hUpf1 being more similar to PcrA.
Figure 1.

Structure of hUpf1hd and its comparison with PcrA and Vasa helicases. The ribbon diagrams are drawn with domains 1A (pink) and 2A (wheat) in the same orientation. Bound nucleotide is shown in stick model and Mg2+ ion in purple sphere. (A) hUpf1hd-AMPPNP. Domains 1B and 1C are colored in lime and slate, respectively. (B) The PcrA/ADPNP/dsDNA complex. Domains 1B and 2B are shown in lime and yellow respectively. Bound DNA is shown in orange cartoon tube. (C) The Vasa/AMPPNP/ssRNA complex. Bound ssRNA is shown in purple cartoon tube.
Subdomains 1B and 1C are structurally unique domains for hUpf1 and not closely related to equivalent regions in any of the previously determined helicase structures. Domain 1B, residues 325–414, is a β-barrel consisting of six anti-parallel β strands that is located above the interface between domains 1A and 2A (Figure 1A). Domain 1C contains three helices formed by residues 556–609 and is situated on domain 1A, making a few contacts with domain 1B (Figure 1A).
Nucleotide-binding site and ATP hydrolysis
AMPPNP is bound in the pocket between domains 1A and 2A (Figure 1A). The interactions of AMPPNP with the protein residues are similar to those observed in the PcrA/DNA/ADPNP complex (Velankar et al, 1999) with some notable differences. As shown in Figure 2A, the adenine base is held in a hydrophobic pocket between Tyr702 and Pro469. Tyr702 is structurally equivalent to Tyr286 of PcrA, whereas Pro469 has no structural counterpart in PcrA. Val500 contacts the ribose ring via hydrophobic interactions. In contrast, Arg39 of PcrA, which corresponds to Val500 in hUpf1, interacts with the ribose ring of ATP through its methylene group. Four residues in hUpf1hd make direct contacts with the γ-phosphate of AMPPNP, namely Lys498, Gln665, Arg703 and Arg865 from conserved motifs I, III, IV and VI, respectively. In contrast, in PcrA, only three residues Gln254, Arg287 and Arg610 interact directly, with the γ-phosphate of the nucleotide, with Lys37 (Lys498 in hUpf1) contacting the β-phosphate. In PcrA, Arg610 acts as an ‘arginine finger', which has been proposed to stabilize the transition state, thus facilitating ATP hydrolysis (Caruthers and McKay, 2002; Velankar et al, 1999). Residue Arg610 in PcrA corresponds to Arg865 in Upf1p, which has a similar interaction with the γ-phosphate and might fulfill the same role. In addition, Gln665 in hUpf1hd appears to act as a ‘γ-phosphate sensor', similar to Gln194 in RecA, which has been proposed to detect the presence or absence of the γ-phosphate of ATP and relay the information to other sites within the molecules through conformational changes (Story and Steitz, 1992).
Figure 2.
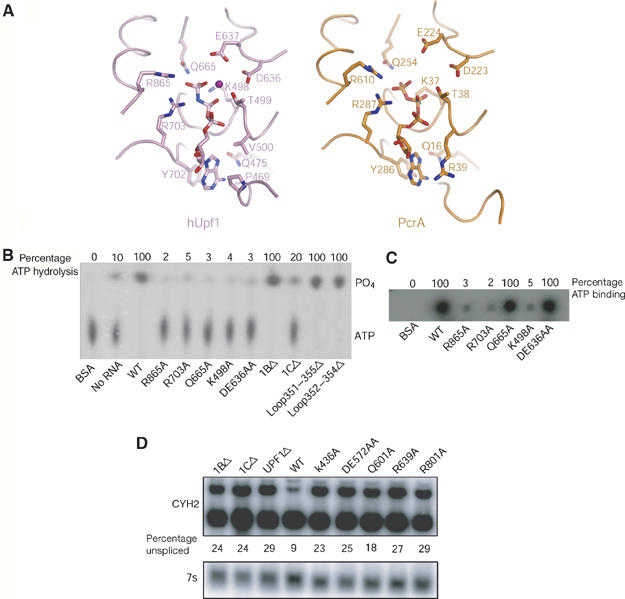
Nucleotide binding and hydrolysis of hUpf1hd. (A) Comparison of the nucleotide-binding sites of hUpf1hd-AMPPNP and PcrA. hUpf1hd-AMPPNP and PcrA in the PcrA/ADPNP/dsDNA complex are colored in pink and orange, respectively. (B) ATPase activities of WT and mutant hUpf1hd proteins (200 nM) in the presence of 500 nM 15-mer poly(C). hUpf1hd showed very weak intrinsic ATPase activity without RNA. (C) ATP-binding activities of WT and mutant hUpf1hd proteins (15 μg). (D) Northern analysis of the yeast CYH2 mRNA from strains expressing UPF1 mutant plasmids. mRNA from a UPF1 deletion strain containing plasmids expressing deletions (1BΔ or 1CΔ), empty vector (UPF1Δ), WT, or point mutations as listed above each lane. The upper panel shows the unspliced precursor (upper band) and spliced CYH2 mRNA (lower band). The % of unspliced mRNA for each strain is given under each lane. The lower panel shows the 7S RNA used for standardization.
Full-length hUpf1 (hUpf1-FL) has been shown to have RNA-dependent ATPase activity (Bhattacharya et al, 2000). In order to find out which residues are critical for ATP hydrolysis, we mutated several residues in the AMPPNP-binding pocket to alanine and examined their effects on RNA-dependent ATPase activity. We observed that hUpf1hd exhibited very low intrinsic ATPase activity (Figure 2B), and poly(C) stimulated this activity to the greatest extent (data not shown). Consistent with previous results of both yeast and hUpf1p, both K498A and DE636AA were strongly defective for ATPase activity (Weng et al, 1996; Bhattacharya et al, 2000; Figures 2B), but only K498A was defective in ATP binding (Figure 2C). In addition, mutants R865A and R703A are defective in both ATP binding and hydrolysis, whereas mutation of Gln665 to Ala nearly eliminated the ATPase activity but had no effect on ATP binding (Figure 2B and C). This result is similar to the observation that mutation of these three residues in PcrA (Gln254, Arg287 and Arg610) to Ala impaired ATPase activity (Velankar et al, 1999). In line with the observation that mutation R865A abolished the ATPase activity of hUpf1hd, the TR800AA form of yeast Upf1p, in which both Thr800 and Arg801 (Arg865 in hUpf1) were mutated to Ala, was defective in RNA-dependent ATPase activity (Weng et al, 1996). These data suggest that the SF1 helicases share a similar ATP-binding and -hydrolysis mechanism irrespective of their DNA or RNA substrates.
To test if these residues were required for Upf1p function in vivo, we took advantage of the fact that Upf1p is a conserved protein with a similar role in NMD from yeast to mammals. Thus, we mutated the corresponding amino acids in S. cerevisiae Upf1p and examined their effect on NMD in yeast cells by examining the steady-state levels of the CYH2 pre-mRNA, which is normally rapidly degraded and thereby reduced in level by NMD (He et al, 1993). Replacement with alanine of Gln601, Arg639 and Arg801, which are analogous to Gln665, Arg703 and Arg865 in hUpf1, respectively, prevented the function of Upf1p in NMD and led to the accumulation of CYH2 pre-mRNA (Figure 2D). This observation indicates that these residues are also required for Upf1p function in vivo.
Conformational change during the Upf1 ATPase cycle
The coupling of conformational changes to NTP binding and hydrolysis is a general mechanism by which helicases and motor proteins catalyze physical transitions. To map out the conformational changes during the ATPase cycle of hUpf1p, we compared the hUpf1hd structures in AMPPNP-, ADP- and phosphate-bound forms. Structural comparison reveals that all four individual domains are similar to one another among the three crystal structures (phosphate-, ADP- and AMPPNP-bound form) with mean pairwise Cα r.m.s.d. ∼0.5 Å. Despite these similarities of the individual domains, the relative orientation of domain 2A with respect to domain 1A differs significantly among all three states of hUpf1hd. When the corresponding domains of hUpf1hd-ADP and hUpf1hd-AMPPNP are compared to each other, the orientations of domain 2A differ by ∼20° (Figure 3A). Similarly, when the corresponding domains of hUpf1hd-AMPPNP and hUpf1hd-phosphate are compared to each other, the orientations of domain 2A differ by ∼10° (Figure 3B).
Figure 3.
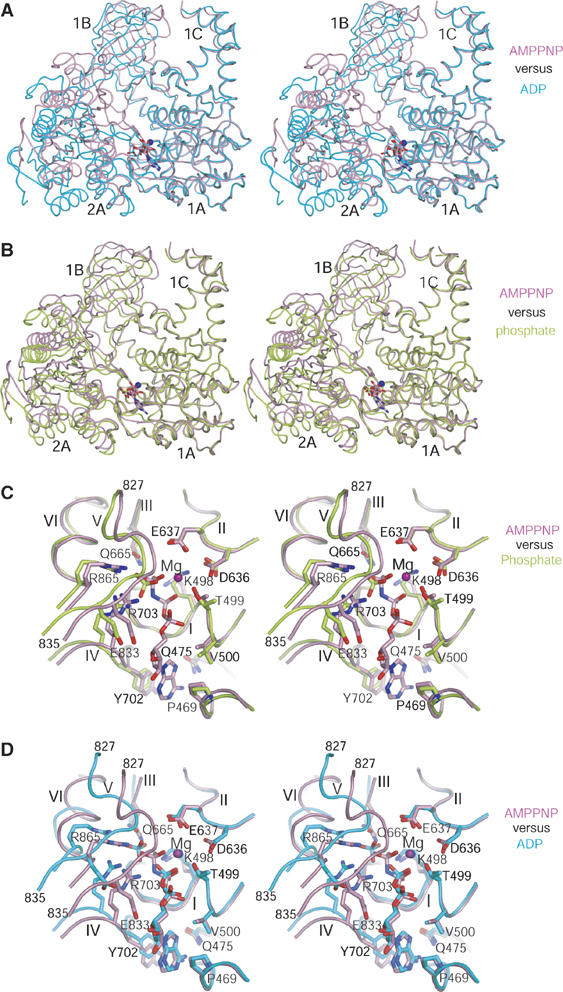
Conformational changes of hUpf1hd upon nucleotide binding and hydrolysis. (A) Stereo view of superposition of domain 1A of hUpf1hd-AMPPNP with that of hUpf1hd-ADP. (B) Stereo view of superposition of domain 1A of hUpf1hd-AMPPNP with that of hUpf1hd-phosphate. (C) Stereo view of superposition of the nucleotide-binding sites of hUpf1hd-phosphate and hUpf1hd-AMPPNP. (D) Stereo view of superposition of the nucleotide-binding sites of hUpf1hd-AMPPNP and hUpf1hd-ADP. The phosphate-, AMPPNP- and ADP-bound forms of hUpf1hd are shown in limon, pink and cyan, respectively. AMPPNP, ADP, phosphate ion and the residues involved in interaction with ligands or Mg2+ coordination are shown in stick models. Mg2+ ion is shown as a purple sphere.
Inspection of the structures shows that the large-scale rigid-body movement of domain 2A relative to domain 1A is caused by closure of the cleft between domains 1A and 2A induced by interactions with the γ-phosphate of AMPPNP. In the phosphate-bound structure, a phosphate ion in the nucleotide-binding pocket occupies the position equivalent to the γ-phosphate of AMPPNP and interacts with Gln665, Arg703 and Arg865 (Figure 3C). These interactions thereby contribute to the phosphate-bound structure being more similar to the AMPPNP-bound structure.
Comparison of the phosphate and AMPPNP-bound states illustrates that the presence of the nucleotide and/or γ-phosphate stabilizes a network of interactions between domains 1A and 2A. Upon AMPPNP binding, motif V (residues 827–835) in domain 2A is positioned ∼2 Å closer to domain 1A, thus narrowing the nucleotide-binding cleft between domains 1A and 2A (Figure 3C). Specifically, residue Glu833 undergoes displacement of 1.7 Å to interact with the O3 group of the ribose ring of the bound nucleotide. Moreover, Arg703 and Arg865 are displaced 1.6 and 1.5 Å, respectively, to interact with the γ-phosphate (Figure 3C). The position of Gln665, a residue acting as a γ-phosphate sensor, is unchanged compared to its corresponding position in the phosphate state (Figure 3C). The closure of the cleft between domains 1A and 2A (Figure 3B and C) and the direct interactions between residues acting as ‘arginine finger' or ‘γ-phosphate sensor' with the γ-phosphate (Figure 2A) suggest that the AMPPNP-bound complex represents the substrate-bound state. In this manner, AMPPNP appears to act as a crosslinking bridge inducing cleft closure, thus bringing domains 1A and 2A into close proximity. A similar role for AMPPNP in cleft closure between domains 1A and 2A has been observed in PcrA (Velankar et al, 1999) with ∼15° rotation of domain 2A relative to domain 1A when the ADPNP- and sulfate-bound forms are compared.
Comparison of the AMPPNP and ADP structures of hUpf1hd reveals that conformational changes would occur with nucleotide hydrolysis. In the ADP state, the interactions of the adenine base, ribose ring and the β-phosphate group with the protein residues are the same as those observed in the AMPPNP state (Figure 3D). Despite these similarities, motif V in the ADP state is displaced 4 Å from its corresponding position in the AMPPNP state (Figure 3D). Consequently, residues Gln665, Arg703 and Arg865 are displaced 3.1, 2.6 and 4.5 Å, respectively, from their respective positions in the AMPPNP state. Such structural changes are most likely induced by the absence of the γ-phosphate group, as these residues make direct contacts with the γ-phosphate group in the AMPPNP-bound structure (Figure 3D). In agreement with this view, extensive analysis of crystal packing showed that the closure and opening of the cleft between domains 1A and 2A are not caused by crystal packing, as the interface of domains 1A and 2A is distant from any symmetry-related molecules in all three structures. Consistent with the importance of the γ-phosphate group, the presence of a phosphate in the same position in the phosphate-bound structure is sufficient, even in the absence of a nucleotide, to partially organize the cleft between the two RecA-like domains, thereby narrowing down the cleft between them. In the ADP state, in addition to the bound ADP, a free phosphate is located 4.8 Å from the β-phosphate of ADP, which is not equivalent to the γ-phosphate group of AMPPNP (Figure 3D). The opening of the nucleotide-binding pocket and the coexistence of ADP with a free phosphate in the nucleotide-binding pocket suggest that the ADP-bound form represents the product-bound state after ATP hydrolysis.
In addition to the changes in the cleft between domains 1A and 2A in the different structures, there are significant movements of domains 1B and 1C that alter their positions relative to each other. In the AMPPNP state, the close packing of domain 2A with domain 1A results in extensive interactions between domain 2A and domain 1B with a buried solvent-accessible surface of 1215.8 Å2. These interactions may stabilize the closed conformation of domains 1A and 2A, and also displace domain 1B toward domain 1C, thereby narrowing the large channel between them by 2.4 Å (Cα–Cα distance between Arg396 and Arg596) as compared to the phosphate-bound structure (Figure 3B). Upon ATP hydrolysis (the ADP state), the opening of the cleft between domains 1A and 2A leads to domain 2A making fewer contacts with domain 1B with a buried solvent-accessible surface of 310.5 Å2, thereby widening the channel between domains 1B and 2A. Consequently, the channel between domains 1B and 1C in the ADP state is widened by 4.7 Å (Cα–Cα distance between Arg396 and Arg596) compared to that in the AMPPNP state (Figure 3A). These differences suggest that ATP hydrolysis will lead to significant conformational changes in domains 1B and 1C, which may have important mechanistic roles in the catalytic mechanism of hUpf1 (see below).
Allosteric effect of ATP binding coupled with RNA binding
The Upf1 protein has been shown to bind ssRNA in a manner modulated by ATP (Czaplinski et al, 1995; Weng et al, 1996, 1998). Mutagenesis combined with biochemical characterization demonstrated that ATP binding rather than hydrolysis promoted dissociation of the Upf1–RNA complex (Weng et al, 1996). However, the mechanism by which ATP modulates the RNA-binding activity remains unclear.
In order to identify the RNA-binding channel in Upf1p, we took advantage of the fact that the two RecA-like domains are very similar among hUpf1hd, PcrA and Vasa (see above; Figure 1). The ssDNA of PcrA overlays very well with the ssRNA of Vasa and shares the same sequence polarity: 5′-to-3′ direction from domains 2A to 1A. Based on these observations, we reasoned that the ssRNA would bind the analogous site in hUpf1, given the high structural similarity among these enzymes. Superposition of domain 1A in either PcrA or Vasa with that of hUpf1hd results in a model in which the binding of ssRNA to hUpf1hd involves domains 1A, 2A and 1B, with the 3′ end pointing to the channel between domains 1B and 1C (Figure 4A and B).
Figure 4.

The channel between domains 1B and 1C is a potential ssRNA-binding site. (A) Distribution of the basic residues in the channel between domains 1B and 1C in hUpf1hd-AMPPNP. Superimposed ssDNA from the PcrA/ADPNP/dsDNA complex and ssRNA from the Vasa/AMPPNP/ssRNA complex are shown in orange and magenta cartoon tubes, respectively, based on the superposition of domains 1A and 2A among hUpf1hd, PcrA and Vasa. (B) Distribution of the basic residues in the channel between domains 1B and 1C in hUpf1hd-ADP. Nucleic acids are generated and shown as in (A). (C) Solvent-accessible surface and electrostatic potentials of hUpf1hd-AMPPNP, hUpf1hd-phosphate and hUpf1hd-ADP. Residues involved in RNA binding are labeled.
As a second method to identify the RNA-binding site and examine how RNA binding relates to the ATPase cycle of Upf1, we also mapped the electrostatic potential on the molecular surface of hUpf1hd in the three structures. The electrostatic potential mapping showed that in both phosphate- and ADP-bound states, a positively charged patch consisting of Arg549 of domain 1A, and Lys599, Arg600, Arg604 of domain 1C, is in close proximity to the outlet of the large channel between domains 1B and 1C (Figure 4C). In the AMPPNP state, this patch is extended to domain 1B owing to nearly complete closure of the channel between domains 1B and 1C (Figure 4C). Notably, the channel between domains 1B and 1C is rich in conserved basic residues (Figure 4A and B), suggesting that domain 1C and this channel may be involved in RNA binding.
To examine biochemically the importance of domain 1C in RNA binding, a variant lacking domain 1C (1CΔ) was expressed in E. coli and purified. Gel filtration showed that 1CΔ behaved similar to the wild-type (WT) protein in solution (Supplementary Figure S2), indicating that removal of domain 1C had no significant effect on the general folding or stability of the hUpf1 protein. Filter ssRNA-binding assays and surface plasmon resonance (SPR) showed that deletion of domain 1C abolished the RNA-binding activity of hUpf1hd (Figure 5A and D). Further evidence that domain 1C is involved in ssRNA binding is that point mutations in this domain reduce RNA binding. Specifically, substitution of both Lys599 and Arg600 to Ala or R604A reduced the RNA binding to hUpf1hd substantially (Figure 5A). These observations argue that domain 1C is essential for RNA binding, which also explains why the RNA-dependent ATPase activity of mutant 1CΔ is substantially reduced (Figure 2B). Moreover, consistent with domain 1C having an important functional role, in vivo a 1CΔ mutant of yeast Upf1p was expressed close to WT levels, but was completely defective in NMD (Figure 2D and Supplementary Figure S3).
Figure 5.

ssRNA-binding activity of WT and mutant hUpf1hd proteins in the presence or absence of nucleotide. (A) Upper panel: filter RNA-binding assay showing the ssRNA-binding activity of WT and mutant hUpf1hd proteins in the absence of ATP. Lower panel: SDS–PAGE gel showing the levels of proteins used in the RNA-binding assay. (B) Sensorgrams of 100 nM WT hUpf1hd binding to ssRNA in the absence or presence of 2 mM ligands (ATP, AMPPNP, ATPγS and ADP). (C) Sensorgrams of 100 nM mutant 1BΔ binding to ssRNA in the absence or presence of 2 mM ATP. (D) Sensorgrams of 100 nM mutant 1CΔ binding to ssRNA in the absence or presence of 2 mM ATP. (E) Sensorgrams of 100 nM mutant loop (Δ351–355) binding to ssRNA in the absence or presence of 2 mM ATP. (F) Sensorgrams of 100 nM mutant loop (Δ352–354) binding to ssRNA in the absence or presence of 2 mM ATP.
An important aspect of Upf1p is how the conformational changes present in the different structures affect the possible RNA-binding channel. We noticed two features, namely the ordering of the loop 349–355 and the closure and opening of the channel between domains 1B and 1C, that could modulate RNA binding. A small loop (residues 349–355) from domain 1B in the AMPPNP state clashes with the modeled ssRNA (Figure 6A), whereas this loop is disordered in the ADP state (Figure 6B). In the phosphate state, although the tip of this loop (residues 352–354) is disordered, most part of this small loop is ordered, which would be involved in steric clashes with ssRNA. Another conformational change altered by nucleotide binding that may affect RNA binding is the status of the channel formed between domains 1B and 1C. In the ADP state, the channel is wide open with a width of ∼10 Å, which would be big enough to accommodate the 3′ end of ssRNA (Figure 4B and C). In contrast, the channel is narrowed down to ∼3 and ∼5 Å for the AMPPNP and phosphate states, respectively, thereby potentially blocking the ssRNA binding (Figure 4A and C).
Figure 6.

Allosteric effects of ATP binding/hydrolysis on RNA binding. (A) The figure depicts the potential location of the ssRNA in hUpf1hd-AMPPNP. Left: hUpf1hd-AMPPNP is shown in ribbon diagram (skyblue) covered with transparency surface. Nucleic acids are generated and shown as in Figure 4A. The loop (349–355) region in domain 1B is shown in red. Right: close-up view of the zoomed region in left. The loop (758–763) and helix α15 are shown in green and yellow, respectively. (B) The potential location of ssRNA in hUpf1hd-ADP. Left: ribbon diagram of ADP-hUpf1hd (slate) covered with transparency surface. Nucleic acids are generated and shown as in Figure 4B. Right: close-up view of the zoomed region in left. The disordered loop (349–355) region is shown in a dotted red line. The color coding for loop (758–763) and helix α15 is as in (A).
Close inspection of the structures of hUpf1hd in three states shows that both the closure of the channel between domains 1B and 1C and the potential blocking of the ssRNA binding by the loop (residues 349–355) are caused by the allosteric effects of nucleotide binding. Specifically, conformational changes induced by ATP binding and hydrolysis are relayed to the RNA-binding site through residues in motif VI. Upon AMPPNP binding, the presence of γ-phosphate in the nucleotide-binding pocket is sensed by the residues in motif VI (Figure 3C), leading to the rigid-body movement of domain 2A toward domain 1A (Figure 3A), which narrows the nucleotide-binding cleft. This large domain movement also leads to helix α15 and the loop (residues 758–763) linking β17 and β18 in domain 2A to make direct contacts with the loop 349–355 of domain 1B (Figure 6A), thereby stabilizing the 349–355 loop and pushing domain 1B toward domain 1C to close the channel between them. In the ADP state, as a consequence of ATP hydrolysis, domain 2A moves away from domain 1A (Figure 3A). Consequently, domain 2A makes very few contacts with domain 1B, resulting in the disorder of loop 349–355 and opening of the channel between domains 1B and 1C (Figure 6B). Thus, these interactions suggest a model wherein the decreased affinity for RNA by Upf1p in the presence of ATP is due to the ordering of the 349–355 loop and closing of the channel between 1B and 1C.
As an initial test of the importance of domain 1B and the 349–355 loop in coupling ATP binding to decreased RNA affinity, we tested the RNA binding with and without ATP of hUpf1hd variants either lacking the entire domain 1B (1BΔ), the entire small loop (Δ351–355) or just the tip of the small loop (Δ352–354). Strikingly, deletion of domain 1B still allowed substantial RNA binding, but, more importantly also slowed down dramatically the dissociation of the hUpf1hd–RNA complex and rendered hUpf1hd insensitive to the effect of ATP binding (Figure 5A and C). Deletion of residues 351–355 or 352–354 in loop 349–355 also did not reduce the binding of ssRNA but made the binding insensitive to ATP binding (Figure 5E and F). Taken together, these results indicate that domain 1B is critical for coupling the binding of ATP to the decreased affinity for RNA. Consistent with an important functional role for domain 1B, in vivo a 1BΔ mutant of yeast Upf1p was completely defective in NMD (Figure 2D and Supplementary Figure S3). However, because the levels of the 1BΔ variant were reduced compared to WT Upf1p, it is possible that the defect in NMD is a combination of the loss of domain 1B and a reduction in Upf1p levels.
The structural changes induced by AMPPNP binding provide a good explanation for the dissociation of the Upf1–RNA complex by ATP binding rather than its hydrolysis, and correlate well with the previous biochemical data (Weng et al, 1996, 1998). In this regard, the AMPPNP state we have observed likely mimics the ATP-binding state. Consistent with this view, SPR assay showed that AMPPNP, ATPγS and ATP reduced the ssRNA binding (association) to hUpf1hd to a similar extent with similar rates (Figure 5B). Interestingly, it seems that ATPγS dissociates RNA much slower than ATP with AMPPNP in between (Figure 5B). The small amount of rapid dissociation by ATPγS at ∼180 s is probably due to free proteins not forming a tight complex with RNA. These results are consistent with the observation that a higher concentration of ATPγS is required to dissociate the Upf1p–RNA complex, and in contrast to the finding that AMPPNP did not promote the dissociation of the Upf1p–RNA complex (Weng et al, 1998). We also note that the SPR assay showed that the binding of ssRNA to hUpf1hd is stronger in the presence of ADP than in the presence of ATP, but is weaker than in the ligand-free state whereby the channel between domains 1B and 1C is conceived to be even wider than the ADP state (Figure 5B).
Differential effects of Upf1p mutants on P-body formation
The above results begin to define a cycle of conformational changes in Upf1p that occur during the process of NMD and are likely to be coupled to and influenced by other protein–protein and protein–RNA interactions. The process of NMD involves the formation of an mRNP that can accumulate within cytoplasmic processing bodies (P-bodies), with Upf1p being required for targeting of aberrant mRNAs to P-bodies, whereas the ATPase activity of Upf1p is required at a later stage of NMD to allow mRNA decapping and degradation (Sheth and Parker, 2006). Thus, the accumulation of P-bodies identifies an intermediate stage in the NMD pathway and thereby can be used to map specific aspects of Upf1p function to upstream or downstream of the formation of mRNP that can accumulate within P-bodies. Given this, we examined how different mutations in Upf1p affected the formation of yeast P-bodies as assessed by the presence of a Dcp2p-GFP reporter protein.
These experiments revealed that mutations (specifically K436A, DE572AA, Q601A and R639A, corresponding to K498A, DE636AA, Q665A and R703A in hUpf1, respectively) in Upf1p that inhibit the ATPase activity of Upf1p (Figure 2B), but still allow RNA binding (Figure 5A), all led to the accumulation of large P-bodies (Figure 7). This is consistent with earlier results with the DE572AA allele and suggests that the ATPase activity of Upf1p functions after the formation of a translationally repressed mRNP, which can accumulate in P-bodies (Sheth and Parker, 2006). In contrast, deletion of domain 1B or 1C, or mutation of Arg801 (Arg865 in hUpf1) to Ala, does not cause enhanced P-body accumulation, even though they inhibited the process of NMD, as assessed by mRNA levels (Figure 2D). These results reveal allele-specific effects on the Upf1p-dependent accumulation of P-bodies. Most notably, the absence of P-body accumulation in the 1CΔ suggests that RNA binding is required either early in the process of NMD before formation of the mRNP that can accumulate and form P-bodies following ATP hydrolysis by Upf1p for stable mRNA accumulation in P-bodies. Interestingly, when the 1CΔ was combined with the DE572AA allele, which blocks the ATPase and leads to the accumulation of P-bodies, we observed an accumulation of P-bodies similar to what was seen in the DE572AA strain. This indicates that the function of domain 1C is not required to form P-bodies and implies that RNA binding to Upf1p occurs after targeting of RNA to P-bodies. This is consistent with a model of Upf1p function wherein RNA binding is important at a late stage of the process following ATP hydrolysis (see Discussion).
Figure 7.

Visualization of DCP2GFP in live yeast strains expressing UPF1 mutants. The panels show microscopic accumulation of a Dcp2p-GFP fusion protein as a marker for P-bodies. Cells are upf1Δ strain expressing either the WT UPF1 protein or specific mutant proteins of UPF1 as designated above each panel.
Discussion
In this work, we have described the structure of the helicase core domain of hUpf1. The structure of hUpf1hd reveals two ‘RecA-like' domains with the relative orientations of these two domains similar to those observed in PcrA, Vasa and Rep helicases. One striking structural feature of hUpf1hd is the existence of two subdomains 1B and 1C in domain 1, which form structural entities above domains 1A and 2A (Figure 1). Subsequent analysis suggests both domains 1B and 1C have important functional roles in NMD. Domain 1C is required for RNA binding and maximal ATP hydrolysis rates in vitro (Figures 2 and 5) and for Upf1p function in NMD in vivo (Figure 2D). Domain 1B is also required for NMD in vivo (Figure 2D). Moreover, domain 1B appears to play a role in modulating the effects of ATP binding to Upf1p on RNA affinity because deletion of domain 1B or smaller deletions within the domain 1B 349–355 loop allow RNA binding to be relatively insensitive to ATP (Figure 5). Interestingly, structural comparisons showed that domains 1B and 1C of hUpf1hd undergo conformational changes due to the closure and opening of the cleft between domains 1A and 2A induced by nucleotide binding and/or hydrolysis.
A common theme in SF1 helicases is that nucleotide cofactors allosterically regulate nucleic acid binding (Korolev et al, 1997; Velankar et al, 1999). However, the details of the underlying mechanisms for hUpf1, PcrA and Rep appear to be somewhat different. In Rep helicase, ADP favors the binding of ssDNA whereas AMPPNP favors simultaneous binding of duplex DNA and ssDNA (Wong et al, 1992). Moreover, the Rep helicase crystal structure showed that the allosteric effect of ADP binding is transduced to the ssDNA-binding site through motif II (Korolev et al, 1997). The conformational change induced by ADPNP binding in PcrA also favors duplex DNA binding, although motif VI rather than motif II appears to mediate the transduction of nucleotide-induced conformational change to the DNA-binding site (Velankar et al, 1999). For Upf1p, biochemical data showed that ATP binding rather than its hydrolysis reduced the affinity for ssRNA (Weng et al, 1998). These differences illustrate how the basic and flexible module of two RecA domains can be coupled to ATP-dependent changes in affinity for different nucleic acids.
The structures of hUpf1hd presented here suggest a mechanistic model for how ATP binding and hydrolysis alter the affinity of Upf1p for RNA. In brief, the binding of ATP to Upf1p would stabilize a closed conformation of domains 1A and 2A. This closed conformation would reduce the width of the channel between domains 1B and 1C, and stabilize the loop 349–355, which would also sterically hinder the Upf1p–RNA interaction. These ATP-induced conformational changes would reduce but not abolish the binding of ssRNA to Upf1, consistent with our SPR data (Figure 5B) and previous biochemical data by Weng et al (1998). Following ATP hydrolysis, the movement of the released phosphate to the position seen in the ADP-bound structure would disrupt interactions with the γ-phosphate and thereby destabilize the network of interactions holding domains 1A and 2A in close proximity. The resulting opening of the cleft between domains 1A and 2A and the disordering of loop 349–355 would thereby allow enhanced RNA binding.
An unresolved issue is how RNA enhances the ATPase activity of Upf1p. One possibility is that transient binding of RNA to the ATP-bound Upf1p triggers ATPase, which could then lead to tighter binding of that RNA to Upf1p complexed with ADP. Alternatively, RNA might stimulate the ATPase activity by keeping the cleft between domains 1A and 2A open, thereby stimulating the ATPase activity by facilitating the release of ADP and free phosphate.
An interesting issue is how ATP hydrolysis by Upf1p and changes in RNA binding are coupled with other events in the pathway of NMD. We have used the accumulation of P-bodies as a marker for an intermediate step in the pathway of NMD to reveal the order of events in NMD. Our results indicate that any mutation that blocks Upf1p ATPase activity leads to increased accumulation of P-bodies, which suggests that ATP hydrolysis by Upf1p is required after targeting of mRNAs to P-bodies. Our results also argue that RNA binding is only required after targeting of RNAs to P-bodies, and are consistent with a model of NMD wherein Upf1p promotes the targeting of mRNA to P-bodies, followed by ATP hydrolysis, leading to enhanced RNA binding and ultimately mRNA decapping. An important goal of future work will be to understand how other protein–protein interactions occurring during NMD either modulate, or are affected by, this cycle of ATP hydrolysis and conformational changes.
Materials and methods
Protein expression and purification
The cDNA encoding the helicase domain of hUpf1 (hUpf1hd, residues 295–914) was cloned into the pGEX-6P-1 vector (Amersham) and expressed as GST-fusion proteins in E. coli. The fusion protein was purified by glutathione affinity column. The GST was released by PreScission protease cleavage and the hUpf1hd fragment was further purified by MonoS and Superdex-200 gel filtration columns (Amersham). The protein was concentrated to ∼10 mg/ml for crystallization. SeMet-substituted hUpf1hd was expressed in a minimal medium containing 20 mg/l SeMet, and purified in the same way as the native protein.
Structure determination
Crystallization was performed for Se-Met-substituted hUpf1hd using hanging-drop vapor diffusion method at 20°C. hUpf1hd-phospahte was crystallized using a buffer containing 100 mM Na/K phosphate, pH 5.8, 6–8% PEG3350 and 10 mM DTT. For co-crystallization with AMPPNP and ADP, 5 mM MgCl2 and 5 mM nucleotide were added to the protein sample and incubated for 30 min. hUpf1hd-AMPPNP was crystallized from a buffer containing 50 mM MES, pH 6.0, 0.5–3% PEG4000, 0.005 mM MgSO4 and 10 mM DTT. Crystals of hUpf1hd-ADP were obtained from a buffer containing 100 mM Tris, pH7.5, 8–13% PEG3350, 140 mM (NH4)3PO4, 10% ethylene glycol and 10 mM DTT. Phosphate- and AMPPNP-bound crystals belong to space group P43212 with one molecule per asymmetric unit (AU). ADP-bound crystals belong to space group P1 with two molecules in the AU. SAD data were collected at ID29, ESRF (Grenoble, France) and processed with the CCP4 package (CCP4, 1994).
SAD phasing was carried out using the data collected from the AMPPNP-bound crystal. Thirteen Se sites were located by using the program SnB (Miller et al, 1994). Refinement of the heavy atom sites and phasing were carried out using SHARP (De la Fortelle and Bricogne, 1997). After density modification and solvent flattening, 60% of the final model was built automatically by RESOLVE (Terwilliger, 2002). The rest of the model was built manually with the program O (Jones et al, 1991). The structure of hUpf1hd-ADP was solved by molecular replacement with Molrep (CCP4, 1994) using the structure of hUpf1hd-AMPPNP as a search model. The structure of hUpf1hd-phosphate was solved by SAD phasing combined with molecular replacement. Crystallographic refinement was performed with the programs CNS (Brunger et al, 1998) and REFMAC5 (Murshudov et al, 1997). Ramachandran plot showed that no outlier is found for hUpf1hd-ADP whereas Leu370 and Glu573, two residues distant from either active site or the disordered regions, are located in the disallowed regions for AMPPNP and phosphate forms, respectively. Statistics for data collection and refinement are summarized in Table I.
Mutagenesis and in vitro ATP binding and ATPase activity
Alleles of hUpf1hd used in the in vitro analysis were created using Quicksite-mutagenesis (Stratagene). Specific point mutants were generated and verified by sequence analysis. ATPase assay was performed in a 20 μl reaction containing the purified 200 nM WT or mutant hUpf1 proteins in 50 mM MES (pH 6.0), 50 mM KAc, 2.5 mM Mg(Ac)2, 1 mM DTT, 500 nM 15-mer poly(C), 0.1 mg/ml BSA, 10 μM cold ATP and 1 μCi [γ-32P]ATP. After incubation for 1 h at 37°C, the reaction was terminated by adding 1 μl 500 mM EDTA. For TLC analysis, 1 μl of the reaction mixture was spotted on a PEI-cellulose plate and developed by 0.3 M K2HPO4 (pH 7.6).
For ATP-binding assay, equal amounts (15 μg) of BSA, WT and mutant hUpf1hd proteins were dot-blotted on PVDF membrane. The membrane was blocked in buffer A (20 mM HEPES, 50 mM KAc, 2.5 mM Mg(Ac)2, 2 mM DTT, 3% BSA, pH 7.0) for 1 h at room temperature. Then, 50 μCi of 5000 Ci/mmol [γ-32P]ATP (Amersham) was added for the binding assay in buffer B (20 mM HEPES, 50 mM KAc, 2.5 mM Mg(Ac)2, 2 mM DTT, 1.5% BSA, pH 7.0). After incubation in buffer B for 30 min, the membrane was washed with 20 ml buffer B twice and quantified using a phosphorimager (Molecular Dynamics). BSA protein serves as a negative control.
In vitro RNA-binding assay
For filter RNA-binding assay, equal amounts (15 μg) of BSA, WT and mutant hUpf1hd proteins were dot-blotted on PVDF membrane. The membrane was blocked in buffer A (20 mM HEPES, 50 mM KAc, 2.5 mM Mg(Ac)2, 2 mM DTT, 3% BSA, pH 7.0) for 1 h at room temperature. ssRNA (5′CGCCCGAGGCTGTGCCGT3′) was purchased from Dharmacon. RNA substrates were labeled with T4 kinase (Invitrogen) in the presence of [γ-32P]ATP (Amersham). A 1 pmol portion of 32P-labeled RNA substrate was added for the binding assay in buffer B (20 mM HEPES, 50 mM KAc, 2.5 mM Mg(Ac)2, 2 mM DTT, 1.5% BSA, pH 7.0). After incubation in buffer B for 1 h, the membrane was washed with 20 ml buffer B three times and quantified using a phosphorimager (Molecular Dynamics). BSA protein serves as a negative control.
SPR was carried out on a Biacore 2000 instrument. The 5′ end biotin-labeled ssRNA (TGTCATTCGAGTACAGTCTGTTCAGCTAGTCTCC) purchased from CureVAc was attached to a streptavidin-coated sensor chip (Biacore). A buffer of 10 mM HEPES, pH 7.0, 150 mM NaCl and 0.005% (v/v) Tween 20 was flowed across the chip until the trace leveled off. The biotin-labeled RNA was then attached to the flow cell 2 by injecting 20 μl of 100 nM RNA in 0.3 M NaCl at flow rate 5 μl/min. After immobilization, flow cell 2 and reference flow cell 1 were blocked with 100 μl of 1 mg/ml biotin at 5 μl/min. A binding buffer of 20 mM HEPES, pH 7.0, 100 mM NaCl, 2 mM MgCl2, 2 mM DTT and 0.002% (v/v) Tween 20 was flowed across flow cells 1 and 2. Typically, 150 μl of 100 nM protein sample was injected into the same buffer across the chip at 50 μl/min. The data were fitted to a 1:1 binding mode mass transfer using BIAevaluation 3.1.
In vivo NMD analysis and P-body formation
The UPF1 plasmids used were on a CEN LEU2 vector: WT, pRP910; DE572AA, pRP912. Additional UPF1 point mutants were created using the Quick Change method (Stratagene) on a UPF1 BglII fragment cloned into pBluescript (pRP1318). Mutations were verified by sequencing and then put back into pRP910. Plasmids created were pRP1319 (Δ261–351), pRP1320 (Δ494–546), pRP1321 (K436A), pRP1324 (Q601A), pRP1325 (R639A) and pRP1329 (R801A). These mutants were analyzed in the yeast strain yRP1864: MATa, ura3-52, his4-539, leu2, lys2, trp1-1, UPF1∷URA3, DCP2GFPNEO. Strains containing the UPF1 plasmids were grown in minimal selective media to mid-log and either harvested for RNA, or visualized microscopically as previously described (Sheth and Parker, 2006). Northern blots were probed either for the 7S RNA (oRP100) or the CYH2 mRNA (oRP1299 and/or 1300).
Coordinates
The coordinates and structure-factor amplitudes for hUpf1hd-AMPPNP, hUpf1hd-ADP and hUpf1hd-phosphate have been deposited in the Protein Data Bank with accession codes 2GJK, 2GK6 and 2GK7, respectively.
Supplementary Material
Figure S1
Figure S2
Figure S3
Acknowledgments
We thank the beamline scientists at ID29 (ESRF, France) for assistance and access to synchrotron radiation facilities. We are grateful to Drs Lynne Maquat and Jens Lykke-Andersen for supplying the hUpf1 cDNA. This work was financially supported by the Agency for Science, Technology and Research (A*Star) in Singapore (HS) and by the Howard Hughes Medical Institute (RP).
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Applequist SE, Selg M, Raman C, Jack HM (1997) Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res 25: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KE, Parker R (2004) Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 16: 293–299 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT (2004) The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell 14: 585–598 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Caruthers JM, McKay DB (2002) Helicase structure and mechanism. Curr Opin Struct Biol 12: 123–133 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E (2005) Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 17: 316–325 [DOI] [PubMed] [Google Scholar]
- Cui Y, Hagan KW, Zhang S, Peltz SW (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev 9: 423–436 [DOI] [PubMed] [Google Scholar]
- Culbertson MR, Leeds PF (2003) Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev 13: 207–214 [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW (1995) Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA 1: 610–623 [PMC free article] [PubMed] [Google Scholar]
- De la Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction method. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E (2003) Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 22: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A (1997) Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol 17: 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A (2003) Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A (1993) Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc Natl Acad Sci USA 90: 7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P (1989) A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S (2006) Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF (2005) Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800 [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE (2005) Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G (1997) Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90: 635–647 [DOI] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE (2005) Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol 17: 309–315 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Mendell JT, ap Rhys CM, Dietz HC (2002) Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422 [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC (2004) Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Miller R, Gallo SM, Khalak HG, Weeks CM (1994) SnB: crystal structure determination viashake-and-bake. J Appl Crystallogr 27: 613–621 [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S (2006) Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125: 287–300 [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE (2001) Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21: 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2006) Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P, Wigley DB (2001) Unwinding the ‘Gordian knot' of helicase action. Trends Biochem Sci 26: 47–54 [DOI] [PubMed] [Google Scholar]
- Story RM, Steitz TA (1992) Structure of the recA protein-ADP complex. Nature 355: 374–376 [DOI] [PubMed] [Google Scholar]
- Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8: 251–262 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2002) Automated structure solution, density modification and model building. Acta Crystallogr D 58: 1937–1940 [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97: 75–84 [DOI] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW (1996) Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW (1998) ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4: 205–214 [PMC free article] [PubMed] [Google Scholar]
- Wong I, Chao KL, Bujalowski W, Lohman TM (1992) DNA-induced dimerization of the Escherichia coli Rep helicase. Allosteric effects of single-stranded and duplex DNA. J Biol Chem 267: 7596–7610 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3


