Abstract
Polymeric immunoglobulins (pIgs) that are present at mucosal surfaces play key roles in both the innate and adaptive immune responses. These pIgs are delivered to the mucosal surface via transcytosis across the epithelium, a process mediated by the polymeric immunoglobulin receptor (pIgR). Previous studies demonstrate that expression of the pIgR is regulated by multiple immunomodulatory factors including interleukin-4 (IL-4) and interferon-γ (IFN-γ). In studies using human intestinal epithelial cells (HT29), multiple inhibitors of the transcription factor nuclear factor-κB (NF-κB), including a dominant negative IκBα-serine mutant, inhibited both IL-4- and IFN-dependent increases in pIgR expression. Under identical conditions, NF-κB inhibitors had no effect on cytokine-dependent increases in expression of the transcription factor interferon regulatory factor-1. Over-expression of the IκBα-serine mutant also inhibited reporter gene expression in response to IL-4, TNF-α, IL-1β, and in some cases IFN-γ using constructs with sequences from the pIgR promoter. Reduced levels of pIgR were observed even when inhibitors were added ≥24 hr after cytokines suggesting that prolonged activation of NF-κB is required. Finally, reporter gene studies with NF-κB enhancer elements indicated that IFN-γ alone and IL-4 in combination with other cytokines activated NF-κB in HT29 cells. Together, these studies provide additional insight into the signalling pathways that contribute to expression of the pIgR, a critical player in mucosal immunity.
Introduction
In mucosal tissues, immunoglobulins are secreted by fully differentiated B cells (plasma cells) present in the lamina propria. Following secretion, polymeric immunoglobulin A pIgA and IgM, as well as pIg-containing immune complexes1 are transported from the submucosal space to the mucosal surface by the polymeric immunoglobulin receptor (pIgR). Transport of pIgs across the epithelium involves binding to the pIgR at the epithelial basolateral membrane, internalization, transcytosis, and release at the apical membrane.2 During transport, disulphide-bond formation and proteolytic cleavage of the pIgR leads to release of a covalent pIg–pIgR complex into the lumen. The portion of the pIgR in this complex is referred to as secretory component (SC). Constitutive transcytosis of the pIgR in the absence of ligand results in release of free SC. In addition to its role in transport, SC increases the half-life of pIgA by protecting it from proteolysis3 and can act as an anti-inflammatory molecule by binding to inflammatory chemokines, thus reducing their chemotactic activity.4
Several immunomodulatory factors increase pIgR expression by human epithelial cells. These factors include transforming growth factor-β (TGF-β),5 tumour necrosis factor-α (TNF-α),6 interleukin-1β (IL-1β),7 interferon-γ (IFN-γ)8 and interleukin-4 (IL-4).9 Studies also demonstrate additivity/synergy between multiple factors9–11 and indicate that vitamin A enhances pIgR expression in IL-4- and IFN-treated HT29 cells.12 The pIgR is also up-regulated by androgens in a tissue-specific manner.13
Increased pIgR protein levels correlate with increased steady state levels of pIgR mRNA suggesting that regulation is caused, in large part, by increased transcription and/or mRNA stability.14 Moreover, increases in pIgR expression are delayed following cytokine addition9,14,15 and IFN-, IL-4- and TNF-dependent increases in pIgR mRNA require protein biosynthesis.14,16,17 Both observations suggest a role for inducible factors. Consistent with these observations, the inducible factor interferon regulatory factor-1 (IRF-1) has been demonstrated to play a role in both IFN- and TNF-dependent pIgR expression.16,18,19
Studies to characterize the mechanisms that regulate pIgR expression have identified promoter elements required for constitutive expression (elements that regulate the consistently high levels observed in vivo)20 and for basal expression (minimal promoter elements),21,22 as well as elements contributing to the androgen-specific response.13 Moreover, nuclear factor-κB (NF-κB) promoter elements have been identified proximal to the start site and in intron 1.17,23 Each may contribute to TNF-dependent pIgR expression. Interestingly, the NF-κB site in intron 1 appears to co-operate with an interferon-stimulated response element (ISRE) in the proximal promoter region.17,23 Intron 1 also contains a STAT6 (signal transducer and activator of transcription-6) site that may play an important role in regulating IL-4-dependent pIgR expression.24
Studies by our laboratory are designed to determine how IL-4 and IFN-γ regulate pIgR expression and focus on identifying key intermediates in signalling pathways. In this respect, numerous studies have shown involvement of NF-κB in gene expression regulated by cytokines such as TNF-α and IL-1. Moreover, as stated above, studies suggest that NF-κB contributes to TNF-dependent regulation of the pIgR.17,23 In contrast, whereas one study reports activation of NF-κB by IFN-β25 and studies in B cells indicate NF-κB is important for IL-4-dependent IgE expression,26 to our knowledge no studies have addressed the role of NF-κB in IL-4- and IFN-dependent pIgR expression.
The effects of NF-κB on protein expression can be either direct (gene of interest) or indirect (expression of regulatory proteins for the gene of interest). Moreover, not only newly activated but also constitutively active NF-κB can contribute to gene expression.26 Thus, as a more global approach to determine whether NF-κB contributes to cytokine-dependent pIgR expression, we tested the effect of NF-κB inhibitors on this response. Our studies, using multiple inhibitors of NF-κB, provide evidence that NF-κB contributes to both IL-4- and IFN-dependent expression of the pIgR. Moreover, we found that IFN-γ alone and IL-4 in combination with other cytokines activated NF-κB in HT29 cells. Finally, reporter gene studies with sequences from the pIgR promoter provide additional insight into the contribution of these sequences to cytokine-dependent increases in pIgR expression.
Materials and methods
Materials
Human recombinant IL-1β, TNF-α, IL-4, and IFN-γ were obtained from R & D Systems (Minneapolis, MN). NF-κB inhibitors were obtained from several sources: Bay 11-7085 (Biomol, Plymouth Meeting, PA), caffeic acid phenethyl ester (CAPE) and MG115 (Calbiochem, San Diego, CA), and sodium selenite (Aldrich Chemical Co., Milwaukee, WI).
Cell culture and cytokine treatment
HT29 (American Type Culture Collection; ATCC HTB38) and HT29 clone 19A27 cells are human epithelial cell lines isolated from an adenocarcinoma of the colon. Cells were cultured in Dulbecco's modified Eagle's minimal essential medium: Ham's F12 (1: 1) supplemented with 10% fetal bovine serum, 2 mm glutamine, and 500 U/ml each of penicillin and streptomycin as previously described.9 The concentrations of cytokines used in these studies were previously found to give maximal increases in pIgR expression.9,14
Inhibitor treatments
Cells were cultured in tissue cultureware until they were 80–90% confluent. Unless indicated otherwise, inhibitors were added directly to the medium at least one day after a medium change, cultures were incubated for 1 hr, and then cytokines were added to the indicated flasks: IL-4 (10 ng/ml), IFN-γ (200 U/ml), TNF-α (10 ng/ml), or IL-1β (10 ng/ml). Preliminary studies were performed with each inhibitor to find the optimal inhibitory concentration that was not cytotoxic to the cells.
RNase protection assay (RPA)
HT29 cells were cultured in T75 flasks. RNA was isolated using TRI REAGENT (Molecular Research Center, Cincinnati, OH) and non-radioactive RPAs were performed as previously described14 using 40–80 µg of total RNA. 28S rDNA was used as a control for recovery of RNA during the assay.14
Northern blot analysis
A 422 base pair probe for IRF-1 was generated as follows. Total cellular RNA was isolated from IFN-γ-treated HT29 clone 19A cells (200 U/ml, 24 hr) using TRI REAGENT. Reverse transcription–polymerase chain reaction (RT–PCR) was performed on 1 µg of total RNA according to standard methods using sense and antisense primers 5′-ACAGTTCCAGCCTA CATGCAG-3′ and 5′-CAGGTGGCATCC ATGTTCTTC-3′, respectively. The resulting DNA was blunt-end cloned into pSTBlue1 (Novagen, Madison, WI). Radiolabelled probe was generated using α[32P]-CTP by transcribing from the T7 promoter using HindIII-linearized plasmid and the Promega Riboprobe system (Promega, Madison, WI). For Northern blot analysis, total RNA was isolated from T75 flasks of control and cytokine-treated (IL-4, 10 ng/ml; IFN-γ, 200 U/ml; 24 hr) cells using TRI REAGENT: samples (20 µg of total RNA/lane) were separated on a 1.2% agarose/formaldehyde gel and transferred overnight to Nytran by capillary action. The RNA was crosslinked by exposure to UV radiation and the blot was incubated with prehybridization buffer (6× standard saline citrate (SSC), 10× Denhart's, 0.5% sodium dodecyl sulphate (SDS), 50% formamide, 50 µg/ml t-RNA) for 4 hr at 42°. The blot was subsequently hybridized overnight at 42° with 1 × 106 c.p.m. of radiolabelled probe in hybridization buffer (6× SSC, 0.5% SDS, 50% formamide, 10% dextran). Blots were washed as follows: 2 × 15 min, RT, 6× SSC + 0.1% SDS; 1 × 30 min, 37°, 2× SSC + 1 µg/ml RNase A; 2 × 15 min, 37°, 1× SSC + 0.5% SDS; and 1 × 30 min, 65°, 0.1× SSC + 0.5% SDS. Blots were exposed to film and processed for autoradiography.
Western blot analysis
Cells were cultured in T25 flasks and treated under the indicated conditions. Cells were washed with phosphate-buffered saline (PBS), scraped into PBS, transferred to a microfuge tube, and pelleted by centrifuging at 10 000 g for 3 min The cells were incubated in PBS with 1% NP-40 and protease inhibitors (Protease Arrest, Pierce, Rockford, IL) for 15 min on ice, centrifuged at 12 000 g for 15 min at 4°, and the supernatant fraction was transferred to a new microfuge tube. An aliquot was taken to measure cellular protein using the micro bicinchoninic acid assay (BCA; Pierce), and 5× gel sample buffer was added to the remainder. Equal amounts of protein (100–250 µg) were separated on 7.5% SDS polyacrylamide gels, transferred to nitrocellulose, and subjected to Western blot analysis using the indicated antibody. Briefly, non-specific binding was blocked by incubating the blots for 1 hr at room temperature with non-fat dry milk (NFDM), 5% NFDM in PBS with 0.05% Tween 20. After each step the blots were washed four to five times for 10 min each with wash buffer (10 mm Tris-HCl, pH 7.3, 150 mm NaCl, 1 mm ethylenediaminetetraacetic acid, 0.05% Tween 20). The blots were incubated overnight at 4° with monoclonal anti-secretory component (1 µg/ml; Sigma), with polyclonal anti-IRF-1 (0.4 µg/ml in NFDM; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), or to verify expression of the dominant negative IκBα-serine mutant with anti-IκBα (0.2 µg/ml in NFDM, Santa Cruz Biotechnology). Blots were then incubated for 1 hr at room temperature with horseradish peroxidase (HRP)-conjugated sheep anti-mouse or donkey anti-rabbit IgG (1: 10 000 in NFDM; Amersham Life Sciences, Piscataway, NJ). Antibody binding was visualized using the Pierce SuperSignal reagent and autoradiography.
Gene transfection studies with adenoviral vectors
An adenoviral construct expressing a dominant negative form of IκBα (Ad5CMVIκB-serine mutant) was generously provided by Dr John Engelhardt (Department of Anatomy and Cell Biology, University of Iowa, Iowa City, IA). A construct without an insert (Ad empty) and one expressing β-galactosidase (Ad5CMVLacZ) were used to control for adenoviral effects. Both were provided through the University of Iowa Gene Therapy Vector Core Facility.
To determine whether over-expressing IκBα-serine mutant inhibited cytokine-dependent increases in pIgR expression, a cell-based enzyme-linked immunosorbent assay (ELISA) method was developed. For this method, 2 × 105 cells were seeded per well of a 24-well culture plate and allowed to attach overnight. Cells were then transfected with Ad5CMVLacZ or Ad5CMVIκB-serine mutant (100 multiplicity of infection (MOI) in culture medium). Twenty-four hr later, cytokine(s) were added directly to the wells and cultures were incubated an additional 24–48 hr: IL-4, 10 ng/ml; IFN-γ, 200 U/ml, TNF-α, 10 ng/ml; IL-1β, 10 ng/ml. Cells were washed once with Dulbecco's PBS with calcium and magnesium (DPBS/Ca2+/Mg2+) and incubated for 30 min with blocking buffer (DPBS/Ca2+/Mg2+/0.1% BSA). Cultures were incubated for 2–4 hr at room temperature or overnight at 4° with monoclonal antihuman secretory component (Sigma Chemical Co.; 1 µg/ml in blocking buffer). Cultures were washed twice with DPBS/Ca2+/Mg2+, once with Blocking Buffer, and then incubated for 1 hr at room temperature with HRP-conjugated goat anti-mouse IgG (Amersham Life Sciences; 1: 2000 in blocking buffer). Cultures were washed extensively with blocking buffer and antibody binding was visualized by adding 100 µl of tetramethylbenzidine (TMB) substrate solution (Sigma Chemical Co.) to each well and allowing colour to develop. The reaction was stopped by addition of 100 µl 0.5 m H2SO4 and the absorbance at 450 nm was determined using a Bio-Rad microplate spectrophotometer.
To verify that over-expressing IκBα-serine mutant inhibited NF-κB activation, cells were seeded at 2 × 105 cells/well in 24-well plates and allowed to attach overnight. Cells were then cotransfected with Ad5CMVLacZ or Ad5CMVIκB-serine mutant (100 MOI) and Lipofectamine 2000 (LF2000): NFKB–SEAP (where NFKB-SEAP is NF-κB promoter elements linked to the gene coding for secretory alkaline phosphatase; see below). Four hr after the start of transfection, medium with 10% serum was added. At 24 hr, cytokine(s) was added: IL-4, 10 ng/ml; IFN-γ, 200 U/ml, IL-1β, 10 ng/ml; TNF-α, 10 ng/ml. The medium was harvested 24 hr later and SEAP activity was measured.
To examine the effect of over-expressing IκBα-serine mutant on IRF-1 expression using Western blot analysis, cultures (2 × 106 cells per 100 mm dish) were infected overnight with Ad5CMVLacZ, Ad empty, or Ad5CMVIκB-serine mutant (250 MOI) and treated for 24–48 hr with cytokines as described above. Cells were then harvested and cell extracts were assayed for IRF-1 and IκBα expression by Western blot analysis.
Densitometry
Quantitation of autoradiographs was performed using the AlphaImager Digital Imaging System 2000 (Alpha Innotech Corp., San Leandro, CA) as previously described.14 Only bands within the linear range of the film and instrument were used for subsequent calculations.
Assay for activation of NF-κB
The Mercury™ Pathway Profiling System was used for these studies (BD Biosciences, Clontech, Palo Alto, CA). Cells were seeded into 24-well plates at 1 × 105 cells/well and allowed to attach overnight. LF2000 (Gibco BRL, Life Technologies, Gaithersburg, MD) was diluted in OptiMEM (Gibco BRL) at 4 µl/ml. A separate plasmid DNA solution was made in OptiMEM: 1 µg CMVβgal (internal control) combined with either 3 µg of pSEAP-control (positive control with SV40 promoter), 3 µg pTal-SEAP (negative promoter-less control) or 3 µg NFKB-SEAP (experimental plasmid) per ml. After 5 min at room temperature, equal volumes of the LF2000 and DNA solutions were combined and incubated for 15 min at room temperature. The medium was removed from the cultures and 100 µl of lipid–DNA complex was added per well. Cells were incubated at 37° for 4–6 hr then 0.4 ml of medium containing 10% serum and the indicated cytokine(s) was added: IL-4, 10 ng/ml; IFN-γ, 200 U/ml, TNF-α, 10 ng/ml, and IL-1β, 10 ng/ml. Cultures were incubated 24–48 hr. Beta-galactosidase activity was measured in the cells using the βgal One Step Assay (Pierce Chemical Co., Rockford, IL) and measuring optical absorbance at 450 nm. To measure SEAP activity in the medium, samples were incubated at 65° for 2 hr and 100 µl of sample was combined with 100 µl of substrate (4-methylumbelliferyl phosphate liquid substrate, 4-MUP, Sigma Chemical Co.) in wells of a 96-well plate. Samples were then incubated at room temperature for 1–24 hr and sample fluorescence (λex/λem, 355/460 nm) was measured using the BMG FluoStar microplate fluorometer (BMG Laboratory Technologies, Inc., Durham, NC).
pIgR promoter studies
Primers for promoter sequences proximal to the transcription start site were designed with HindIII and XhoI sites at the 5′- and 3′- end, respectively. Polymerase chain reaction (PCR) was run with the primers using as a template a clone of the pIgR 5′ regulatory region (−6300 to +1151), a generous gift of Dr Charlotte Kaetzel (University of Kentucky). PCR settings were 94° for 1 min followed by 30 cycles of 94°, 30 seconds; 55°, 1 min; 72°, 3 min and a final cycle of 72° for 20 min The PCR products were separated on a 1% low-melting point (LMP) agarose gel, purified with the Promega PCR purification kit and sequenced. Nucleotide sequences were then blunt end cloned into pST blue1 and re-sequenced. Inserts were cut out of the plasmid using HindIII and XhoI, separated on a 1% LMP gel, purified with the Promega kit and cloned into SEAP basic (Clontech). Positive clones were identified by sequencing.
For promoter sequences surrounding the putative STAT6 site in intron 1, primers were designed and PCR was performed using blood DNA as a template. Products were separated on a 1% LMP gel, purified, sequenced and blunt end cloned into pSTBlue 1. Clones with the desired sequences were identified and the insert was cut out using EcoRI and cloned into SEAP basic. All plasmid preparations were purified by repeated caesium chloride gradient ultracentrifugation.
For experiments, cells were seeded at 2 × 105 cells per well of a 24-well plate and allowed to attach overnight. Cells were transfected with plasmid DNA (1 µg/well) with and without 100 MOI/well of Ad empty or of Ad5CMVIκB-serine mutant using LF2000 as described above. 24 h later, cytokines were added directly to the wells and cultures were incubated for 24 hr. The medium was assayed for SEAP activity.
Statistical analysis
Combined data from multiple experiments were expressed as the mean ± standard error of the mean (SEM). Statistically significant differences were defined as P < 0.05 using Student's t-test.
Results
Effect of NF-κB inhibitors on cytokine-dependent increases in pIgR mRNA
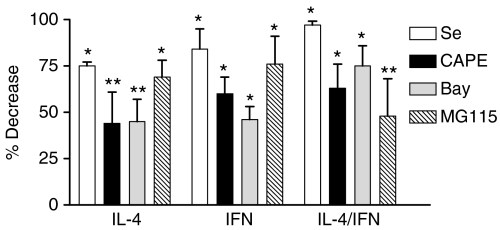
To determine the effect of chemical NF-κB inhibitors on steady state levels of pIgR mRNA, cells were pretreated with increasing concentrations of the antioxidants sodium selenite28 or CAPE29 the IKK kinase inhibitor Bay 11-7085, 30 or the 26S proteosome inhibitor MG115 31 for 1 hr – see Discussion for a description of inhibitor mechanisms. Cytokines were then added and cultures were incubated for 24 hr. Steady-state levels of pIgR mRNA were determined using a non-radioactive RPA as previously described. 14 All of the inhibitors tested reduced both IL-4- and IFN-dependent increases in pIgR mRNA in a concentration-dependent manner (data not shown). Quantitation of autoradiographic band intensities was then performed for those bands within the linear range of the film. Combined data for maximal concentrations of each inhibitor were expressed as percent decrease and results are summarized in Fig. 1. All four inhibitors significantly inhibited cytokine-dependent increases in pIgR mRNA (P < 0.01 for each, n = 3–6) in response to IL-4 and IFN-γ alone and in combination. In contrast to these results, a selenium-based antioxidant ebselen that does not inhibit NF-κB 32 had no effect at concentrations less than 200 μm (data not shown).
Figure 1.
Effect of NF-κB inhibitors on cytokine-dependent increases in pIgR mRNA. HT29 cells were preincubated for 1 hr without and with 5 μm Na2SeO3 (Se), 30 μg/ml CAPE, 50 μm Bay 11-7085 (Bay), or 5 μm MG115 and then with 10 ng/ml IL-4, 200 U/ml IFN-γ or both together for 24 hr. Total RNA was isolated and pIgR mRNA was determined by RPA. Densitometric analysis of autoradiographs was performed as described in Materials and Methods and the percent decrease relative to cultures without inhibitor treatment was determined. Values represent the mean ± SEM for combined data from three to six independent experiments for each condition. A significant decrease in steady state mRNA levels was observed in inhibitor-treated cells relative to controls: *P < 0.001, **P < 0.01.
Effect of NF-κB inhibitors on cytokine-dependent increases in pIgR protein
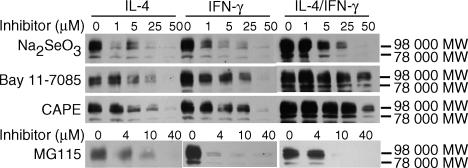
In parallel experiments, the effect of chemical inhibitors on cytokine-dependent increases in pIgR protein levels was determined using a monoclonal antibody against secretory component and Western blot analysis. As previously described 9 a major band with an apparent molecular weight of 98 000 MW was detected in all samples (Fig. 2). In some experiments, a second band with an apparent molecular weight of 78 000 MW was observed. Previous studies by our laboratory identified these two bands as the full-sized receptor and free secretory component, respectively. 9 No non-specific bands were previously observed with the monoclonal antibody used for these studies. 9 Other minor bands, when observed, likely reflect limited proteolysis that occurred despite our addition of protease inhibitors. For each inhibitor tested, a similar concentration response was observed when parallel cultures were stimulated with IL-4 alone, IFN-γ alone, or both cytokines together (Fig. 2). These results suggest that each inhibitor may act via a similar mechanism to inhibit the effects of IL-4 and IFN-γ.
Figure 2.
Effect of NF-κB inhibitors on cytokine-dependent increases in pIgR protein measured by Western blot analysis. HT29 cells were preincubated without and with the indicated concentration of inhibitor for 1 hr. The indicated cytokines were then added and cultures were incubated for 30 hr at 37°: IL-4, 10 ng/ml; IFN-γ, 200 U/ml; or both together (IL-4/IFN-γ). At the end of the incubation period, whole cell extracts were prepared and pIgR protein was determined by Western blot analysis using a monoclonal antibody against human secretory component. The 98 000 MW band represents the full-sized receptor and the 78 000 MW band represents secretory component. Similar results were seen in three or more independent experiments for each condition.
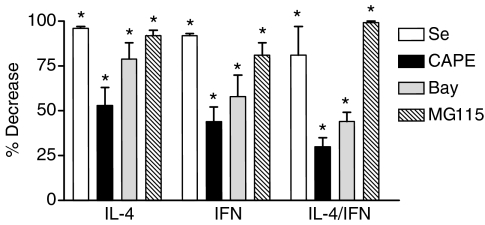
For quantitation purposes, the combined intensity of the 98 000 and 78 000 MW bands was determined. Data were expressed as percentage decrease, and combined data from these experiments is summarized in Fig. 3. As both the RPAs and Western blots are semiquantitative, caution should be used in comparing the relative effects of inhibitors using each approach. However, as with steady state mRNA levels, all four inhibitors significantly inhibited cytokine-dependent increases in pIgR protein (P < 0.01 for each, n = 3–5).
Figure 3.
Effect of NF-κB inhibitors on cytokine-dependent increases in pIgR protein. Densitometric analysis of autoradiographs from experiments like those shown in Figure 2 was performed as described in Materials and Methods for cultures treated without and with 5 μm sodium selenite (Se), 30 μg/ml CAPE, 50 μm Bay 11-7085 (Bay), or 5 μm MG115. The percentage decrease relative to the corresponding cultures without inhibitor treatment was then determined. Values represent the mean ± SEM for combined data from three to six independent experiments for each condition. A significant decrease in pIgR protein was observed in inhibitor-treated cells relative to controls: *P < 0.001.
Effect of over-expressing a dominant negative mutant of IκBα
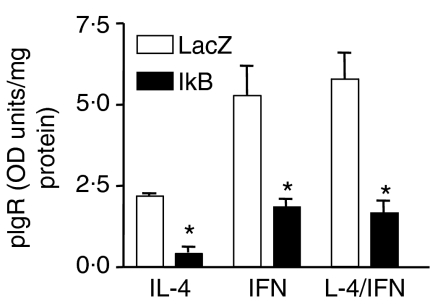
Studies using chemical inhibitors of NF-κB were consistent with a role for NF-κB in IL-4- and IFN-dependent pIgR expression. However, while the use of several inhibitors – each of which acts by a distinct mechanism – strengthens this conclusion, concerns about inhibitor specificity remained. To address these concerns, experiments were performed using an adenoviral construct to over-express a dominant negative form of IκBα. 33 To facilitate these studies, we developed a cell-based ELISA that measured surface expression of the pIgR (see Materials and methods). In preliminary studies, we determined that a multiplicity of infection (MOI) of 100 MOI had maximal effects (data not shown). We found that over-expressing the IκBα-serine mutant significantly decreased surface-expressed pIgR in response to IL-4 and IFN-γ alone and both cytokines together when compared to cells transfected with a control adenoviral vector (AdLacZ). An example of these results is shown in Fig. 4. Combined data from three independent experiments showed that over-expressing the mutant inhibited the response to IL-4, IFN-γ, and to both cytokines together by 46 ± 10%, 45 ± 10%, and 49 ± 11% (mean ± SEM, P < 0.001 in each case), respectively, after correcting for changes in basal expression. Western blot analysis also demonstrated inhibition by over-expressing the IκBα-serine mutant (data not shown).
Figure 4.
Effect of over-expressing a dominant negative mutant of IκBα on cytokine-dependent increases in pIgR surface expression. HT29 cells were transfected with 100 MOI of an adenoviral vector expressing β -galactosidase (LacZ, controls for adenoviral effects) or IκBα-serine mutant (IκB) overnight. The indicated cytokine was then added (IL-4, 10 ng/ml; IFN-γ, 200 U/ml; both together) and cultures were incubated for 48 hr. Surface expression of the pIgR was quantitated using a cell-based ELISA as described in Materials and Methods. Data represent the mean ± standard deviation for triplicate samples. A significant decrease was observed with IκBα-serine mutant relative to the LacZ control; *P < 0.001. Similar results were seen in three independent experiments.
Effect of NF-κB inhibitors on cytokine-dependent induction of IRF-1
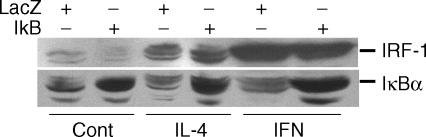
Previous studies demonstrate that the inducible transcription factor IRF-1 contributes to cytokine-dependent increases in pIgR expression. 16 Because potential NF-κB binding sites are present in the IRF-1 promoter and some studies have argued for a role for NF-κB in IRF-1 expression 34 studies were designed to determine whether NF-κB inhibitors prevented cytokine-dependent increases in pIgR expression by blocking induction of IRF-1 in our cell model system. For these studies, samples were blotted with monoclonal anti-SC and then were re-blotted with polyclonal anti-IRF-1. Under these conditions, no apparent effect on IRF-1 induction was observed with any of the chemical inhibitors tested (data not shown). Similarly, no change in IRF-1 induction was observed in cells over-expressing the IκBα-serine mutant relative to cells transfected with a control adenoviral vector (AdLacZ) (Fig. 5, upper panel). Over-expression of IκBα-serine mutant in these studies was verified using Western blot analysis (Fig. 5, lower panel).
Figure 5.
Effect of overexpressing the IκBα-serine mutant on induction of IRF-1. HT29 cells were transfected with 100 MOI of an adenoviral vector expressing β -galactosidase (LacZ, controls for adenoviral effects) or IκBα-serine mutant (IκB) overnight. Medium without (Cont) or with IL-4 (10 ng/ml) or IFN-γ (200 U/ml) was then added and cultures were incubated for 24 hr at 37°. Whole cell extracts were prepared and IRF-1 protein was measured by Western blot analysis (upper panel). Blots were stripped and reblotted with antibody against IκBα (lower panel).
Experiments were subsequently designed to determine whether the lack of effect on IRF-1 protein levels reflected a lack of effect on steady state IRF-1 mRNA levels. In parallel cultures, the inhibitors had no effect on IFN-dependent increases in IRF-1 protein or mRNA levels, as measured by Western blot (100 μg total cell protein per lane) and Northern blot (internal control 28S rRNA) analysis, respectively (data not shown).
Prolonged activation of NF-κB is required for maximal pIgR expression
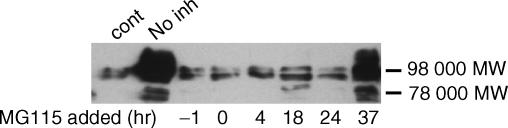
Activation of NF-κB can be transient or prolonged. To test whether prolonged activation of NF-κB is required for continuing increases in pIgR expression, MG115 was added at several times before and after the addition of cytokines. Figure 6 demonstrates that the proteosome inhibitor markedly reduced cytokine-dependent increases in pIgR protein, even if it was added as late as 24 hr after cytokine addition. In these studies, all cultures were harvested at 48 hr. Some reduction relative to the ‘no inhibitor’ control was also observed if inhibitor was added as late as 37 hr after cytokine addition. A similar pattern was observed with Bay 11-7085 (data not shown). These results suggest that prolonged activation of NF-κB is required for continued induction of pIgR expression.
Figure 6.
Effect of adding NF-κB inhibitor at varying times on cytokine-dependent increases in pIgR protein. HT29 cells were incubated without (Cont) or with IL-4 (10 ng/ml) and IFN-γ (200 U/ml) together. MG115 was added at the indicated time relative to cytokine addition (0 hr). All cultures were harvested at the same time (48 hr) after cytokine addition. Whole cell extracts were prepared and pIgR protein was measured by Western blot analysis using a monoclonal antibody against human secretory component. The 98 000 MW band represents the full-sized receptor and the 78 000 MW band represents secretory component.
Effect of cytokines on activation of NF-κB
In addition to exploring the role of NF-κB in IL-4- and IFN-dependent pIgR expression, we wished to determine whether IL-4 and/or IFN-γ activated NF-κB in HT29 cells. As a preliminary approach, we used the electrophoretic mobility shift assay (EMSA). We did not see consistent increases in response to either cytokine using this assay. However, we were concerned that this method was limited in its ability to detect small but potentially significant increases in NF-κB activation. Moreover, EMSA is not quantitative. To overcome these limitations, studies were performed using reporter gene constructs containing consensus NF-κB consensus sequences upstream of SEAP (NFKB-SEAP). As negative controls, cells were transfected with the parent plasmid lacking the NF-κB elements (pTal-SEAP). As a positive control, cells were transfected with plasmid containing the SV40 viral promoter sequence upstream of SEAP (pSEAP-control).
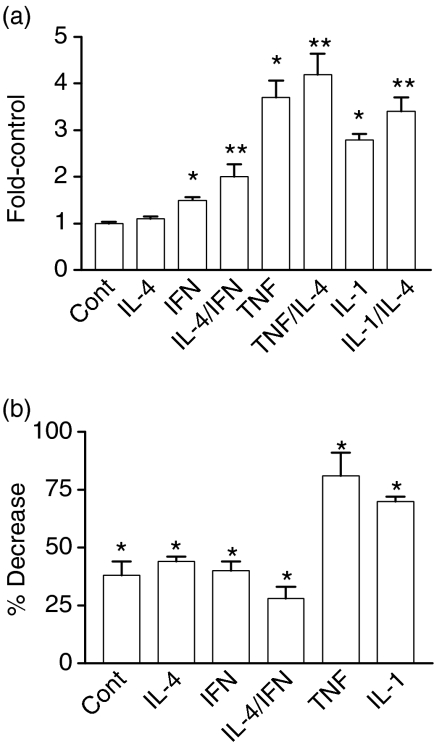
The SV40-SEAP positive control yielded consistently high values while the negative control (pTal-SEAP) in the presence and absence of cytokine(s) was indistinguishable from reagent blank (data not shown). To combine data from independent experiments, the fold-increase over controls, where controls were values for unstimulated cells, was determined (Fig. 7a, mean ± SEM, n ≥ 12). Measurable levels of SEAP activity were observed in media from unstimulated HT29 cells suggesting a basal level of NF-κB activation. Basal activity is not surprising since NF-κB regulates numerous genes related to growth, as well as to inflammation and immunity. IFN-γ (1.5 ± 0.06), TNF-α (3.7 ± 0.36) and IL-1β (2.8 ± 0.12) all significantly increased activation of NF-κB (P < 0.001). IL-4 alone had little effect on NF-κB activation, but did cause a further increase when combined with IFN-γ, TNF-α or IL-1: P < 0.05 for each combination relative to the individual cytokines. Together, these data suggest that IL-4 may activate pathways that can contribute to NF-κB activation in the context of additional cytokine stimulation.
Figure 7.
Cytokine-dependent activation of NF-κB as measured using reporter gene constructs. (a) HT29 cells were cotransfected with CMV-β gal (transfection efficiency control) and NFKB-SEAP as described in Materials and Methods. Four hours later medium with the indicated cytokine(s) was added to the cells: IL-4, TNF-α, and IL-1β, each 10 ng/ml; IFN-γ, 200 U/ml. Cultures were incubated for an additional 18–20 hr. Culture medium was harvested for SEAP assays and cells were assayed for β -galactosidase activity. SEAP activity was normalized to β -galactosidase activity. To combine data from multiple experiments, the fold increase over control, where control values were for unstimulated cells, was determined. Values represent the mean ± SEM for combined data (n ≥ 9; *P < 0.001 relative to controls; **P < 0.05 for cultures treated with IL-4 and the indicated cytokine relative to the indicated cytokine alone). (b) HT29 cells were cotransfected with CMV-β gal, NFKB-SEAP, and 100 MOI of an adenoviral vector expressing β -galactosidase (LacZ) or IκBα-serine mutant (IκB) overnight. Medium without (Cont) or with the indicated cytokine was then added and cultures were incubated for 18 hr at 37°. The percent decrease in NF-κB activation for AdIκB-serine mutant-transfected cells relative to LacZ-transfected cells was then determined. Values represent the mean ± SEM, n = 12. A significant decrease was observed with the IκBα-serine mutant relative to the LacZ control; *P < 0.001.
We also used the SEAP reporter gene assay system to verify that over-expressing the IκBα-serine mutant inhibited cytokine-dependent NF-κB activation. For these studies, cultures were cotransfected with the NFKB-SEAP plasmid and the adenoviral construct expressing either β-galactosidase (LacZ control) or IκBα-serine mutant. Over-expression of the serine mutant inhibited constitutive levels of activation as well as the response to each cytokine tested (P < 0.001 in each case). The percent inhibition (mean ± SEM, n = 8) was calculated using combined data from two independent experiments (Fig. 7b). Incomplete inhibition of NF-κB-mediated SEAP expression in control, IL-4- and IFN-treated cells suggests either that some cells in the culture were not expressing the mutant protein and/or that there is a basal level of NF-κB activation that is not readily inhibited using this technique. However, the potent inhibition of the response to TNF-α clearly indicates the effectiveness of this approach and provides evidence that over-expressing the mutant significantly inhibits cytokine-dependent NF-κB activation.
Reporter gene studies with pIgR promoter sequences
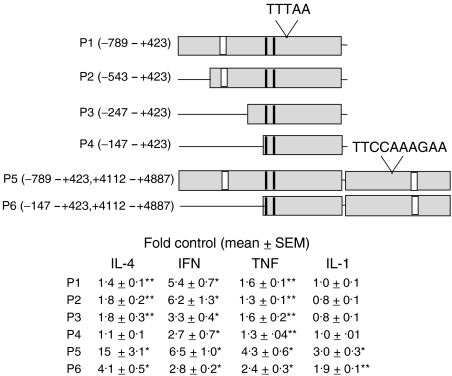
As a final approach to address the role of NF-κB in pIgR expression, we created SEAP reporter plasmid constructs with pIgR promoter sequences proximal to the transcription start site either alone (P1-4) or in combination with sequences surrounding a STAT6 site in intron 1 24 (P5-6). A diagram of these promoter sequences is shown in Fig. 8. The diagram indicates the TATA box (TTTAA), as well as the putative STAT-6 site (TTCCAAAGAA) in intron 1. Also indicated are NF-κB sites (vertical white bars) in the proximal promoter region (−461 to −440) and in intron 1 (+4391 to +4411) that were reported to play a role in TNF-dependent reporter gene expression 17,23 as well as the ISREs (vertical black bars) (−138 to −128; −103 to −94) that are reported to contribute to IFN- and TNF-dependent expression. 18
Figure 8.
Effect of cytokines on reporter gene expression using pIgR promoter sequences. The diagram represents pIgR promoter sequences that were linked to the reporter gene for SEAP. Included in the diagram are the TATA box (TTTAA), the STAT6 site (TTCCAAAGAA) in intron 1, identified κB sites (white boxes), and identified interferon-stimulated response elements (ISREs, black boxes). HT29 cells were transfected with the indicated plasmid (P1-P6) as described in Materials and Methods and then treated for 48 hr without and with cytokine: IL-4, TNF-α, and IL-1β, each 10 ng/ml; IFN-γ, 200 U/ml. SEAP activity was then measured in the medium and data are expressed as fold control where the controls were unstimulated cells. Values represent the mean ± SEM for combined data from multiple experiments with quadruplicate samples in each experiment (n ≥ 12). A significant increase was observed with some cytokine treatments relative to controls; *P < 0.001, **P < 0.05.
First, to determine the effect of cytokines on reporter gene expression using these constructs, cells were transfected for 4 hr with plasmid and then stimulated for 48 h with and without cytokine. Media from the cultures were assayed for SEAP activity and fold-increase over controls was determined for combined data from three or more independent experiments. Values for the mean ± SEM (n ≥ 9) are shown in Fig. 8.
As previously reported18,23 sequences in the proximal promoter region (P1-4) significantly increased IFN- and TNF-dependent reporter gene expression. We also found that sequences in this region increased IL-4-dependent expression. Interestingly, no increase in expression was observed with IL-1β. In parallel samples, the absolute values for SEAP expression, both basal and cytokine-stimulated, were greater for P2 than for P1 (data not shown) consistent with a previously reported negative regulatory element.18 When sequences from −543 to −148 were deleted (P2 versus P4), expression in response to both IL-4 and IFN-γ decreased. Note that the decrease for IL-4 involved deletion of sequences from −247 to −148 (P3 versus P4), whereas the major decrease for IFN-γ involved deletion of sequences from −543 to −248 (P2 versus P3). Furthermore, whereas a response to IFN-γ and to TNF-α was observed with the smallest sequence tested (P4) (likely caused by the ISREs), IL-4-responsiveness was lost. This suggests either that these ISREs do not contribute to the response to IL-4 or that sequences upstream of the ISREs are required for a contribution by the ISREs in the context of these studies.
Also as previously reported, sequences in intron 1 appear to play an important role in IL-4- and in TNF-dependent expression (compare P1 and P5). Regulatory elements for IL-1β also appear to be present in intron 1. Conversely, addition of intron 1 sequences did not significantly increase the response to IFN-γ. Removal of upstream sequences from −789 to −148 significantly reduced the response to all cytokines, including IL-1β. This suggests cooperation between proximal and intron 1 elements for IL-4 and for IL-1β, as was observed previously for TNF-α.23
Next, to determine whether NF-κB activity contributed to reporter gene expression under these conditions, cells were transfected with plasmid (P1, 4, 5, or 6) together with 100 MOI of an empty adenoviral vector control (Ad empty) or the IκBα-serine mutant vector (AdIκB). Cytokines were then added to the cells and the media were collected 48 h later for SEAP assay. The percentage remaining SEAP activity from AdIκB-transfected relative to Ad blank-transfected cultures was determined. Combined data (mean ± SEM, n ≥ 9) from three or more independent experiments is shown in Fig. 9.
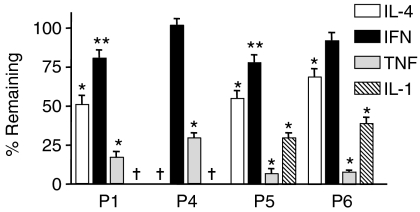
Figure 9.
Effect of over-expressing the IκBα-serine mutant on cytokine-dependent reporter gene expression. HT29 cells were cotransfected with the indicated plasmid and 100 MOI of an empty adenoviral vector or one expressing the IκBα-serine mutant overnight. Cultures were then treated with the indicated cytokine for 48 hr: IL-4, TNF-α, and IL-1β, each 10 ng/ml; IFN-γ, 200 U/ml. SEAP activity was measured in the medium. The percentage SEAP activity remaining in medium from cells transfected with the IκBα-serine mutant relative to cells transfected with empty adenovirus was determined. Values represent the mean ± SEM (n ≥ 12) for combined data from three or more independent experiments with quadruplicate samples per experiment. A significant decrease was observed with the IκBα-serine mutant relative to the empty vector control; *P < 0.001, **P < 0.05. †No cytokine-dependent increases in SEAP expression under these conditions.
Not surprisingly, the greatest effect of over-expressing the IκBα-serine mutant was observed with TNF- and IL-1-treated cells. This effect for TNF-α was due, at least in part, to elements in the proximal promoter sequences and for IL-1β for elements in intron 1. IL-4-dependent expression was also reduced in cultures transfected with the serine mutant, and this was slightly more pronounced for P1- than for P4-containing plasmids. Conversely, the effect of over-expressing the mutant on IFN-γ was small (P1, 5) or insignificant (P4, 6).
Discussion
To our knowledge, our inhibitor studies, including over-expressing a dominant negative IκBα, provide the first evidence that NF-κB contributes to both IL-4- and IFN-dependent regulation of pIgR expression and suggest that long-term activation is required for maximal expression. In contrast to their effects on pIgR expression, none of the NF-κB inhibitors, including the IκBα-serine mutant, prevented cytokine-dependent induction of IRF-1. This suggests that NF-κB activation is not upstream of IRF-1 in signalling pathways activated by IL-4 and IFN-γ. It further suggests that inhibition of pIgR expression by these inhibitors was not caused by effects on IRF-1. Finally, a lack of effect on IL-4- and IFN-dependent induction of IRF-1 provides indirect evidence that the inhibitors did not inhibit STAT1 and STAT6 pathways, both of which contribute to IRF-1 expression, or that the inhibitors have nonspecific effects on protein biosynthesis.
For these studies, several inhibitors were used each of which acts by a different mechanism. Sodium selenite has been shown to increase levels of glutathione peroxidase (GPx), a cytosolic enzyme that consumes H2O2. Increased GPx activity by selenium supplementation decreases NF-κB activation in response to numerous agonists.28 In addition, sodium selenite may directly inhibit NF-κB by oxidizing sulphydryl residues essential for NF-κB/DNA binding.35 Finally, sodium selenite may indirectly inhibit NF-κB by oxidizing and inactivating thioredoxin and thioredoxin reductase, proteins required for maintaining NF-κB's sulphydryl residues in the reduced state. CAPE inhibits TNF-dependent activation of NF-κB.29 This inhibition occurs at the level of NF-κB/DNA binding and is reversed by reducing agents again suggesting a role for critical sulphhydryl groups in NF-κB activation. In these CAPE studies, no effect was observed on several other transcription factors including the oxidant-sensitive transcription factor AP-1, as well as TFIID and Oct-1. These latter observations suggest a relative selectivity in CAPE's effects. The two remaining chemical inhibitors act by blocking steps that lead to release of NF-κB from its inhibitory protein IκB. Bay 11-7085 does so by inhibiting phosphorylation of IκB,30 and MG115 inhibits degradation of IκB by the 26S proteasome.31 Finally, in addition to studies using chemical inhibitors of NF-κB, experiments were performed in which an adenoviral vector was used to overexpress a dominant negative mutant form of IκBα.33 This mutant lacks serine residues that are phosphorylated in response to cellular agonists. Without serine phosphorylation, dissociation from NF-κB and degradation of IκBα by the 26S proteasome does not occur.
It should be noted that these inhibitor studies do not distinguish between a direct contribution, binding of NF-κB to the pIgR promoter, and an indirect contribution, NF-κB-dependent expression of proteins that regulate pIgR expression. The major advantage of using inhibitors, rather, is that they can provide more global insight into the importance of NF-κB to protein expression. Moreover, the possibility exists that NF-κB contributes to cytokine-dependent effects on pIgR expression by more than one mechanism.
Studies reported herein also provide the first evidence that IFN-γ activates NF-κB (∼50%) in HT29 cells. IL-4 alone was a poor stimulus but contributed significantly to activation when combined with IFN-γ, TNF-α, or IL-1β. It remains unclear whether the low level of NF-κB activation by IL-4 alone (∼10%) contributes significantly to IL-4-dependent increases in pIgR. It should be noted that NF-κB may be constitutively bound to the pIgR promoter and that it is this constitutive binding that plays a significant role in cytokine-dependent pIgR expression. An example of this for IL-4 was seen in B cells where constitutively bound NF-κB played an essential role in IL-4-dependent expression of IgE by interacting with STAT6.26
To date, only a few studies have examined the role of transcription factors in pIgR expression. IRF-1 was first determined to play a role in IFN-dependent pIgR expression16,18 and later was shown to be coordinately up-regulated with pIgR in response to TNF-α and IL-1β as well.19 IL-4 also induces IRF-1 in HT29 cells.14 Two ISREs, one of which binds IRF-1, were identified in the proximal promoter region18 and were shown to contribute to IFN- and to TNF-dependent increases in expression.18,23 We also observed cytokine-dependent increases in reporter gene expression using constructs containing these ISREs (Fig. 8, P1-4). For TNF-α and IFN-γ, the minimal construct tested (P4) was sufficient for increased expression. Upstream elements further increased the response to IFN-γ but not to TNF-α. Additionally, while not ruling out a role for IRF-1 in IL-4-dependent pIgR expression, our data suggest that the ISREs are not sufficient for a response to IL-4 but rather that elements upstream of the ISREs are required for the response to this cytokine. and finally, although IL-1β and TNF-α activate similar transcription factors, including NF-κB, no IL-1-dependent expression was observed using these proximal promoter constructs.
In addition to the proximal promoter elements, regulatory elements were identified in intron 1. This included a STAT6 site that contributes significantly to IL-4-dependent reporter gene expression.24 Using constructs that included the intron 1 sequences, we also observed a large increase in the response to IL-4 (Fig. 8, P1 versus P5). Moreover, our studies suggest that proximal promoter elements co-operate with intron 1 elements to increase IL-4-dependent expression, as removal of these sequences (−789 to −148) markedly reduced the response (three- to fourfold). Similarly, co-operativity between a κB site in intron 1 and an ISRE in the proximal promoter region was observed in TNF-stimulated cells.23 Our findings with TNF-α are consistent with this report. However, in contrast, our data suggest that co-operativity involves elements other than and/or in addition to the ISREs.
We also found that elements in intron 1 contributed to IL-1-dependent promoter gene expression. Moreover, although insufficient alone, elements in the proximal promoter region increased IL-1-dependent expression when intron 1 sequences were present. Unlike the other cytokines tested, there was little or no increase in the response to IFN-γ when the intron 1 sequences were added. Also of note, although IL-4 and IFN-γ are additive/synergistic in the context of the whole cell, no additivity and/or synergy was observed in our reporter gene studies (data not shown). This suggests that there are additional promoter elements that mediate this effect.
With respect to a role for NF-κB, a number of putative NF-κB sites have been proposed in the 5′ regulatory region.34 Two of these sites (κB1, 759 to −738; κB2, −461 to −440) when mutated, reduced reporter gene expression in response to TNF-α.17 The κB2 sequence in EMSA studies was shown to bind p65 and p50. Because of the variability and magnitude of the effects, it was difficult for us to ascertain whether a similar decrease was observed in the response to TNF-α when these κB sequences were removed. In contrast to these earlier studies, when the κB2 site was mutated in the context of a larger promoter construct that included sequences from intron 1.23 TNF-dependent reporter gene expression was enhanced. The basis for these observed differences remains to be determined.
Over-expressing the IκBα-serine mutant inhibited cytokine-dependent reporter gene expression for constructs with proximal promoter elements with a relative potency of TNF-α > IL-4 > IFN-γ (Fig. 9, P1). Over-expressing the mutant also inhibited TNF-, but not IFN-dependent expression from the smallest construct tested (Fig. 9, P4).
NF-κB sites have also been described in sequences surrounding the STAT6 site in intron 1.24 As stated above, one of these κB sites (+4396 to +4404) contributes to TNF-dependent reporter gene expression and co-operates with an ISRE that is over 4 kb upstream.23 Activation and binding of p65 to this κB sequence was rapid whereas binding of p50 was weaker and delayed. Over-expressing the IκBα-serine mutant also inhibited cytokine-dependent expression for intron 1-containing constructs with a relative potency, similar to before, of TNF-α > IL-1β > IL-4 > IFN-γ.
Thus, reporter gene studies provide data in support of inhibitor studies suggesting a role for NF-κB in IL-4- and IFN-dependent pIgR expression. Specifically, they suggest that one or more NF-κB binding sites in the proximal promoter region and/or in intron 1 may contribute to increased expression in response to TNF-α, IL-1β, IL-4 and IFN-γ. Alternatively or in addition to this, NF-κB may increase expression of or promote binding of other factors to these promoter regions. Given the complexity of the pIgR promoter, it seems highly likely that additional regulatory sites will be identified, including additional sites regulated directly or indirectly by NF-κB, for cytokines that regulate the pIgR.
Numerous studies indicate that the pIgR and free secretory component play important roles in both innate and adaptive immunity.3,4,36–39 Furthermore, results with pIgR knockout mice suggest that the pIgR plays an important role in maintaining homeostasis in the gut.40 Thus, a critical need for high levels of the pIgR may explain its up-regulation by multiple immunomodulatory factors including IL-4 and IFN-γ. Data support the hypothesis that regulation by these factors is complex and involves multiple transcriptional regulators. Understanding the mechanisms by which host factors regulate pIgR expression will provide valuable insight regarding the basic mechanisms that regulate immunity and inflammation at mucosal surfaces.
Acknowledgments
Support for these studies was provided by grants to Dr Denning from the Office of Research and Development, Medical Research Service, Department of Veterans Affairs and from the American Heart Association, Heartland Affiliate. We acknowledge the Cell Fluorescence Core facility at the VA Medical Center for the use of their equipment in performing SEAP and β-galactosidase assays.
Abbreviations
- AP-1
activator protein-1
- CAPE
caffeic acid phenethyl ester
- EMSA
electrophoretic mobility shift assay
- GPx
glutathione peroxidase
- IFN-γ
interferon-γ
- IRF-1
interferon regulatory factor-1
- IL-1β
interleukin-1β
- IL-4
interleukin-4
- NF-κB
nuclear factor-κB
- LF2000
Lipofectamine 2000
- pIgs
polymeric immunoglobulins
- pIgA
polymeric immunoglobulin A
- pIgR
polymeric immunoglobulin receptor
- RPA
RNase protection assay
- sIgA
secretory immunoglobulin A
- TGF-β
transforming growth factor-β
- TNF-α
tumour necrosis factor-α
References
- 1.Kaetzel CS, Robinson JK, Chintalacharuvu KR, Vaerman JP, Lamm ME. The polymeric immunoglobulin receptor (secretory component) mediates transport of immune complexes across epithelial cells: a local defense function for IgA. Proc Natl Acad Sci U S A. 1991;88(19):8796. doi: 10.1073/pnas.88.19.8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mostov KE, Blobel G. Biosynthesis, processing, and function of secretory component. Methods Enzymol. 1983;98:458–66. doi: 10.1016/0076-6879(83)98173-9. [DOI] [PubMed] [Google Scholar]
- 3.Crottet P, Corthesy B. Secretory component delays the conversion of secretory IgA into antigen-binding competent F(ab′)2: a possible implication for mucosal defense. J Immunol. 1998;161:5445–53. [PubMed] [Google Scholar]
- 4.Marshall L, Perks B, Ferkol T, Shute J. IL-8 released constitutively by primary bronchial epithelial cells in culture forms an inactive complex with secretory component. J Immunol. 2001;167:2816–23. doi: 10.4049/jimmunol.167.5.2816. [DOI] [PubMed] [Google Scholar]
- 5.McGee D, Aicher W, Eldridge J. Transforming growth factor-β enhances secretory component and major histocompatibility complex class I. Cytokine. 1991;3:543–50. doi: 10.1016/1043-4666(91)90480-2. [DOI] [PubMed] [Google Scholar]
- 6.Kavale D, Lovhaug D, Sollid L, Brandtzaeg P. Tumor necrosis factor-α up-regulates expression of secretory component, the epithelial receptor for polymeric Ig. J Immunol. 1988;140:3086–9. [PubMed] [Google Scholar]
- 7.Hayashi M, Takenouchi N, Asano M, Kato M, Tsurumachi T, Saito T, Moro I. The polymeric immunoglobulin receptor (secretory component) in human intestinal epithelial cell lines is up-regulated by interleukin-1. Immunology. 1997;92:220–5. doi: 10.1046/j.1365-2567.1997.00341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sollid LM, Kvale D, Brandtzaeg P, Markussen G, Thorsby E. Interferon-gamma enhances expression of secretory component, the epithelial receptor for polymeric immunoglobulins. J Immunol. 1987;138(12):4303–6. [PubMed] [Google Scholar]
- 9.Denning G. IL-4 and IFN-γ synergistically increase total polymeric IgA receptor levels in human intestinal epithelial cells. J Immunol. 1996;156:4807–14. [PubMed] [Google Scholar]
- 10.Phillips JO, Everson MP, Moldoveanu Z, Lue C, Mestecky J. Synergistic effect of IL-4 and IFN-gamma on the expression of polymeric Ig receptor (secretory component) and IgA binding by human epithelial cells. J Immunol. 1990;145(6):1740–4. [PubMed] [Google Scholar]
- 11.Loman S, Jansen H, Out T, Lutter R. Interleukin-4 and interferon-γ synergistically increase secretory component gene expression, but are additive in stimulating secretory immunoglobulin A release by Calu-3 airway epithelial cells. Immunology. 1999;96:537–43. doi: 10.1046/j.1365-2567.1999.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar J, Gangopadhyay N, Moldoveanu Z, Mestecky J, Stephensen C. Vitamin A is required for regulation of polymeric immunoglobulin receptor (pIgR) expression by interleukin-4 and interferon-γ in a human intestinal epithelial cell line. J Nutr. 1998;128:1063–9. doi: 10.1093/jn/128.7.1063. [DOI] [PubMed] [Google Scholar]
- 13.Verrijdt G, Schoenmakers E, Alen P, Haelens A, Peeters B, Rombauts W, Claessens F. Androgen specificity of a response unit upstream of the human secretory component gene is mediated by differential receptor binding to an essential androgen response element. Mol Endocrinol. 1999;13:1558–70. doi: 10.1210/mend.13.9.0347. [DOI] [PubMed] [Google Scholar]
- 14.Ackermann L, Wollenweber L, Denning G. IL-4 and IFN-γ increase steady state levels of polymeric Ig receptor mRNA in human airway and intestinal epithelial cells. J Immunol. 1999;162:5112–8. [PubMed] [Google Scholar]
- 15.Nilsen E, Johansen F-E, Kvale D, Krajci P, Brandtzaeg P. Different regulatory pathways employed in cytokine-enhanced expression of secretory component and epithelial HLA class I genes. Eur J Immunol. 1999;29:168–79. doi: 10.1002/(SICI)1521-4141(199901)29:01<168::AID-IMMU168>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Piskurich JF, France JA, Tamer CM, Willmer CA, Kaetzel CS, Kaetzel DM. Interferon-γ induces polymeric immunoglobulin receptor mRNA in human intestinal epithelial cells by a protein synthesis dependent mechanism. Mol Immunol. 1993;30:413–21. doi: 10.1016/0161-5890(93)90071-i. [DOI] [PubMed] [Google Scholar]
- 17.Takenouchi-Ohkubo N, Takahashi T, Tsuchiya M, Mestecky J, Moldoveanu Z, Moro I. Role of nuclear factor-κB in the expression by tumor necrosis factor-α of the human polymeric immunoglobulin receptor (pIgR) gene. Immunogenetics. 1999;51:289–95. doi: 10.1007/s002510050622. [DOI] [PubMed] [Google Scholar]
- 18.Piskurich J, Youngman K, Phillips K, Hempen P, Blanchard M, France J, Kaetzel C. Transcriptional regulation of the human polymeric immunoglobulin receptor gene by interferon-γ. Mol Immunol. 1997;34:75–91. doi: 10.1016/s0161-5890(96)00079-x. [DOI] [PubMed] [Google Scholar]
- 19.Blanch V, Piskurich J, Kaetzel C. Cutting edge. Coordinate regulation of IFN regulatory factor-1 and the polymeric Ig receptor by proinflammatory cytokines. J Immunol. 1999;162:1232–5. [PubMed] [Google Scholar]
- 20.Johansen F-E, Bosloven B, Krajci P, Brandtzaeg P. A composite DNA element in the promoter of the polymeric immunoglobulin receptor regulates its constitutive expression. Eur J Immunol. 1998;28:1161–71. doi: 10.1002/(SICI)1521-4141(199804)28:04<1161::AID-IMMU1161>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Solorzano- Vargas, Sergio R, Wang J, et al. Multiple transcription factors in 5′-flanking region of human polymeric Ig receptor control its basal expression. Am J Physiol. 2002;283:G415–G25. doi: 10.1152/ajpgi.00420.2001. [DOI] [PubMed] [Google Scholar]
- 22.Hempen P, Phillips K, Conway P, Sandoval K, Schneeman T, Wu H-J, Kaetzel C. Transcriptional regulation of the human polymeric Ig receptor gene: Analysis of basal promoter elements. J Immunol. 2002;169:1912–21. doi: 10.4049/jimmunol.169.4.1912. [DOI] [PubMed] [Google Scholar]
- 23.Schjerven H, Brandtzaeg P, Johansen FE. A novel NF-kappa B/Rel site in intron 1 cooperates with proximal promoter elements to mediate TNF-alpha-induced transcription of the human polymeric Ig receptor. J Immunol. 2001;167(11):6412–20. doi: 10.4049/jimmunol.167.11.6412. [DOI] [PubMed] [Google Scholar]
- 24.Schjerven H, Brandtzaeg P, Finn-Erik J. Mechanism of IL-4-mediated up-regulation of the polymeric Ig receptor: Role of STAT6 in cell type-specific delayed transcriptional response. J Immunol. 2000;165:3898–906. doi: 10.4049/jimmunol.165.7.3898. [DOI] [PubMed] [Google Scholar]
- 25.Kirchhoff S, Wilhelm D, Angel P, Hauser H. NFκB activation is required for interferon regulatory factor-1-mediated interferon β induction. Eur J Biochem. 1999;261:546–54. doi: 10.1046/j.1432-1327.1999.00308.x. [DOI] [PubMed] [Google Scholar]
- 26.Messner B, Stutz A, Albrecht B, Peiritsch S, Woisetschlager M. Cooperation of binding sites for STAT6 and NFκB/rel in the IL-4-induced up-regulation of the human IgE germline promoter. J Immunol. 1997;159:3330–7. [PubMed] [Google Scholar]
- 27.Augeron C, Laboisse CL. Emergence of permanently differentiated cell clones in a human colonic cancer cell line in culture after treatment with sodium butyrate. Cancer Res. 1984;44(9):3961–9. [PubMed] [Google Scholar]
- 28.Piette J, Piret B, Bonizzi G, Schoonbroodt S, Merville M-P, Legrand-Poels S, Bours V. Multiple redox regulation in NF-κB transcription factor activation. J Biol Chem. 1997;378(11):1237–45. [PubMed] [Google Scholar]
- 29.Natarajan K, Singh S, Burke T, Grunberger D, Aggarwal B. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-κB. Proc Natl Acad Sci U S A. 1996;93:9090–4. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierce J, Schoenleber R, Jesmok G, Best J, Moore S, Collins T, Gerritsen M. Novel inhibitors of cytokine-induced IκBα phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 31.Lee D, Goldberg A. Selective inhibitors of the proteosome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:27280–4. doi: 10.1074/jbc.271.44.27280. [DOI] [PubMed] [Google Scholar]
- 32.de-Mello M, Flodstrom M, Eizirik D. Ebselen and cytokine-induced nitric oxide synthase expression in insulin-producing cells. Biochem Pharmacol. 1996;52:1703–9. doi: 10.1016/s0006-2952(96)00520-5. [DOI] [PubMed] [Google Scholar]
- 33.Iimuro Y, Nishiura T, Hellerbrand C, Behrns K, Schoonhoven R, Grisham J, Brenner D. NFκB prevents apoptosis and liver dysfunction during liver regeneration. J Clin Invest. 1998;101:802–11. doi: 10.1172/JCI483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verrijdt G, Swinnen J, Peeters B, Verhoeven G, Rombauts W, Claessens F. Characterization of the human secretory component gene promoter. Biochim Biophys Acta. 1997;1350:147–54. doi: 10.1016/s0167-4781(96)00214-x. [DOI] [PubMed] [Google Scholar]
- 35.Kim I, Stadtman T. Inhibition of NF-κB DNA binding and nitric oxide induction in human T cells and lung adenocarcinoma cells by selenite treatment. Proc Natl Acad Sci U S A. 1997;94:12904–7. doi: 10.1073/pnas.94.24.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazanec MB, Kaetzel CS, Lamm ME, Fletcher D, Nedrud JG. Intracellular neutralization of virus by immunoglobulin A antibodies. Proc Natl Acad Sci U S A. 1992;89(15):6901–5. doi: 10.1073/pnas.89.15.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mestecky J, Killian M. Immunoglobulin A (IgA) Methods Enzymol. 1985;116:37–75. doi: 10.1016/s0076-6879(85)16005-2. [DOI] [PubMed] [Google Scholar]
- 38.Holmgren J, Czerkinsky C, Lycke N, Svennerholm AM. Mucosal immunity: implications for vaccine development. Immunobiology. 1992;184:157–79. doi: 10.1016/S0171-2985(11)80473-0. [DOI] [PubMed] [Google Scholar]
- 39.Corthesy B, Spertini F. Secretory immunoglobulin A. from mucosal protection to vaccine development. J Biol Chem. 1999;380:1251–62. doi: 10.1515/BC.1999.160. [DOI] [PubMed] [Google Scholar]
- 40.Finn-Eirik J, Pekna M, Norderhaug I, Haneberg B, Hietala M, Krajci P, Betsholtz C, Brandtzaeg P. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin/secretory component-deficient mice. J Exp Med. 1999;190:915–21. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]