Abstract
Upon DNA damage, a complex called the PIDDosome is formed and either signals NF-κB activation and thus cell survival or alternatively triggers caspase-2 activation and apoptosis. PIDD (p53-induced protein with a death domain) is constitutively processed giving rise to a 48-kDa N-terminal fragment containing the leucine-rich repeats (LRRs, PIDD-N) and a 51-kDa C-terminal fragment containing the death domain (DD, PIDD-C). The latter undergoes further cleavage resulting in a 37-kDa fragment (PIDD-CC). Here we show that processing occurs at S446 (generating PIDD-C) and S588 (generating PIDD-CC) by an auto-processing mechanism similar to that found in the nuclear pore protein Nup98/96 and inteins. Auto-cleavage of PIDD determines the outcome of the downstream signaling events. Whereas initially formed PIDD-C mediates the activation of NF-κB via the recruitment of RIP1 and NEMO, subsequent formation of PIDD-CC causes caspase-2 activation and thus cell death. A non-cleavable PIDD mutant is unable to translocate from the cytoplasm to the nucleus and loses both activities. In this way, auto-proteolysis of PIDD might participate in the orchestration of the DNA damage-induced life and death signaling pathways.
Keywords: auto-proteolysis, caspase-2, DNA damage, NF-κB, PIDD
Introduction
PIDD (p53-induced protein with a death domain), also known as LRDD (leucine-rich repeat and death domain-containing protein), was discovered during a bioinformatics screen for novel death domain (DD)-containing proteins using the DD of RIP1 as bait (Telliez et al, 2000). Independently, PIDD was identified as a novel p53-inducible gene in an erythroleukemia cell line expressing a temperature-sensitive p53 mutant allele, and shown to be implicated in p53-dependent apoptosis (Lin et al, 2000). PIDD contains seven leucine-rich repeats (LRRs) at the N-terminus, followed by two ZU-5 domains and a C-terminal DD.
Recent studies have shown that PIDD plays a critical role in the activation of caspase-2 (Tinel and Tschopp, 2004; Berube et al, 2005; Ren et al, 2005; Seth et al, 2005). As caspase-2 shares characteristics of both effector and initiator caspases, its activation mechanism remained enigmatic for a long time. An important breakthrough was made by showing that caspase-2 was activated in a high-molecular-weight complex, reminiscent of the apoptosome and other initiator caspase-activating platforms (Read et al, 2002). The recruitment and activation of caspase-2 within this complex is entirely dependent on the presence of an adaptor protein called RAIDD and on PIDD, which together with caspase-2 constitute the so-called PIDDosome (Tinel and Tschopp, 2004; Berube et al, 2005). These results were particularly intriguing as PIDD, just like caspase-2, had been suspected to participate in DNA damage-induced apoptosis (Robertson et al, 2002; Castedo et al, 2004; Giagkousiklidis et al, 2005; Zhivotovsky and Orrenius, 2005; Vakifahmetoglu et al, 2006).
PIDD also plays an essential role in NF-κB activation induced by DNA damage. Indeed, genotoxic stress triggers the formation of an alternative PIDDosome, consisting of PIDD, RIP1 and NEMO (Hur et al, 2003; Janssens et al, 2005). NEMO is recruited to the PIDDosome via RIP1 and appears to be modified immediately upon recruitment to the activated PIDDosome by sumoylation (Janssens et al, 2005). This is the beginning of a sequential series of modifications on NEMO, in which the nuclear protein kinase ATM plays an essential role, and which alter the protein's subcellular localization, providing a means to link a nuclear signal (DNA damage) to the activation of a cytosolic NF-κB-activating complex (the IKK complex) (Huang et al, 2003; Wu et al, 2006).
Regulation of protein activation can be achieved in many different ways. Apart from a tightly controlled transcriptional response as a DNA damage-dependent response gene, PIDD activation appears to be controlled by a second, unusual mechanism of post-translational cleavage. Full-length PIDD has a predicted molecular mass of 100 kDa, but is constitutively processed at two sites giving rise to an N-terminal fragment of 48 kDa and a C-terminal fragment of 51 kDa, which is further cleaved, resulting in a 37 kDa fragment (Telliez et al, 2000; Tinel and Tschopp, 2004). The current study aims at characterizing the molecular mechanisms underlying PIDD cleavage and determining its functional consequences.
Results
PIDD is constitutively processed at S446 and S588
As previously reported, full-length PIDD (PIDD-FL, 910 aa) with an apparent molecular weight of 100 kDa is constitutively cleaved into three main fragments, an N-terminal fragment of approximately 48 kDa (PIDD-N) and two C-terminal fragments of 51 kDa (PIDD-C) and 37 kDa (PIDD-CC) (Tinel and Tschopp, 2004). This processing is well documented for cells in which PIDD is transiently or stably overexpressed. In order to exclude an artifact of overexpression, it was however important to determine whether these cleavage events also occurred at the level of the endogenous protein. A monoclonal antibody directed against the DD of PIDD was generated, which was able to detect endogenous PIDD. In all cell lines tested, the PIDD-C and the PIDD-CC fragments formed the major species, whereas full-length endogenous PIDD was hardly detectable or even absent (Figure 1A and Supplementary Figure 1A). Interestingly, several species were detected around 51 kDa, possibly reflecting the expression of different isoforms, as all species disappeared when the cells were transfected with siRNA directed against PIDD (Supplementary Figure 1B). The ratio between the PIDD-C and PIDD-CC fragments varied from cell line to cell line, but was approximately 2:1 in most cases. As PIDD is a p53-inducible gene, we investigated whether p53 influences the extent of PIDD processing. Although both fragments were generated in the presence and absence of p53, the amount of PIDD-C appeared to be slightly higher in p53-proficient cells (Figure 1A).
Figure 1.
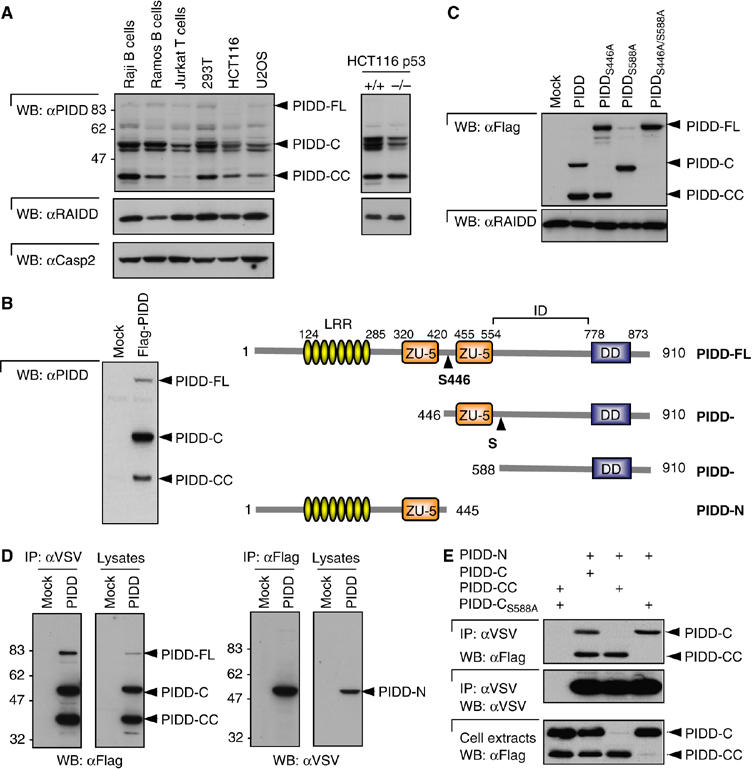
PIDD is constitutively processed. (A) Detection of endogenous PIDD. Western blot analysis of cell extracts of various cell lines probed with a monoclonal anti-PIDD antibody recognizing the death domain of PIDD. (B) Schematic representation of the PIDD fragments and cleavage sites. PIDD is processed at Ser446 giving rise to the PIDD-C fragment and at Ser588 resulting in the PIDD-CC fragment. FL, full-length; DD, death domain; ZU-5, ZU-5 domain (domain present in ZO-1 and Unc5-like netrin receptors); ID: intermediate domain; LRR, leucine-rich repeat. Immunoblot analysis of extracts of HEK293T cells stably expressing PIDD shows the various fragments. (C) Mutants at the cleavage site are no longer processed. Mutant forms of PIDD were stably expressed in HEK293T cells and cell extracts were analyzed by Western blotting. (D) HEK293T cells stably expressing PIDD with an N-terminal VSV and a C-terminal Flag tag were lysed and anti-VSV and anti-Flag immunoprecipitates analyzed by Western blots. (E) HEK293T cells were transiently transfected with the indicated constructs and an anti-VSV immunoprecipitation was performed. PIDD-N is VSV-tagged and PIDD-C and PIDD-CC constructs are Flag-tagged.
In order to map the two processing sites, C-terminally Flag-tagged PIDD was immunoprecipitated from human embryonic kidney 293T (HEK293T) cells stably expressing PIDD. The purified protein was trypsinized and subjected to tandem mass spectrometry. Protein fragments detected in the digests of PIDD-C and PIDD-CC and not in PIDD-FL were identified as SWFLVVSR and SWYWLWYTTK, respectively. This allowed us to map the sites as F445/S446 (PIDD-C) and F587/S588 (PIDD-CC) (Figure 1B). The mapping of the cleavage sites was confirmed by mutational analysis, as S446A and S588A mutations inhibited the generation of the different fragments (Figure 1C). PIDD S446A was still cleaved at the S588 site and PIDD S588A was also cleaved at the S446 site. However, a substantial amount of PIDD-FL was detected in cells stably expressing the PIDD S446A mutant, but not PIDD S588A mutant, suggesting that the processing was not as efficient in the PIDD S446A mutant as in PIDD wildtype (wt) or PIDD S588A mutant (see also further).
Importantly, PIDD fragments remained associated after processing (Figure 1D). Immunoprecipitates of a PIDD construct containing both an N-terminal VSV and a C-terminal Flag tag revealed that a considerable portion of the respective fragments remained bound to each other (Figure 1D). PIDD-N appears to bind equally to PIDD-C and PIDD-CC (Figure 1E). Additional mapping experiments indicated that the intramolecular interaction was mediated by the regions flanking the two ZU-5 domains (Supplementary Figure 1C–E).
PIDD cleavage occurs by auto-processing
A close examination of the sequence surrounding the two cleavage sites revealed high sequence similarity, suggesting that one distinct protease was responsible for both PIDD processing events (Figure 2A). Interestingly, the two surrounding sequences also showed strong similarity to a sequence found in Nup98 (Figure 2A), a component of the nuclear pore (Fabre et al, 1994; Fontoura et al, 1999). Nup98 contains a highly conserved HFS motif, which corresponds to the site of auto-processing that is essential for the assembly of Nup98 in the nuclear pore complex (Rosenblum and Blobel, 1999; Hodel et al, 2002). The proposed self-cleavage mechanism for Nup98 is similar to that of other auto-proteolytic enzymes including the self-splicing protein inteins, hedgehog, EMR2 and the N-terminal nucleophile (NTN) family of enzymes (Krasnoperov et al, 1997; Paulus, 2000; Perler and Adam, 2000; Nechiporuk et al, 2001; Chang et al, 2003). All classes of enzymes utilize acyl shift chemistry, with the side chains of cysteine, threonine or serine acting as the nucleophile. In addition, the inteins and the NTN enzymes contain a conserved histidine residue positioned two residues N-terminal from the catalytic C/T/S side chain (Figure 2A). Analysis of the Nup98 structure revealed that the HFS motif resides in a hydrophobic pocket, which is closed by a loop composed of residues N-terminal to the HFS motif (Hodel et al, 2002). This results in the requirement of a sequence of approximately 120 aa at the N-terminus of the HFS motif but only a short sequence (approximately 8 aa) at the C-terminus of Nup98 for successful auto-processing. Mutants of PIDD with deletions leaving around 40–60 aa on each side from the S446 site (PIDDΔ1–379 and PIDDΔ490–554) were generated and stably expressed in HEK293T cells (Figure 2B). Only mutant PIDDΔ490–554 was processed at S446, indicating that for auto-processing, a longer sequence N-terminal to the HFS motif and a shorter C-terminal region are required, as is the case with Nup98. In agreement with this result, PIDD-CC was not generated in mutant PIDDΔ490–554, strengthening the fact that both sites are processed by the same unusual mechanism.
Figure 2.
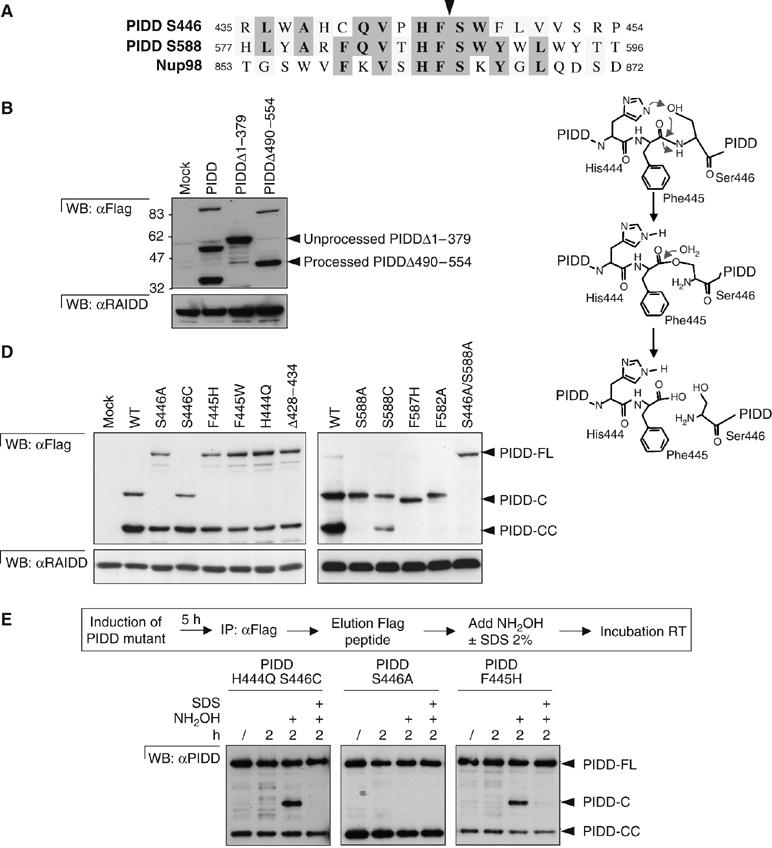
PIDD undergoes auto-processing at Ser446 and Ser588. (A) Alignment of amino-acid sequences surrounding the two PIDD cleavage sites together with the Nup98 cleavage site. The arrow indicates the cleavage site. (B) Deletion mutants of PIDD (Δ1–379 and Δ490–554) or the full-length protein were stably expressed in HEK293T cells and processing was analyzed by Western blotting. (C) Proposed mechanism for the autoproteolytic process of PIDD at the PIDD-C cleavage site, based on the mechanism proposed by Rosenblum and Blobel (1999). An identical mechanism is also proposed for the PIDD-CC cleavage site. (D) PIDD-FL containing mutations in amino acids preceding or following the PIDD-C (left panel) or PIDD-CC (right panel) cleavage sites was analyzed for its capacity to auto-process. (E) In vitro processing of inactive H444Q/S446C, S446A and F445H mutants induced by the nucleophile NH2OH. Expression of the Flag-tagged PIDD mutants was induced by doxocycline treatment of HEK293Trex during 5 h and purified on an anti-Flag affinity column. Immunoprecipitates were eluted using Flag peptides. The various purified PIDD mutants were then incubated with NH2OH in the presence or absence of denaturing SDS. PIDD cleavage was analyzed by Western blotting using the monoclonal anti-PIDD antibody.
The sequence resemblance between Nup98 and the two PIDD cleavage sites therefore suggested that auto-processing of PIDD was a more likely mechanism than cleavage by exogenous proteases. Self-proteolysis reactions preceding serines, cysteines or threonines involve a nucleophilic attack by the hydroxyl or thiol group of the respective amino acids on the preceding peptide bond (Rosenblum and Blobel, 1999), resulting in the replacement of the peptide bond by an ester or a thioester bond (Figure 2C). These bonds are more reactive than peptide bonds and can then be attacked by a second nucleophile and broken. This model implies that serine, cysteine or threonine (in the case of PIDD, a serine) is essential for the reaction, and that they are interchangeable with only limited effects on catalytic activity. In contrast, nonhydroxyl-containing amino acids are predicted to inactivate the enzymatic activity. As expected, mutating the active site S446 or S588 to Ala completely inhibited the generation of the PIDD-C and PIDD-CC fragments, respectively, whereas mutating S446 and S588 to cysteine still permitted cleavage and almost equivalent amounts of the PIDD-C or PIDD-CC fragment were detectable (Figure 2D). The importance of the conserved HFS motif was further investigated by mutating F445 to Trp or His. Both mutations led to the ablation of the activity, indicating delicate structural requirements (the analogous Phe → Trp change in Nup98 conserves the activity). In agreement with the proposed role of the His in the HFS motif, acting to deprotonate the OH group of Ser (Figure 2C), replacement of H444 with Gln resulted in inactivation of the proteolytic activity (Figure 2D, left panel). Analogous mutations in the second HFS motif also led to the disappearance of the PIDD-CC fragment (Figure 2D, right panel). To definitively prove that cleavage of the PIDD precursor is a self-catalyzed process, we purified PIDD from HEK293T cells that stably expressed PIDD mutants unable to spontaneously generate the PIDD-C fragment. On the basis of mutations in Nup98 shown to hydrolyze very slowly in the absence of exogenously added nucleophiles (Rosenblum and Blobel, 1999), the purified non-cleavable mutants H444Q/S446C and F445H were exposed to hydroxylamine (NH2OH), which in both cases caused auto-processing as evidenced by the appearance of the PIDD-C fragment (Figure 2E and Supplementary Figure 2). Processing was not observed in the presence of denaturing sodium dodecyl sulfate (SDS) or with the S446A mutant, indicating that cleavage induced by NH2OH indeed occurred at the S446 site. Taken together, the above results indicate that PIDD is one of the few known human proteins where auto-processing occurs in an intein-like manner.
PIDD-N, a regulatory fragment
In order to investigate the functional consequences, if at all, of PIDD processing, we expressed the individual fragments on their own and measured their capacity to interact with molecules known to be present in the PIDDosome. We first concentrated on the PIDD-N fragment. Unlike PIDD-FL or PIDD-C, PIDD-N did not interact with RAIDD, and thus did not activate caspase-2 (Figure 3A). Similarly, PIDD-N-expressing cells did not significantly alter NF-κB activation in response to DNA damage (Figure 3B). In contrast, cells expressing a PIDD mutant lacking the N-terminal LRR showed enhanced activation of NF-κB (Figure 3B). These data suggest that PIDD-N somehow represses PIDD functions and that its deletion leads to increased activation of PIDD. This is reminiscent of proteins of other initiator caspase-activating platforms, such as the apoptosome (Hu et al, 1998; Srinivasula et al, 1998), and the NOD-like receptor family, such as the NODs or NALPs, where the LRRs exert a strong auto-inhibitory effect (Poyet et al, 2001; Martinon and Tschopp, 2005). PIDD-N, however, does not seem to act as a dominant negative interfering with PIDD's ability to induce caspase-2 (Supplementary Figure 3A) or NF-κB activation (Supplementary Figure 3B).
Figure 3.
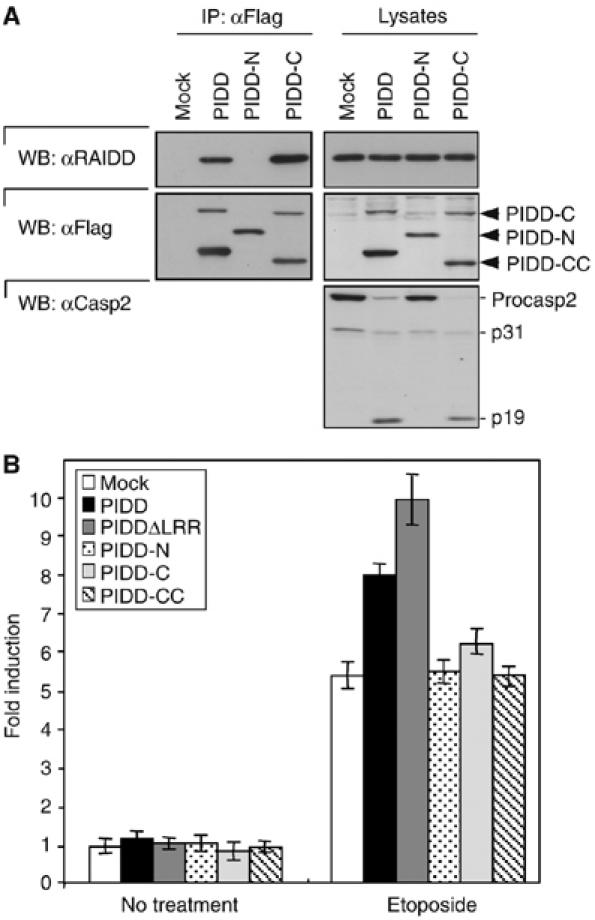
PIDD-N is a regulatory fragment of PIDD. (A) PIDD-N is not required for RAIDD interaction. FL PIDD, PIDD-N and PIDD-C were stably expressed in HEK293T cells and their interaction with RAIDD and their capacity to activate caspase-2 assayed. (B) HEK293T cells stably expressing the indicated constructs were tested for their NF-κB-activating capacity induced by DNA damage (40 μM etoposide) using a luciferase-based reporter gene assay.
Cleavage at the S588 site is required for caspase-2 activation
We next evaluated the functions of the PIDD-C and PIDD-CC fragments. We first compared the caspase-2-activating capacity of wt PIDD with the different mutant forms of PIDD that are unable to generate PIDD-C, PIDD-CC or both of these fragments. Interestingly, transient overexpression of the PIDD-CC-deficient S588A mutant failed to activate caspase-2, whereas the PIDD-C-deficient S446A mutant was still competent (Figure 4A). As expected from these data, the double-PIDD-C/PIDD-CC-deficient S446A/S588A mutant also lost its caspase-2-activating capacity. Caspase-2 activation by PIDD is mediated by RAIDD. Whereas wt and PIDD-C-deficient S446A mutant bound to RAIDD, the PIDD-CC-deficient S588A mutant or the double mutant did not, explaining at the molecular level the absence of caspase-2 activation (Figure 4B). These data suggest that the generation of the PIDD-CC fragment is required for PIDD-induced activation of caspase-2. To further strengthen this finding, we established populations of HeLa cells stably expressing the different mutants and treated them with doxorubicin (Figure 4C). Only wt PIDD and PIDD-C-deficient S446A mutant sensitized the cells to doxorubicin-induced apoptosis. The low level of sensitization observed with the PIDD-CC-deficient S588A mutant is probably owing to overexpression and residual caspase-2 activation. In order to determine which fragment of PIDD is the active fragment involved in caspase-2 activation, HEK293T cells stably expressing both Flag-PIDD and VSV-RAIDD were generated. These cells exhibited constitutive caspase-2 activation owing to elevated expression of PIDD (Tinel and Tschopp, 2004). When RAIDD was immunoprecipitated, the PIDD-CC fragment, but not PIDD-C, was detectable in the complex (Figure 4D and Supplementary Figure 4A), confirming the importance of PIDD-CC generation for caspase-2 activation. The PIDD-CC fragment requirement for activation of the caspase-2 pathway was also seen in HEK293Trex cell lines, which express PIDD-wt, PIDD-C or PIDD-CC in an inducible manner. Induction of expression of the various PIDD fragments showed that caspase-2 was most efficiently activated in the clone expressing preformed PIDD-CC, whereas in cells expressing wt and PIDD-C, caspase-2 activation was slower and followed the appearance of the PIDD-CC fragment (Figure 4E). Immunoprecipitation studies performed on these cells allowed us to correlate increased caspase-2 activation with increased RAIDD and caspase-2 recruitment (Supplementary Figure 4B). Thus, PIDD-CC expression is necessary and sufficient for the activation of caspase-2. Importantly, stable expression of PIDD-DD (PIDD 776–910) failed to activate caspase-2 (Supplementary Figure 4C), indicating that yet another region of PIDD is required for caspase-2 activation in addition to PIDD-DD.
Figure 4.
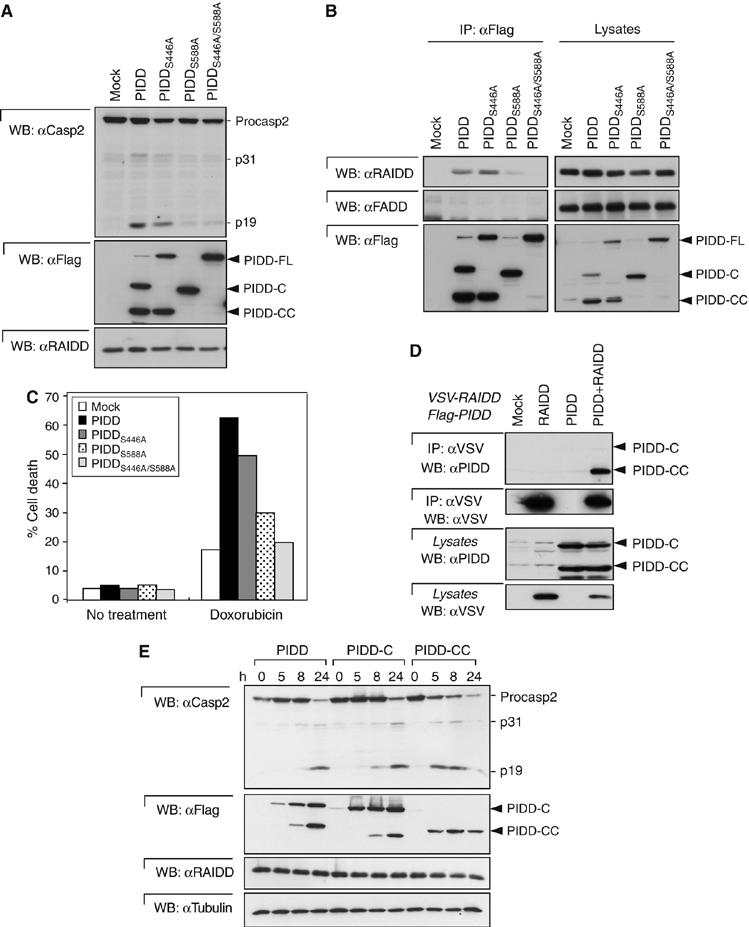
PIDD-CC recruits RAIDD and activates caspase-2. (A) The different PIDD mutants were transiently expressed in HEK293T cells and caspase-2 activation determined by monitoring cleavage of caspase-2. (B) Flag-tagged PIDD mutants stably expressed in HEK293T cells and immunoprecipitated using anti-Flag antibodies were analyzed for their capacity to interact with RAIDD. (C) HeLa cells stably expressing the indicated constructs were treated with doxorubicin (5 μg/ml) and the dead cells were counted by Trypan blue. Representative of three independent experiments is shown. (D) HEK293T cells, stably expressing wt Flag-PIDD, VSV-RAIDD or both, were lysed and anti-VSV-RAIDD immunoprecipitates analyzed for the presence of PIDD fragments. (E) HEK293Trex cells expressing FL-PIDD, PIDD-C or PIDD-CC were induced by doxycycline for the indicated times and caspase-2 activation was monitored.
Cleavage at S446 is required for NF-κB activation
PIDD constitutes not only a platform that, upon DNA damage, results in caspase-2 activation, but is also an essential protein for the DNA-damage-triggered NF-κB response (Janssens et al, 2005). We previously showed that, in response to DNA damage, PIDD translocates to the nucleus (Janssens et al, 2005). To address how PIDD processing affects nuclear translocation, we analyzed the localization of several processing-defective PIDD mutants upon treatment with camptothecin (CPT), a DNA-damaging drug. When DNA damage was induced, a substantial portion of PIDD was detectable in the nucleus (Figure 5A and B), whereas in non-treated cells, PIDD was mostly cytoplasmic, in agreement with previous results (Janssens et al, 2005). In contrast, a PIDD-C-deficient S446A or double S446A/S588A mutant no longer translocated to the nucleus, whereas the lack of cleavage before S588 in the PIDD-CC-deficient S588A mutant only slightly affected accumulation in the nucleus upon genotoxic stress (Figure 5A and B). These results therefore indicate that generation of the PIDD-C fragment is a prerequisite for PIDD nuclear translocation upon DNA damage. Indeed, expression of the different PIDD fragments confirmed that PIDD-C was able to translocate to the nucleus, whereas PIDD-CC alone showed an exclusively cytoplasmic localization (see Supplementary Figure 5).
Figure 5.
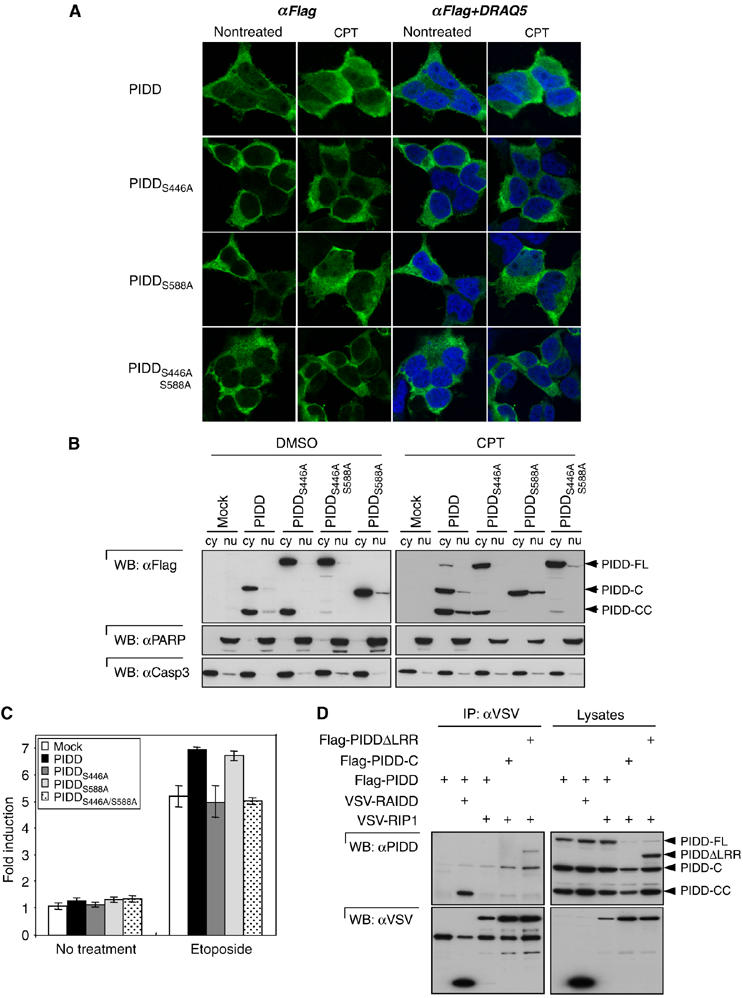
PIDD-C recruits RIP1 and activates NF-κB in response to DNA damage. (A) PIDD mutants unable to generate PIDD-C do not translocate to the nucleus in response to genotoxic stress. PIDD- or mutant PIDD-expressing HEK293T cells were left untreated or stimulated for 2 h with 50 μM CPT. Confocal images show nuclear (blue, DRAQ5) or PIDD (green, Alexa 488) staining. (B) Subcellular fractionation of HEK293T cells stably expressing the different PIDD mutants. The cells were treated with DMSO or with 50 μM CPT. (C) PIDD-FL and PIDD mutant proteins stably expressed in HEK293T cells were tested for their NF-κB-activating capacity induced by DNA damage (40 μM etoposide) using a luciferase-based reporter gene assay. (D) HEK293T cells were cotransfected with VSV-RAIDD or VSV-RIP1 in combination with different Flag-tagged PIDD constructs and immunoprecipitated with an anti-VSV antibody. The presence of the different PIDD fragments in the eluates was monitored.
To check if the altered translocation capacity also affected PIDD's ability to activate NF-κB, we analyzed HEK293T cells stably expressing the various PIDD mutants using an NF-κB-dependent luciferase reporter gene assay (Figure 5C). We previously demonstrated that in comparison to mock-transfected cells, cells stably expressing PIDD showed a moderate but significant enhancement of the NF-κB response upon DNA damage (Janssens et al, 2005). In keeping with our translocation data, we observed that the enhanced NF-κB response was only detectable for the wt PIDD and PIDD-CC-deficient mutant (S588A), whereas the PIDD-C-deficient mutants (S446A and S446A/S588A) were inactive. An identical tendency could be observed when phosphorylation of IκB was used as a readout for NF-κB activation (Supplementary Figure 6).
We next determined which fragment of PIDD interacted with the downstream adaptor RIP1 (Figure 5D). RIP1 mediates recruitment of NEMO to PIDD, resulting in NEMO sumoylation and subsequent NF-κB activation (Janssens et al, 2005). In contrast to RAIDD that interacted with PIDD-CC, RIP1 recruited exclusively the PIDD-C fragment.
Thus, the generation of the PIDD-C fragment is crucial for the activation of NF-κB but not of caspase-2. Moreover, cleavage at S446 is a prerequisite for PIDD translocation to the nucleus in response to DNA damage, showing an intimate link between these two processes.
The two PIDD fragments are formed in a sequential manner
The results of Figure 1C, in which we evaluated which fragments were formed in the different PIDD cleavage mutants, already suggested that the cleavage of the two fragments occurred in a sequential manner. Thus, the generation of PIDD-CC appeared much more efficient in molecules that were initially cut at S446. We confirmed this sequential order of events by monitoring the kinetics of PIDD fragment formation in stable cell lines expressing PIDD in an inducible manner (Figure 6A). HEK293 Flp-In TRex (HEK293Trex) cells were induced to express PIDD by addition of doxycycline. Four hours following the induction, only PIDD-C was detectable, whereas the PIDD-CC form was absent. Minor amounts of PIDD-FL were also detected, but PIDD-FL levels did not increase, confirming that PIDD-FL was directly processed after expression. PIDD-CC was detectable only after 6–8 h. The ratio of PIDD-CC to PIDD-C then steadily increased to attain comparable levels 16 h after induction. The notion that PIDD processing occurs constitutively in our stable cells was further supported by an experiment in which protein synthesis was blocked with cycloheximide (CHX) (Figure 6B). Together with PIDD-FL, the amount of PIDD-C fragment steadily diminished, whereas the relative amount of PIDD-CC slightly increased. That the disappearance of PIDD-C fragment represented a genuine conversion to PIDD-CC and not proteasome-dependent degradation was apparent based on the observation that (1) it also disappeared in the presence of the proteasome inhibitor MG-132 and (2) in the case of the PIDD-CC-deficient S588A mutant, the PIDD-C fragment was stable up to 16 h following CHX addition. Of note, PIDD-FL was rapidly and completely processed into PIDD-C and PIDD-N. No intermediary PIDD-N fragment (1–587) was detected (Figure 1D and Supplementary Figure 6), suggesting that PIDD-CC does not normally arise from PIDD-FL processing. Addition of CHX did not result in further processing of PIDD-N (Supplementary Figure 7). A sequential processing of PIDD-FL to PIDD-N and PIDD-C and then to PIDD-CC is therefore likely to occur.
Figure 6.
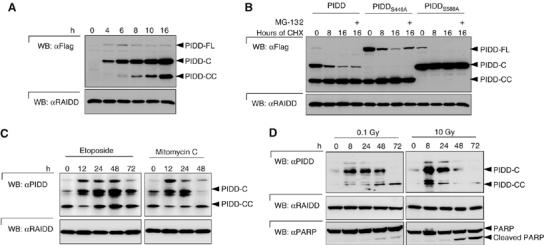
PIDD-C and PIDD-CC are induced with differential kinetics. (A) Induction of PIDD expression in HEK293Trex cells by doxycycline during the indicated times. Processing of PIDD was monitored by Western blot analysis of the cell extracts. (B) HEK293T cells stably expressing the indicated PIDD mutants were treated for 0, 8 and 16 h with 10 μg/ml CHX in the presence or absence of the proteasome inhibitor MG-132 (10 μM). Processing of PIDD was monitored by Western blot. (C) HCT116 cells were treated with 10 μM etoposide or 10 μM mitomycin C for the indicated times and PIDD expression levels were monitored. (D) HCT116 cells were exposed to a single dose of γ-irradiation (0.1 or 10 Gy), whereafter the cells could recuperate. Endogenous PIDD expression levels and PARP cleavage were monitored.
That this mechanism is of physiological relevance was supported by monitoring PIDD expression in response to DNA damage. In agreement with previous reports (Lin et al, 2000), we found that DNA damage such as that induced by etoposide or mitomycin C treatment led to a general increase of PIDD expression in HCT116 cells (Figure 6C). Interestingly, PIDD-C levels increased before PIDD-CC levels in response to both treatments.
We then investigated the possibility that distinct doses of DNA damage insults would differentially induce the two PIDD fragments upon de novo synthesis. To this end, we exposed the cells to a single dose of γ-irradiation, which, in contrast to continuous treatment with topoisomerase inhibitors, allows the cells to recuperate from the DNA damage insult. We exposed HEK293T cells to different amounts of γ-irradiation, ranging from 0.1 to 10 Gy (only the results for 0.1 and 10 Gy are depicted in the figure). As can be seen in Figure 6D, PIDD-C was the first fragment to be induced and was present from the 8 h time point onwards. We have never been able to detect PIDD-FL, suggesting that the first cleavage event is indeed constitutive. PIDD-CC increase occurred much more slowly, the actual kinetics being strongly dependent on the dose of treatment used. Whereas low doses of γ-irradiation, with only a mild effect on cell death, induced significant levels of PIDD-CC after 48 h, high doses, which induced strong apoptosis, evoked an immediate induction of both PIDD-C and PIDD-CC (Figure 6D). Interestingly, at 72 h, PIDD-C was no longer present, suggesting that all PIDD-C was converted into PIDD-CC. These data therefore indicate that both PIDD fragments are not induced with the same kinetics upon DNA damage, and may reflect a regulated activity of the second cleavage site in response to this stimulus.
Discussion
Protein splicing is a form of post-translational processing that is the protein equivalent of RNA splicing. There is now formal evidence that these reactions exist in several protein maturation pathways, including hedgehog protein maturation (Porter et al, 1996), nucleoporin processing (Rosenblum and Blobel, 1999; Hodel et al, 2002) and GPCR processing (Lin et al, 2004). PIDD is constitutively cleaved into three main fragments: PIDD-N (1–445), PIDD-C (446–910) and PIDD-CC (588–910). Although our results do not provide direct evidence for the exact reaction mechanism involved in PIDD processing, they are consistent with auto-proteolysis, similar to the previously described mechanisms, as they include: (i) the cleavage site before a serine residue, (ii) requirement for a hydroxyl or thiol group at the cleavage position, and (iii) the sensitivity of in vitro cleavage of the H444Q/S446C mutant to hydroxylamine. Thus, PIDD is auto-processed at two sites, F445/S446 and F587/S588, that are highly conserved and contain the HFS tripeptide.
An almost identical auto-processing site is found in Nup98, for which the three-dimensional structure has been determined (Rosenblum and Blobel, 1999; Hodel et al, 2002). Auto-processing of precursor Nup98 into Nup98 and the 8-kDa tail fragment results in only a small molecular rearrangement and the interaction of the mature Nup98 with the 8-kDa tail fragment persists (Hodel et al, 2002). Non-covalent interaction between the different fragments generated appears to be a rule for proteins that undergo auto-processing (Brannigan et al, 1995; Hodel et al, 2002; Chang et al, 2003; Hsieh et al, 2003). Similarly, we found that fragments generated by both cleavage events in PIDD remained associated in a complex containing PIDD-N and PIDD-C or PIDD-CC. The observation that RAIDD preferentially immunoprecipitates PIDD-CC and not PIDD-C suggests that PIDD-C and PIDD-CC may not be present simultaneously in the same complex, or that dissociation of the different fragments may occur upon activation. The fate of the 14-kDa fragment (446–587) generated upon cleavage at S588 is currently unknown.
Our data unambiguously demonstrate an important role of PIDD processing with respect to the initiation of downstream signaling events. We previously demonstrated that PIDD is implicated in the activation of both caspase-2 and NF-κB upon DNA damage. Intriguingly, both functions appear to be controlled by different PIDD fragments generated by autoproteolysis (Figure 7). PIDD-C is able to interact with RIP1, thereby initiating the NEMO modifications that are required for NF-κB activation (Janssens et al, 2005). Interestingly, the PIDD-C-deficient S446A mutant does not translocate into the nucleus in response to DNA damage, which is a prerequisite for NEMO modification to occur (Huang et al, 2003; Janssens et al, 2005; Wu et al, 2006). Theoretically, processing at S446 may unmask a nuclear localization site or a site that allows binding to a nuclear shuttling protein, thereby explaining the need for the first processing. Although the PIDD-C fragment and the PIDD-CC-deficient S588A mutant have the capacity to translocate and induce NF-κB activation in response to DNA damage, they neither recruit RAIDD nor activate caspase-2. The latter functions are apparently triggered only by the RAIDD-interacting PIDD-CC fragment, which is generated at a later stage. It is noteworthy that the mere presence of PIDD-C and PIDD-CC fragments found to be constitutively expressed in a number of cell lines is insufficient to trigger caspase-2 or NF-κB activation in the absence of DNA damage. This suggests that under normal conditions, the activities associated with PIDD-C and PIDD-CC fragments are suppressed by an unknown inhibitor.
Figure 7.
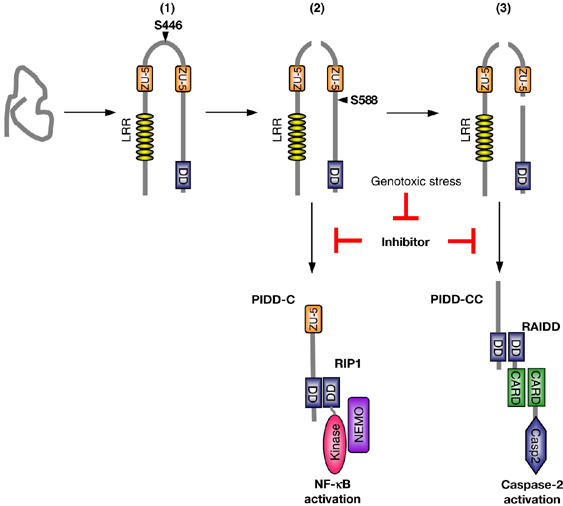
Model of PIDD maturation and its impact on the selection of signaling pathways. Upon synthesis, PIDD folds into a conformation in which one of the potential splicing sites, S446, is immediately active, leading to the generation of PIDD-N and PIDD-C fragments (1). Cleavage at the PIDD-C proteolytic site (2) presumably induces a slight conformational change at the PIDD-CC cleavage site, allowing the proteolytic activity of this site to become active as well, although this second cleavage event appears to be regulated by yet another mechanism, coupled to the severity of the DNA damage (3). The two cleavage events in PIDD lead to the generation of two fragments with distinct functions. Cleavage at S446 generates PIDD-C, which is able to bind RIP1 and NEMO and as such triggers an NF-κB response. Cleavage at S588 generates PIDD-CC, which binds to RAIDD and therefore recruits and activates caspase-2. These two signaling pathways seem to be constitutively inhibited and become activated only upon genotoxic stress.
We have previously shown that PIDD can act as a switch allowing to either activate an NF-κB-dependent cell survival pathway or trigger apoptosis by activation of caspase-2 (Janssens et al, 2005). The strength with which these two signaling pathways are engaged will certainly be modulated by the presence of other regulators. However, we also propose that the regulation of these alternate responses to different stimuli might result from modulation of PIDD processing. Indeed, the relative ratio of PIDD-C and PIDD-CC varies in cells exposed to different DNA-damaging stimuli. Whereas the introduction of a limited amount of DNA breaks initially only leads to the induction of the NF-κB-activating fragment PIDD-C, more severe DNA damage concomitantly induced PIDD-C and PIDD-CC, correlating with a strong apoptotic response of the cells. Thus, the temporarily distinct generation of the PIDD-C and PIDD-CC fragments might govern the fate of the cell, leading by default to the activation of a cell survival pathway and, only when the cell repair process fails, to the activation of the pro-apoptotic caspase-2 pathway.
In conclusion, we have shown that auto-processing of PIDD generates PIDD-C and PIDD-CC fragments that are involved in the activation of NF-κB and caspase-2, respectively. Induction of PIDD upon DNA damage together with regulated auto-processing may function to selectively direct signaling pathways.
Materials and methods
Cell culture and biological reagents
HEK293T cells, HEK293Trex cells (Invitrogen), U2OS human osteosarcoma cells, HCT116 p53+/+ and p53−/− cells (gift from Dr B Vogelstein, John Hopkins University, Baltimore, MD, USA) and 293T cells stably expressing different constructs of PIDD were grown in DMEM+Glutamax (Life Technologies), supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Raji B cells, Ramos B cells and Jurkat T cells were grown in RPMI (Life Technologies) containing the same supplements. The generation of cells stably expressing PIDD or RAIDD was described elsewhere (Tinel and Tschopp, 2004). Camptothecin (CPT), mitomycin C, MG-132, etoposide (Eto) and Ratjadone C were purchased from Alexis. Cells were γ-irradiated with a 137Cs source in a single pulse. Doxycyline, hydroxylamine, CHX, and Flag peptides were obtained from Sigma. Hygromycin was purchased from Invitrogen.
Expression vectors
pCR3-PIDD-Flag, pMSCV-PIDD-Flag, pCR3-RAIDD-VSV and pCR3-RIP1-VSV have been described previously (Martinon et al, 2000; Tinel and Tschopp, 2004). HEK293Trex cells expressing different constructs of PIDD were generated as described by the manufacturer (Invitrogen). Briefly, PIDD constructs were subcloned in the pcDNA5/FRT vector (Invitrogen) and cotransfected with the Flp recombinase-encoding plasmid (pOG44, Invitrogen) into HEK293Trex cells. Clones were then selected with hygromycin. All PIDD constructs are C-terminally Flag-tagged. NFconluc, containing the luciferase reporter gene driven by a minimal NF-κB responsive promoter, was a gift from Dr A Israel (Institut Pasteur, Paris, France). Pactβgal, containing the β-galactosidase gene under the control of the β actin promoter, was obtained from Dr Inoue (Institute of Medical Sciences, Tokyo, Japan).
Transient transfections and NF-κB reporter gene assays
HEK293T cells stably expressing PIDD or PIDD mutants were transiently transfected using the DNA–calcium phosphate precipitation method with 100 ng pNFconluc, 100 ng pactβgal and different concentrations of specific expression plasmids. The total amount of DNA was kept constant by adding empty vector up to 1 μg DNA per well of a six-well plate. NF-κB activity was determined by measuring the luciferase activity present in cell extracts. Luciferase values were normalized for differences in transfection efficiency on the basis of β-galactosidase activity in the same extracts, and expressed as fold induction values relative to the unstimulated empty vector control. Where needed, cells were stimulated 48 h after transfection for 16 h with 40 μM Eto or left untreated.
Mass spectrometry
The samples were loaded on an 8% polyacrylamide gel and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The gel was then incubated in a solution of 10% acetic acid and 40% ethanol for 30 min and stained for 15 min with a solution of 10% acetic acid containing 0.25 g/l Coomassie brilliant blue G 250. The gel was destained with a solution of 10% acetic acid until the required coloration was reached. Bands corresponding to full-length PIDD constructs or their cleavage products were excised and manually in-gel digested with trypsin according to a described protocol (Bienvenut et al, 1999). Tryptic peptides were recovered in the supernatant of the digestion. Samples were mixed 1:10 with α-cyano-4-hydroxycinnamic acid matrix (Sigma) dissolved in a solution containing 50% acetonitrile and 0.1% TFA at a concentration of 5 mg/ml. A 1 μl aliquot of the sample–matrix mixture was deposited onto a single well of a 100-well, stainless-steel, matrix-assisted laser desorption ionization (MALDI) target plate and analyzed on a MALDI TOF/TOF AB 4700 Proteomics Analyzer (Applied Biosystems) in both the MS and MS/MS modes.
PIDD in vitro processing
HEK293T-Rex cells expressing PIDD mutants were induced for 5 h with 0.01 μg/ml doxycycline. The cells were then harvested, lysed and an anti-Flag immunoprecipitation was performed. Immunoprecipitated PIDD was eluted with Flag peptides at 100 μg/ml in 50 mM HEPES (pH 7.4) and 150 mM NaCl. The eluates were then incubated in the presence of 200 mM NH2OH at room temperature for the indicated time.
Co-immunoprecipitation and Western blotting
Co-immunoprecipitation experiments with transfected proteins were performed in lysis buffer containing 1% NP-40, 20 mM Tris, pH 7.4, 250 mM NaCl, 1 mM EDTA, 5% glycerol and a protease inhibitor cocktail. After lysis, the extracts were incubated with anti-Flag antibody for 2 h. After incubation, the beads were washed four times with lysis buffer and analyzed by SDS–PAGE and immunoblotting. The antibodies used for Western blotting were anti-phospho-IκB (Cell Signalling), anti-caspase-2 11B4 (Alexis), rabbit anti-RAIDD (Alexis) and anti-FADD (Transduction Labs). A monoclonal anti-PIDD antibody (Anto-1) was raised against PIDD-DD (776–910) according to standard procedures. AL249, a rabbit anti-PIDD polyclonal antibody raised against a peptide covering amino acids 148–174, was purchased from Alexis.
Confocal microscopy
HEK293T cells stably expressing the different PIDD mutants were cultured on sterile glass coverslips in six-well plates and fixed with 3% paraformaldehyde for 15 min. Cells were permeabilized with 0.3% saponin for 10 min and treated with 2% normal goat serum (NGS)/0.5% BSA/0.1% saponin as a blocking reagent. Flag antibody (M2, Sigma) was used at a dilution of 1/500 in 0.1% saponin/0.1%BSA/PBS; a secondary Alexa 488 anti-mouse IgG1 (Molecular Probes) was used at a dilution of 1/300 in 0.1% saponin/0.1%BSA/PBS. Cells were mounted in FluorSave (Calbiochem) containing a 1/1000 dilution of DRAQ5 (Alexis) for nuclear counterstain. Images from immunostaining were acquired by using a Zeiss inverted laser scanning confocal microscope LSM 510 with a × 63 oil-immersion objective.
Biochemical fractionation
Cells were lysed in a cytosolic lysis buffer containing 10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 300 mM sucrose, 0.5% NP-40, 10 mM KCl, supplemented with DTT and protease inhibitors. The lysates were centrifuged and the supernatant constituted the cytosolic fraction. The pellet was then lysed in RIPA buffer and subsequently sonicated. The supernatant constituted the nuclear-enriched fraction.
Supplementary Material
Supplementary Figures
Supplementary Figure Legends
Acknowledgments
We thank S Hertig for technical support and H Everett, E Meylan, D Muruve, M Thome and P Schneider and the whole Tschopp group for discussions and critical reading of the manuscript. This work was supported by grants from the Swiss National Science Foundation. SJ is supported by an EMBO fellowship and EL by an FRM fellowship.
References
- Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, Yeh WC, Mak TW, Benchimol S (2005) Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci USA 102: 14314–14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenut WV, Sanchez JC, Karmime A, Rouge V, Rose K, Binz PA, Hochstrasser DF (1999) Toward a clinical molecular scanner for proteome research: parallel protein chemical processing before and during Western blot. Anal Chem 71: 4800–4807 [DOI] [PubMed] [Google Scholar]
- Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378: 416–419 [DOI] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Roumier T, Valent A, Raslova H, Yakushijin K, Horne D, Feunteun J, Lenoir G, Medema R, Vainchenker W, Kroemer G (2004) Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene 23: 4362–4370 [DOI] [PubMed] [Google Scholar]
- Chang GW, Stacey M, Kwakkenbos MJ, Hamann J, Gordon S, Lin HH (2003) Proteolytic cleavage of the EMR2 receptor requires both the extracellular stalk and the GPS motif. FEBS Lett 547: 145–150 [DOI] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC (1994) Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell 78: 275–289 [DOI] [PubMed] [Google Scholar]
- Fontoura BM, Blobel G, Matunis MJ (1999) A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol 144: 1097–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagkousiklidis S, Vogler M, Westhoff MA, Kasperczyk H, Debatin KM, Fulda S (2005) Sensitization for gamma-irradiation-induced apoptosis by second mitochondria-derived activator of caspase. Cancer Res 65: 10502–10513 [DOI] [PubMed] [Google Scholar]
- Hodel AE, Hodel MR, Griffis ER, Hennig KA, Ratner GA, Xu S, Powers MA (2002) The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol Cell 10: 347–358 [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Cheng EH, Korsmeyer SJ (2003) Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 115: 293–303 [DOI] [PubMed] [Google Scholar]
- Hu Y, Ding L, Spencer DM, Nunez G (1998) WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J Biol Chem 273: 33489–33494 [DOI] [PubMed] [Google Scholar]
- Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S (2003) Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115: 565–576 [DOI] [PubMed] [Google Scholar]
- Hur GM, Lewis J, Yang Q, Lin Y, Nakano H, Nedospasov S, Liu ZG (2003) The death domain kinase RIP has an essential role in DNA damage-induced NF-kappa B activation. Genes Dev 17: 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J (2005) PIDD mediates NF-kappaB activation in response to DNA damage. Cell 123: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, Chepurny OG, Little AR, Plotnikov AN, Wu D, Holz RW, Petrenko AG (1997) alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron 18: 925–937 [DOI] [PubMed] [Google Scholar]
- Lin HH, Chang GW, Davies JQ, Stacey M, Harris J, Gordon S (2004) Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem 279: 31823–31832 [DOI] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S (2000) Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet 26: 122–127 [DOI] [PubMed] [Google Scholar]
- Martinon F, Holler N, Richard C, Tschopp J (2000) Activation of a pro-apoptotic amplification loop through inhibition of NF-kappaB-dependent survival signals by caspase-mediated inactivation of RIP. FEBS Lett 468: 134–136 [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J (2005) NLRs join TLRs as innate sensors of pathogens. Trends Immunol 26: 447–454 [DOI] [PubMed] [Google Scholar]
- Nechiporuk T, Urness LD, Keating MT (2001) ETL, a novel seven-transmembrane receptor that is developmentally regulated in the heart. ETL is a member of the secretin family and belongs to the epidermal growth factor-seven-transmembrane subfamily. J Biol Chem 276: 4150–4157 [DOI] [PubMed] [Google Scholar]
- Paulus H (2000) Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem 69: 447–496 [DOI] [PubMed] [Google Scholar]
- Perler FB, Adam E (2000) Protein splicing and its applications. Curr Opin Biotechnol 11: 377–383 [DOI] [PubMed] [Google Scholar]
- Porter J, Ekker S, Park W, von Kessler D, Young K, Chen C, Ma Y, Woods A, Cotter R, Koonin E (1996) Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86: 21–34 [DOI] [PubMed] [Google Scholar]
- Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES (2001) Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem 276: 28309–28313 [DOI] [PubMed] [Google Scholar]
- Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S (2002) A novel Apaf-1-independent putative caspase-2 activation complex. J Cell Biol 159: 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Shi M, Liu R, Yang QH, Johnson T, Skarnes WC, Du C (2005) The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci USA 102: 565–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JD, Enoksson M, Suomela M, Zhivotovsky B, Orrenius S (2002) Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J Biol Chem 277: 29803–29809 [DOI] [PubMed] [Google Scholar]
- Rosenblum JS, Blobel G (1999) Autoproteolysis in nucleoporin biogenesis. Proc Natl Acad Sci USA 96: 11370–11375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R, Yang C, Kaushal V, Shah SV, Kaushal GP (2005) p53-dependent caspase-2 activation in mitochondrial release of apoptosis-inducing factor and its role in renal tubular epithelial cell injury. J Biol Chem 280: 31230–31239 [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell 1: 949–957 [DOI] [PubMed] [Google Scholar]
- Telliez JB, Bean KM, Lin LL (2000) LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta 1478: 280–288 [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304: 843–846 [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B (2006) Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene 25: 5683–5692 [DOI] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S (2006) Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S (2005) Caspase-2 function in response to DNA damage. Biochem Biophys Res Commun 331: 859–867 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Figure Legends


